Abstract
C4 photosynthesis is a fascinating example of parallel evolution of a complex trait involving multiple genetic, biochemical and anatomical changes. It is seen as an adaptation to deleteriously high levels of photorespiration. The current scenario for C4 evolution inferred from grasses is that it originated subsequent to the Oligocene decline in CO2 levels, is promoted in open habitats, acts as a pre-adaptation to drought resistance, and, once gained, is not subsequently lost. We test the generality of these hypotheses using a dated phylogeny of Amaranthaceae s.l. (including Chenopodiaceae), which includes the largest number of C4 lineages in eudicots. The oldest chenopod C4 lineage dates back to the Eocene/Oligocene boundary, representing one of the first origins of C4 in plants, but still corresponding with the Oligocene decline of atmospheric CO2. In contrast to grasses, the rate of transitions from C3 to C4 is highest in ancestrally drought resistant (salt-tolerant and succulent) lineages, implying that adaptation to dry or saline habitats promoted the evolution of C4; and possible reversions from C4 to C3 are apparent. We conclude that the paradigm established in grasses must be regarded as just one aspect of a more complex system of C4 evolution in plants in general.
Keywords: C4 photosynthesis, dependent evolution, drought tolerance, molecular clock, salt tolerance, succulence
1. Introduction
The evolutionary importance of changing environmental conditions is exemplified in the evolution of photosynthetic pathways in plants. Most plants sequester carbon using a process—C3 photosynthesis—that originated when the concentration of atmospheric CO2 was considerably higher than it is today. The major photosynthesis enzyme, ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO), can act with either CO2 or O2 as substrate (depending on their relative concentrations). C3 plants compensate for the oxygenase activity of RuBisCO by employing an auxiliary metabolic process, photorespiratory carbon oxidation (C2 cycle or photorespiration), at the cost of re-releasing CO2. Photorespiration increases under warmer, drier and CO2-depleted conditions [1]. Consequently, when atmospheric CO2 decreased dramatically at around 30 Ma [2,3], the cost of photorespiration is likely to have increased. It was subsequent to this environmental change that the C4 photosynthetic pathway, a carbon concentrating mechanism, is inferred to have originated [4], more than 60 times independently in angiosperms [5]. Rather than a single adaptation, C4 photosynthesis represents a syndrome of complex genetic, biochemical and anatomical modifications [6,7]. Low atmospheric CO2 (less than ca 500 ppm) is currently seen as an environmental precondition for its evolution.
Identifying the selective pressures that led to the evolution of C4 is however more complicated than this apparently straightforward scenario might suggest. Increasing evidence [4] that most C4 lineages emerged much more recently than 30 Ma and the long-standing observation that C4 lineages are concentrated in hot and dry climates suggests that further environmental changes were needed to trigger the evolution of the C4, over and above CO2-depleted conditions [8–10]. Heat, aridity and salinity have classically been viewed as promoting C4 [11], but in fact, all environmental conditions that increase the level of photorespiration might have driven its evolution [7]. Furthermore, the transition from C3 to C4 involved several phases of major anatomical, genetic and biochemical changes [12], which might take millions of years to accumulate [4]. Each of the C3/C4 intermediate stages must represent a physiologically stable evolutionary step [1,13,14]. During these different evolutionary phases, various environmental factors might have influenced the further evolution in direction of full C4 syndrome, particularly given the diverse genetic background of the distantly related plant lineages involved.
The challenge of inferring the conditions that led to the evolution of C4 photosynthesis is exemplified by the profound differences between the two plant lineages representing the oldest and greatest numbers of C4 lineages: Amaranthaceae s.l. (including Chenopodiaceae) and Poaceae [4,5,15], which show a range of both convergent and unique modes of C4 evolution. From this, we might hypothesize that ancestral traits and selective pressures, which together facilitated the frequent evolution of C4, may be equally diverse.
With ca 750 C4 species in ca 15 independent C4 lineages Amaranthaceae s.l. comprise the largest number of C4 species and C4 lineages among eudicot families [5,16–20]. The evolution of C4 leaves from various flat and succulent C3 leaf anatomies led to an unmatched variety of C4 leaf anatomies, especially in Chenopodiaceae s.s., including the striking single-cell C4 anatomies of Bienertia and Suaeda aralocaspica [16,20,21]. C4 is found in various life forms, such as annuals, subshrubs, long-lived shrubs and small trees and the majority of species grow in open, warm, often arid and/or saline habitats [16].
Around 4600 species of grasses [5], representing at least 22 independent lineages [15], photosynthesize using the C4 pathway. Of C4 plants, grasses have received the most attention by researchers because they dominate the highly productive C4 grasslands, constitute important crops such as maize and sugarcane, and because there is a great interest in engineering C4 into C3 crops such as rice and wheat. In Poaceae, C4 species show classical Kranz anatomy with slight variation [21], are mostly herbaceous and never succulent. The C4 lineages of grasses are presumably of tropical ancestry [22,23] with their closest C3 relatives occurring in the shaded understory of tropical forest environments [22]. Using phylogenetic comparative analyses, Osborne and Freckleton [24] found that the transition from C3 to C4 in grasses was significantly faster in clades confined to open habitats than in those growing in the shade. Unexpectedly, they also found that clades confined to mesic habitats showed equal likelihood of evolving C4 to that of clades in water-logged, arid or saline habitats. In other words, this supported a long-standing view that growing in an open habitat with high irradiance is a precondition for the evolution of C4 [11] but no statistical evidence was found for an overall dependence of C4 evolution on aridity or salinity. Nevertheless, shifts to arid habitats occurred at higher rates in C4 than in C3 lineages. Therefore, the evolution of C4 was interpreted as a pre-adaptation that facilitated the colonization of arid and saline environments [24].
However, results from grasses may not be readily transferable to other plant lineages. Even within related lineages, a detailed knowledge of the phylogeny is crucial for the unbiased interpretation of comparative physiological or ecological studies [25]. Thus, it remains an open question whether this sequence of evolution—first C4, and then drought resistance—applies to the phylogenetically diverse other plant groups that evolved C4.
Here, we use a dated phylogeny of Amaranthaceae s.l. based on 169 species; two cp markers (the rbcL gene and atpB-rbcL spacer) and ancestral state reconstruction techniques to address the following questions. Did C4 lineages evolve subsequent to the dramatic decrease in CO2 levels ca 30 Ma? Is the rate of C4 evolution higher in ancestrally salt or drought tolerant lineages or did C4 instead evolve prior (as a pre-adaptation to) salt and drought tolerance? Might factors, such as habitat preference or life-history strategy be important in the evolution of C4?
2. Material and methods
(a). Taxon sampling and molecular markers
The data matrix comprised 169 species (20 Amaranthaceae s.s. representing the major clades of the family [19], 147 Chenopodiaceae s.s. and two Achatocarpaceae as outgroups) and 2284 nucleotides (1343 rbcL gene and 941 atpB-rbcL spacer). Thirty-four species in the rbcL matrix and 16 species in the atpB-rbcL spacer matrix are coded as missing data (electronic supplementary material, appendix S1). Species sampling for this analysis was designed to include all major branches of all tribes of Chenopodiaceae s.s., representing the known phylogenetic diversity of C3 and C4 lineages (electronic supplementary material, table S3). Voucher information and GenBank accession numbers are given in the electronic supplementary material, table S4. Thirteen sequences of the rbcL gene and 33 of the atpB-rbcL spacer were newly generated for this study according to the protocols outlined in Kadereit et al. [16,26].
(b). Phylogenetic analysis and relaxed-clock molecular dating
Phylogenetic trees and node ages were generated using Bayesian evolutionary analysis by sampling trees (BEAST v. 1.5.4; [27,28]). The BEAST xml input files (available from the corresponding author upon request) were created with BEAUti v. 1.5.4 [27]. Two representatives of Achatocarpaceae were chosen as outgroups [16]. Monophyly of the ingroup (Amaranthaceae s.l.) was constrained in order to root the tree. The substitution model parameters were set to GTR + G based on JModelTest [29] with four categories for G. A relaxed-clock model was implemented in which rates for each branch are drawn independently from a lognormal distribution [30], and a birth and death demographic model was assumed.
We used two macrofossils to constrain the ages of the stem nodes of clades with which they share one or more synapomorphies: Salicornites messalongoi (stem fragment, 35.4–23.3 Myr ago; [31]) associated with the crown of Salicornioideae [26] and Parvangula randeckensis (seeds, 23.3–16 Myr ago [32]) associated with the stem of Chenopodieae I [16]. The corresponding prior age constraints used in the analyses were 23.3 and 16 Ma for crown of Salicornioideae and stem of Chenopodieae I, respectively, assuming in each case an exponential distribution with offset equal to the minimum bounds (i.e. allowing the posterior distribution to be older, but not younger than the constraint, as influenced by other parameters of the model).
The Markov chain Monte Carlo (MCMC) was initiated on a random starting tree. Two independent runs of 20 000 000 iterations were performed with a sampling frequency of 1000. Topological convergence was confirmed using Awty [33], and convergence of model parameters was confirmed using Tracer [28]. Burn-in values were determined empirically from the likelihood values and posterior probability (PP) clade support was calculated together with the medians and 95% confidence limits for ages of the nodes. The post-burn-in tree sample was also sub-sampled to obtain ca 1000 trees equally spaced throughout the two runs for use in character optimisations (see later text). In addition, clade support was estimated under parsimony and maximum likelihood using bootstrapping in PAUP* v. 4.10b [34] and RAxML [35], respectively.
(c). Ecological data scoring
For each species sampled, we compiled the type of photosynthesis (C3 or C4), habitat data (tolerance of saline conditions and occurrence in coastal or inland habitats), life form and succulence, from published ∂13C isotope data, flora treatments, revisions and other publications (see the electronic supplementary material, table S4). The data matrix comprised the following traits and character states: (i) photosynthesis (C3 = 0; C4 = 1), (ii) salt tolerance (non-tolerant or slightly salt-tolerant = 0, salt-tolerant = 1), (iii) occurrence in coastal habitats (inland habitats = 0, coastal habitats = 1), (iv) succulence (non-succulent = 0, succulent = 1), and (v) life form (perennial = 0, annual/biennial = 1).
(d). Ancestral character state reconstruction
To assess the ancestral states of the four traits, we used Fitch parsimony (FP) implemented in Mesquite v. 2.74 [36] and a hierarchical Bayesian method as implemented in BayesTraits [37], using the tree sample thinned from the Beast analysis. Outgroups and Amaranthaceae s.s. (including Polycnemoideae) were pruned, leaving Chenopodiaceae s.s. as the ingroup. Amaranthaceae s.s. includes both C3 and C4 species, but insufficient data are available to infer a phylogeny that is comparably well sampled and resolved as that for Chenopodiaceae. In our analyses, Amaranthaceae s.s. (including Polycnemoideae) is sister to Chenopodiaceae s.s. (PP: 0.99). We assume that excluding it from analyses will have no impact on our results, other than to remove a likely source of sampling bias.
For BayesTraits analyses, reversible-jump Markov chain Monte Carlo (RJ-MCMC) was used to sample between models with different numbers of parameters. The prior for models was uniform and that for the rate coefficient was exponentially distributed, with the variance drawn from a uniform hyperprior. Runs were of 100 million generations, sampling every 10 000, with a burn-in of one million (electronic supplementary material, table S2). Ninety-five per cent PP distributions of rates and PP per state per node of ancestral state reconstructions were summarized from the resulting output using Tracer v. 1.5 [28].
(e). Model testing
To test for correlated and/or directional evolution we used the discrete mode of BayesTraits with the same approach as for ancestral state reconstruction. Pairs of characters (C3 versus C4 with each other character in turn) were tested for correlated evolution under discrete dependent and independent modes under RJ-MCMC. In addition, reversible versus irreversible models of C4 evolution were tested using standard MCMC with a single rate parameter (one non-zero rate C3 ⇒ C4 versus identical rates C3 ⇔ C4). RJ-MCMC was performed as mentioned earlier, and both RJ-MCMC and standard MCMC were performed with two independent runs each of 1.5 × 109 generations with burn-in of 1 × 109 generations, sampling every 1.5 × 105, in order to ensure consistent and stable estimates of the harmonic mean of the likelihood. The harmonic means were then compared using Bayes factors (BFs): BF = 2(−LnL better fitting model − −LnL worse fitting model). BF ≥ 3 is considered significant. Finally, models of character-associated diversification were tested using BiSSE [38], as implemented in Mesquite using the maximum clade credibility tree, in order to test whether ancestral state reconstruction and model testing might be sensitive to differential rates of speciation or extinction in lineages depending on the inferred state of any of the characters assessed.
3. Results
(a). Phylogeny and ages
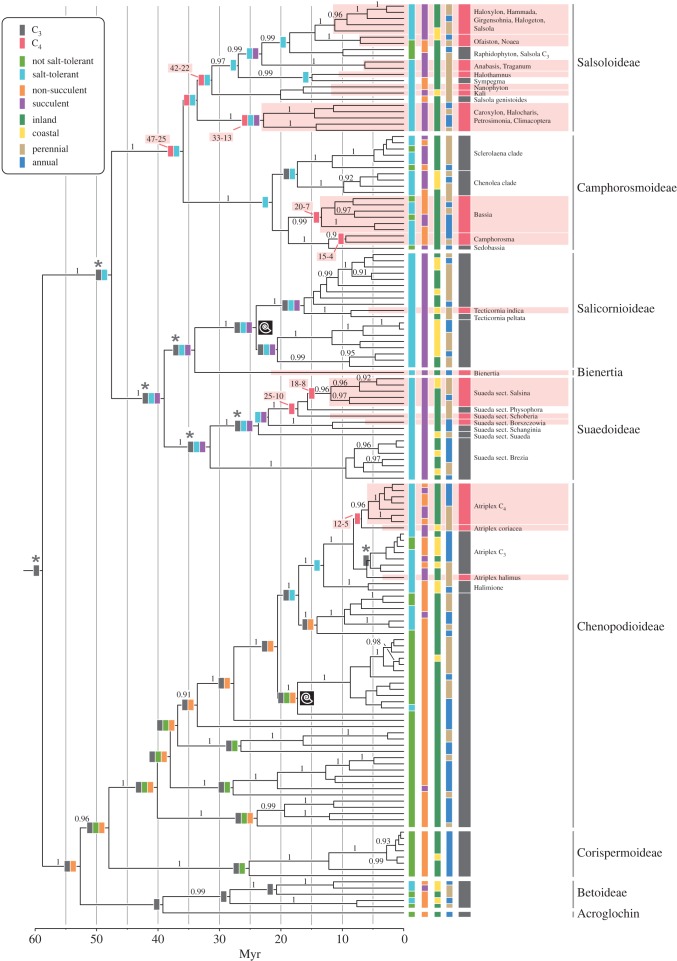
Phylogenetic inference under parsimony, likelihood and relaxed-clock Bayesian methods resulted in consistent tree topologies. Sixteen (not necessarily independent) lineages comprising exclusively C4 species are indicated in figure 1. Calibration of the relaxed molecular clock with two internal fossils (also indicated in figure 1) resulted in a stem age of Amaranthaceae s.l. of 87–47 Myr ago (95% CIs for ages are reported unless otherwise indicated). The chronogram with clade support is presented in figure 1 and the electronic supplementary material, figure S1.
Figure 1.
(Overleaf) Maximum clade credibility phylogenetic tree from BEAST analysis (branch lengths proportional to time; median values for node heights presented) with node support (Bayesian PP; BEAST) above the branches and ancestral states subtending selected nodes. Ancestral states that were unambiguous under FP but equivocal under RJ-MCMC are indicated with an asterisk. Clades of exclusively C4 species are highlighted and ages (millions of years; 95% CI) are given for deeper nodes inferred to be ancestrally C4. The nodes constrained to calibrate the relaxed-clock analyses are indicated with an ammonite symbol.
(b). Ancestral states
Ancestral states were estimated for five binary coded characters: (i) C3/C4; (ii) not salt-tolerant/salt-tolerant; (iii) inland/coastal; (iv) not succulent/succulent; (v) perennial/annual or biennial; using both FP and RJ-MCMC. The results of the two methods were consistent, but, as is to be expected, those under FP were more decisive. In the two characters with the highest overall rates of state changes inferred under RJ-MCMC (iii and v; see electronic supplementary material, table S1), ancestral state reconstructions were largely equivocal under RJ-MCMC, implying uncertainty associated with multiple changes along internal branches. In addition, BiSSE results indicated a state-dependent increased rate of extinction for character iii (see later). Because FP cannot model such phenomena, the FP reconstructions are disregarded for these characters; otherwise we report the most parsimonious solutions for deeper nodes where RJ-MCMC results are consistent, but subject to PP < 0.95 (figure 1). A more complex two-rate model was supported for character iii only: transitions from inland to coastal habitats occurred significantly less frequently than transitions from the coast inland (two-rate models sampled in greater than 95% of the RJ-MCMC). Both forward and reverse rates for all characters were greater than 0 (lower bounds of the 95% posterior distributions did not include zero).
For characters i, ii and iv, a summary of ancestral states recovered in greater than or equal to 95 per cent of trees under FP and/or with greater than or equal to 0.95 PP under RJ-MCMC is presented in figure 1a. Detailed results with PP values for reconstructions at selected nodes are presented in the electronic supplementary material, table S1. The central 95% range of numbers of transitions between C3 and C4 inferred under FP over the sample of BEAST trees is 5–12 gains and three to eight losses.
(c). Model testing
We tested for correlation between the evolution of C4 and the four other binary characters. In all but one case (that of annual/biennial versus perennial life history), dependent models better fitted the data than independent models (see the electronic supplementary material, table S2). These corresponded to two-rate models in which the rate of gain of C4 is higher in lineages (i) with salt tolerance; (ii) in coastal environments; and (iii) with succulence (figure 2). Dependent models for C4 and life-history strategy also scored slightly better than independent models, but the difference was subject to a BF of less than 3, thus not significant. The rate of reversals to C3 was not dependent on any of these factors, neither were transitions between these character states dependent on the mode of photosynthesis. The non-reversible model of C4 evolution was rejected with BF of 8.4. With a single exception, BiSSE models describing character-associated differential rates of diversification were not supported: constraining the rates of extinction or speciation to be equal given the different character states resulted in −LnL values that were not significantly different to those obtained when they were allowed to vary (electronic supplementary material, table S2). The better fitting model for inland versus coastal habitat implied both higher extinction at the coast (9.62 × 10−6 versus 6.53 × 10−6) and, similar to the RJ-MCMC model for this character, a higher transition rate from the coast inland than that from inland to the coast (0.124 versus 0.026).
Figure 2.
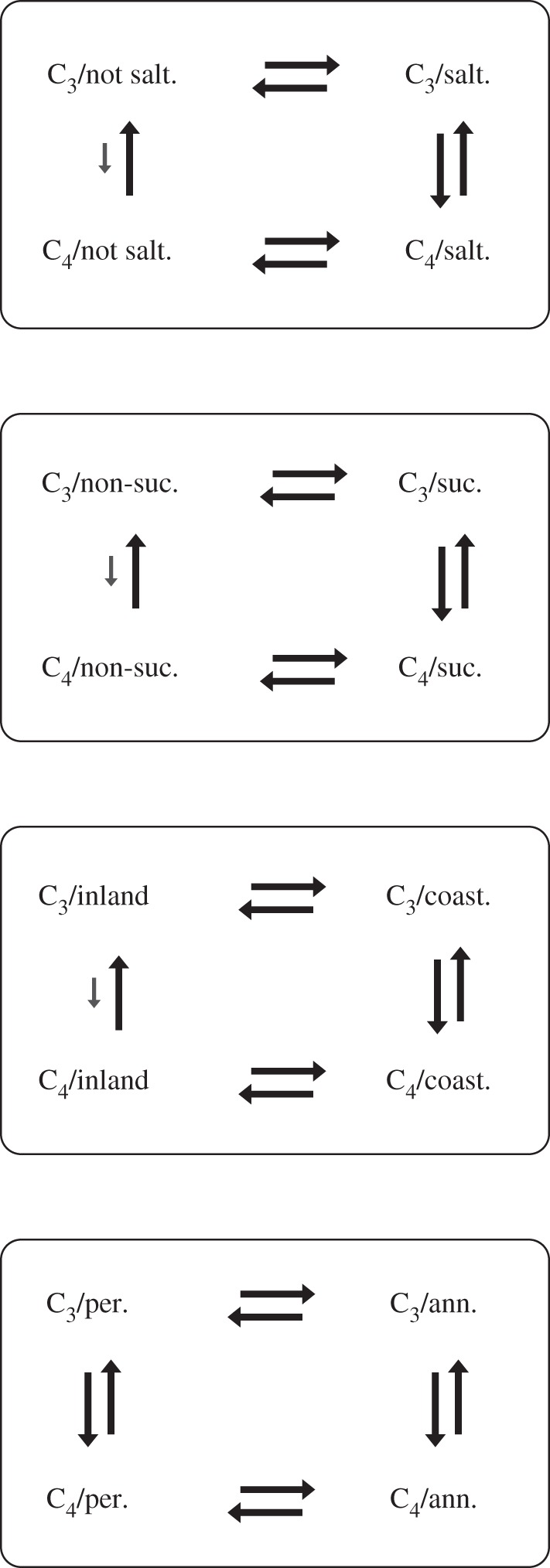
Models of character evolution inferred under RJ-MCMC. Rates of change between character states are represented by larger and smaller arrows (faster and slower rates, respectively).
4. Discussion
(a). Multiple gains and losses of C4 in chenopods
Given the numbers of independent origins of C4 in plants, it is intriguing that phylogenetic studies of clades showing both photosynthetic pathways have to date largely ruled out secondary loss of C4 (but see differences in the interpretation of C4 evolution in the grass genus Alloteropsis; [13,15,39]). The lack of conclusive phylogenetic evidence for reversals is mirrored by an absence of genetic evidence for a former C4 function in C3 lineages closely related to C4 lineages in grasses and sedges [40,41]. To explain this phenomenon, Christin et al. [13] argued that reversal from C4 to C3 is not simply a matter of loss of function of genes and structures involved in the CO2-concentrating pump, but instead would involve a complex restoration of the C3 condition, especially of those genes that were modified directly (without prior duplication) and lost their C3 function [42]. The evolution of C3 from a C4 ancestor might therefore represent a process that is just as complex as the gain of C4 in the first place. To further explain unresolved relationships of C4 lineages and their closest C3 relatives, Christin et al. [13] proposed that ancestral C3–C4 intermediate lineages that realized initial anatomical and biochemical changes on the way to a full C4 syndrome might have given rise to both C4 and C3 lineages by further genetic and biochemical modifications and loss of function, respectively.
Our analyses provide phylogenetic evidence for at least three reversals to C3 from within ‘full’ C4 lineages. Of the various candidate lineages (given uncertainty in both phylogenetic and ancestral state reconstruction) most can be placed within the Salsoloideae/Camphorosmoideae, which was reconstructed as ancestrally C4 and represents the oldest C4 clade within Chenopodiaceae (figure 1). This result may buck the apparent trend (at least in grasses), but it is not entirely without precedent. Carolin et al. [43] indicated a possible loss of C4 in the chenopod subfamily Salsoloideae on the basis of leaf anatomy, and previous phylogenetic analyses of Salsoloideae and Camphorosmoideae [5,18,44] did not rule out secondary loss. Interestingly, some species in Salsoloideae show the C3 pathway in their cotyledons before switching to C4 in their adult leaves [45]. If a fully functional C3 pathway is present in the seedling stage, then there seems little reason why C3 should not also be possible later in the life history. This dual photosynthetic strategy might have facilitated the reversal to a full C3 physiology in adult leaves, especially in cool semi-desert areas of Central Asia at montane to subalpine elevation where C3 appears to be competitive [46]. Further studies of cotyledon anatomy and their photosynthetic pathway as well as denser sampling within the relatively C3-rich Salsoleae (Salsoloideae) are needed for a more precise picture of possible reversions from C4 to C3 within this lineage. This in turn would allow a targeted search within C3 Salsoleae for genetic evidence for a former C4 function.
(b). The earliest known origins of C4 in plants
Our age estimate for Chenopodiaceae is based on fossil evidence unambiguously associated with nodes within the clade. The result for the stem age of Amaranthaceae s.l. (87–47 Myr ago) is consistent with the two oldest pollen fossils that have been assigned to Amaranthaceae s.l. (Polyporina cribaria: 86–65 Myr ago [47]; and Chenopodipollis multiplex: 65–56.5 Myr ago [48]), although these lack the unequivocal synapomorphies that would justify their inclusion in the dating analyses. Our age estimate is distinctly older than that of Wikström et al. [49] (basal split in core Caryophyllales 47–39 Myr ago), while falling within the bounds of more recent estimates for the crown of Caryophyllales (ca 94.5 Ma [50]).
Christin et al. [4] showed that the long-standing view of C4 eudicots having arisen later (mainly during the Pleistocene) than C4 monocots [51] is not correct. Comparison of C3/C4 transition models between eudicots and monocots suggested an increase of the probability of C4 evolution around 28 Ma for both groups [4]. In our analyses, the earliest inferred gain of C4, at the crown of Salsoloideae/Camphorosmoideae, is dated to between 47 and 26 Ma. Crown Salsoloideae, which is also inferred as C4 and comprises a majority of C4 species with similar anatomies, dates to 42–22 Myr ago (figure 1). These estimates are as old or older than that of the oldest C4 grass lineage (25 ± 4 Ma crown of core Chloridoideae; [52]). Nevertheless, the more recent bounds of the age range for C4 origins in Chenopodiaceae also include the pronounced drop of atmospheric CO2 at the Eocene/Oligocene boundary [2], and are therefore consistent with a scenario in which this drop in CO2 was necessary for the origin of C4.
The timing of C4 origins across angiosperms has been attributed to increased photorespiration at atmospheric CO2 levels lower than 500 ppm ([11] and references therein). However, the obvious success of C3 under current conditions as well as the occurrence of reversals to C3 suggests a more complex scenario in which further factors must be important in driving the evolution of C4 [10]. For example, the pronounced drop of atmospheric CO2 at the Eocene/Oligocene boundary [2] was accompanied by a distinct climate change dated to ca 35–33 Myr ago from warm and moist (tropical) to slightly cooler and more seasonal conditions [53,54]. Thus, in Chenopodiaceae, this first emergence of a more seasonal climate, in combination with the CO2 decrease, might have favoured C4 disproportionately in drought-prone and saline habitats.
(c). C4 evolved more often in salt-tolerant and succulent lineages
Saline soils decrease water availability to plants (physiological drought) and cause the accumulation of toxic concentrations of ions [55]. As a result, some plant species have evolved salt tolerance which involves multiple physiological adaptations [56] that are often also advantageous when the species is exposed to other environmental stresses such as drought and flooding [55,57,58]. C4 photosynthesis represents one such adaptation, improving water use efficiency relative to C3 and reducing ionic stress owing to reduced transpiration. Salinity stress may therefore promote the evolution of C4.
Many species of Chenopodiaceae are listed as highly salt-tolerant [56] or even salt requiring [59], and many are also succulent with distinct water storage tissues in leaves and/or stems. Our analyses show that salt tolerance and succulence evolved earlier than C4 and that C4 evolved significantly more often in salt-tolerant and succulent (and hence generally drought tolerant) lineages. The common ancestor of Salicornioideae, Suaedoideae, Camphorosmoideae and Salsoloideae, which dates back to the Eocene (crown group 61–35 Myr ago), was probably salt-tolerant (figure 1). All four subfamilies spread worldwide but originated in Eurasia [18,20,26]. At this time, the Eurasian climate was largely warm and moist [54], and saline habitats existed only on or near to the coast. We infer a high rate of change between coastal and inland habitats that hampers the ancestral state reconstruction. However, the rate of colonization of inland habitats from the coast was higher than the other way around. Through time, the coastal habitat is likely to have been smaller in extent and more dynamic (with changing coastlines) than inland regions. The coast might therefore be characterized by both a lower carrying capacity for species and a higher rate of extinction (the latter also implied by our BiSSE results). In this context, our results suggest a scenario in which adaptation to arid conditions (succulence, salt tolerance and indeed C4 photosynthesis) at the coast served as important pre-adaptations for species to enter dry environments such as steppes and deserts of the continental interior (the first origins of which also date back to the Eocene/Oligocene boundary). There they persisted longer than in the original coastal habitat.
We conclude that drought tolerance achieved by physiological adaptations such as salt tolerance and morphological/anatomical adaptations such as succulence are pre-adaptations that enhanced the evolution of C4 photosynthesis in Chenopodiaceae. This evolutionary sequence of events is in stark contrast to that inferred for grasses, where gain of C4 is seen as a pre-adaptation to enter drier habitats [22,24]. The ecological factors that triggered the evolution of C4 in grasses in the first place remain unclear in these studies. Our data suggest that C4 chenopod lineages are derived from C3 lineages that were already adapted in various ways (including different forms of succulence) to dry and/or saline habitats. These diverse origins led to a range of fundamentally different C4 anatomies in different lineages and might also be responsible for the fact that, in contrast to C4 grasses, C4 chenopods can be found in extremely dry environments. Indeed, a model that describes C4 evolution in chenopods might better explain C4 origins in exclusively arid lineages such as Mollugo cerviana and Mollugo fragilis [60], Chamaesyce [61] or Portulaca [62] than might one appropriate for grasses. In general, these results illustrate the critical importance of ancestral states and past environmental conditions in generating the high diversity of C4 syndromes observed across monocots and dicots. The findings are consistent with the widely held assumption that high levels of photorespiration pose the strongest selective pressure for the evolution of C4 photosynthesis [1,7,11,58]. However, they suggest that the current evolutionary paradigm based on grasses—including C4 as (irreversible) pre-adaptation to drought—may be an inadequate proxy for C4 evolution in general.
Acknowledgements
The Johannes Gutenberg-University Mainz (grant to G.K.) financially supported this study. M.D.P. acknowledges support from a Claude Leon Postdoctoral Fellowship. The authors thank B. Gehrke, J.W. Kadereit and two anonymous reviewers for commenting on the manuscript.
References
- 1.Bauwe H. 2011. Photorespiration: the bridge to C4 photosynthesis. In Advances in photosynthesis and respiration, vol. 32 (eds Raghavendra A. S., Sage R. F.), pp. 81–108 Berlin, Germany: Springer [Google Scholar]
- 2.Pagani M., Zachos J. C., Freeman K. H., Tipple B., Bohaty S. 2005. Marked decline in atmospheric carbon dioxide concentrations during the Paleogene. Science 309, 600–603 10.1126/science.1110063 (doi:10.1126/science.1110063) [DOI] [PubMed] [Google Scholar]
- 3.Beerling D. J., Royer D. L. 2011. Convergent Cenozoic CO2 history. Nat. Geosci. 4, 418–420 10.1038/ngeo1186 (doi:10.1038/ngeo1186) [DOI] [Google Scholar]
- 4.Christin P.-A., Osborne C. P., Sage R. F., Arakaki M., Edwards E. J. 2011. C4 eudicots are not younger than C4 monocots. J. Exp. Bot. 62, 3171–3181 10.1093/jxb/err041 (doi:10.1093/jxb/err041) [DOI] [PubMed] [Google Scholar]
- 5.Sage R. F., Christin P. A., Edwards E. J. 2011. The C4 plant lineages of planet Earth. J. Exp. Bot. 62, 3155–3169 10.1093/jxb/err048 (doi:10.1093/jxb/err048) [DOI] [PubMed] [Google Scholar]
- 6.Monson R. K. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164, S43–S54 10.1086/368400 (doi:10.1086/368400) [DOI] [Google Scholar]
- 7.Sage R. F. 2004. The evolution of C4 photosynthesis. New Phytol. 161, 341–370 10.1046/j.1469-8137.2004.00974.x (doi:10.1046/j.1469-8137.2004.00974.x) [DOI] [PubMed] [Google Scholar]
- 8.Tipple B. J., Pagani M. 2007. The early origins of terrestrial C4 photosynthesis. Annu. Rev. Earth Planet. Sci. 35, 435–461 10.1146/annurev.earth.35.031306.140150 (doi:10.1146/annurev.earth.35.031306.140150) [DOI] [Google Scholar]
- 9.Osborne C. P., Beerling D. J. 2006. Nature's green revolution: the remarkable evolutionary rise of C4 plants. Phil. Trans. R. Soc. B 361, 173–194 10.1098/rstb.2005.1737 (doi:10.1098/rstb.2005.1737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roalson E. H. 2008. C4 photosynthesis: differentiating causation and coincidence. Curr. Biol. 18, R167–R168 10.1016/j.cub.2007.12.022 (doi:10.1016/j.cub.2007.12.022) [DOI] [PubMed] [Google Scholar]
- 11.Sage R. F. 2001. Environmental and evolutionary preconditions for the origin and diversification of the C4 photosynthetic syndrome. Plant Biol. 3, 202–213 10.1055/s-2001-15206 (doi:10.1055/s-2001-15206) [DOI] [Google Scholar]
- 12.Gowik U., Westhoff P. 2011. The path from C3 to C4 photosynthesis. Plant Physiol. 155, 56–63 10.1104/pp.110.165308 (doi:10.1104/pp.110.165308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christin P.-A., Freckleton R. P., Osborne C. P. 2010. Can phylogenetics identify C4 origins and reversals? Trends Ecol. Evol. 25, 403–409 10.1016/j.tree.2010.04.007 (doi:10.1016/j.tree.2010.04.007) [DOI] [PubMed] [Google Scholar]
- 14.McKown A. D., Moncalvo J. M., Dengler N. G. 2005. Phylogeny of Flaveria (Asteraceae) and inference of C4 photosynthesis evolution. Am. J. Bot. 92, 1911. 10.3732/ajb.92.11.1911 (doi:10.3732/ajb.92.11.1911) [DOI] [PubMed] [Google Scholar]
- 15.Aliscioni S. & Grass Phylogeny Working Group II 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 193, 304–312 10.1111/j.1469-8137.2011.03972.x (doi:10.1111/j.1469-8137.2011.03972.x) [DOI] [PubMed] [Google Scholar]
- 16.Kadereit G., Borsch T., Weising K., Freitag H. 2003. Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164, 959–986 10.1086/378649 (doi:10.1086/378649) [DOI] [Google Scholar]
- 17.Kadereit G., Mavrodiev E. V., Zacharias E. H., Sukhorukov A. P. 2010. Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): implications for systematics, biogeography, flower and fruit evolution, and the origin of C4 photosynthesis. Am. J. Bot. 97, 1664–1687 10.1098/ajb.1000169v1 (doi:10.1098/ajb.1000169v1) [DOI] [PubMed] [Google Scholar]
- 18.Kadereit G., Freitag H. 2011. Molecular phylogeny of Camphorosmeae (Camphorosmoideae, Chenopodiaceae): implications for biogeography, evolution of C4-photosynthesis and taxonomy. Taxon 60, 51–78 [Google Scholar]
- 19.Sage R. F., Sage T. L., Pearcy R. W., Borsch T. 2007. The taxonomic distribution of C4 photosynthesis in Amaranthaceae sensu stricto. Am. J. Bot. 94, 1992–2003 10.3732/ajb.94.12.1992 (doi:10.3732/ajb.94.12.1992) [DOI] [PubMed] [Google Scholar]
- 20.Schütze P., Freitag H., Weising K. 2003. An integrated molecular and morphological study of the subfamily Suaedoideae Ulbr. (Chenopodiaceae). Plant Syst. Evol. 239, 257–286 10.1007/s00606-003-0013-2 (doi:10.1007/s00606-003-0013-2) [DOI] [Google Scholar]
- 21.Edwards G. E., Voznesenskaya E. V. 2011. C4 photosynthesis: Kranz forms and single-cell C4 in terrestrial plants. In C4 photosynthesis and related CO2 concentrating mechanisms (eds Raghavendra A. S., Sage R. F.), pp. 29–61 Berlin, Germany: Springer [Google Scholar]
- 22.Edwards E. J., Smith S. A. 2010. Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl Acad. Sci. USA 107, 2532–2537 10.1073/pnas.0909672107 (doi:10.1073/pnas.0909672107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards E. J., Still C. J. 2008. Climate, phylogeny and the ecological distribution of C4 grasses. Ecol. Lett. 11, 266–276 10.1111/j.1461-0248.2007.01144.x (doi:10.1111/j.1461-0248.2007.01144.x) [DOI] [PubMed] [Google Scholar]
- 24.Osborne C. P., Freckleton R. P. 2009. Ecological selection pressures for C4 photosynthesis in the grasses. Proc. R. Soc. B 276, 1753–1760 10.1098/rspb.2008.1762 (doi:10.1098/rspb.2008.1762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor S. H., Hulme S. P., Rees M., Ripley B. S., Ian Woodward F., Osborne C. P. 2010. Ecophysiological traits in C3 and C4 grasses: a phylogenetically controlled screening experiment. New Phytol. 185, 780–791 10.1111/j.1469-8137.2009.03102.x (doi:10.1111/j.1469-8137.2009.03102.x) [DOI] [PubMed] [Google Scholar]
- 26.Kadereit G., Mucina L., Freitag H. 2006. Phylogeny of Salicornioideae (Chenopodiaceae): diversification, biogeography, and evolutionary trends in leaf and flower morphology. Taxon 55, 617–642 10.2307/25065639 (doi:10.2307/25065639) [DOI] [Google Scholar]
- 27.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambaut A., Drummond A. J.2003. Tracer v. 1.5. (1.3 ed.) See http://beast.bio.ed.ac.uk/Tracer .
- 29.Posada D. 2008. JModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 10.1093/molbev/msn083 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 30.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Principi P. 1926. La flora oligocenica de Chiavon e Salcedo. Mem. Carta. Geol. d'Italia 10, 1–127 [Google Scholar]
- 32.Gregor H. J. 1982. Die» Parvangulae «und» Guttulae «Hiltermann & Schmitz 1968 aus dem Randecker Maar—Samenreste von Centrospermae. Palaeontol. Z. 56, 11–18 [Google Scholar]
- 33.Wilgenbusch J. C., Warren D. L., Swofford D. L. 2004. AWTY: a system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. See http://king2.scs.fsu.edu/CEBProjects/awty/awty_start.php. [DOI] [PubMed]
- 34.Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4 Sunderland, MA: Sinauer Associates [Google Scholar]
- 35.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 36.Maddison W. P., Maddison D. R. 2009. Mesquite: a modular system for evolutionary analysis. v. 2.72. See http://mesquiteproject.org
- 37.Pagel M., Meade A. 2006. BayesTraits. See www.evolution.rdg.ac.uk
- 38.Maddison W. P., Midford P. E., Otto S. P. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701. 10.1080/10635150701607033 (doi:10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim D. G., Burke T., Ripley B. S., Osborne C. P. 2009. A molecular phylogeny of the genus Alloteropsis (Panicoideae, Poaceae) suggests an evolutionary reversion from C4 to C3 photosynthesis. Ann. Bot. 103, 127–136 10.1093/aob/mcn204 (doi:10.1093/aob/mcn204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Besnard G., Muasya A. M., Russier F., Roalson E. H., Salamin N., Christin P.-A. 2009. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol. Biol. Evol. 26, 1909–1919 10.1093/molbev/msp103 (doi:10.1093/molbev/msp103) [DOI] [PubMed] [Google Scholar]
- 41.Christin P.-A., Salamin N., Savolainen V., Duvall M. R., Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. 17, 1241–1247 10.1016/j.cub.2007.06.036 (doi:10.1016/j.cub.2007.06.036) [DOI] [PubMed] [Google Scholar]
- 42.Tanz S. K., Tetu S. G., Vella N. G. F., Ludwig M. 2009. Loss of the transit peptide and an increase in gene expression of an ancestral chloroplastic carbonic anhydrase were instrumental in the evolution of the cytosolic C4 carbonic anhydrase in Flaveria. Plant Physiol. 150, 1515–1529 10.1104/pp.109.137513 (doi:10.1104/pp.109.137513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carolin R. C., Jacobs S. W. L., Vesk M. 1975. Leaf structure in Chenopodiaceae. Bot. Jahrb. Syst. 95, 226–255 [Google Scholar]
- 44.Akhani H., Edwards G., Roalson E. H. 2007. Diversification of the Old World Salsoleae s.i. (Chenopodiaceae): molecular phylogenetic analysis of nuclear and chloroplast data sets and a revised classification. Int. J. Plant Sci. 168, 931–956 10.1086/518263 (doi:10.1086/518263) [DOI] [Google Scholar]
- 45.Pyankov V. I., Ziegler H., Kuz'min A., Edwards G. 2001. Origin and evolution of C4 photosynthesis in the tribe Salsoleae (Chenopodiaceae) based on anatomical and biochemical types in leaves and cotyledons. Plant Syst. Evol. 230, 43–74 10.1007/s006060170004 (doi:10.1007/s006060170004) [DOI] [Google Scholar]
- 46.Pyankov V. I., Voznesenskaya E. V., Kondratschuk A. V., Black C. C. 1997. A comparative anatomical and biochemical analysis in Salsola (Chenopodiaceae) species with and without a Kranz type leaf anatomy: a possible reversion of C4 to C3 photosynthesis. Am. J. Bot. 84, 597–606 10.2307/2445895 (doi:10.2307/2445895) [DOI] [PubMed] [Google Scholar]
- 47.Srivastava S. K. 1969. Assorted angiosperm pollen from the Edmonton formation (Maestrichtian), Alberta, Canada. Can. J. Bot. 47, 975–989 10.1139/b69-138 (doi:10.1139/b69-138) [DOI] [Google Scholar]
- 48.Nichols D. J., Traverse A. 1971. Palynology, petrology, and depositional environments of some early Tertiary lignites in Texas. Geosci. Man 2, 37–48 10.1080/00721395.1971.9989707 (doi:10.1080/00721395.1971.9989707) [DOI] [Google Scholar]
- 49.Wikström N., Savolainen V., Chase M. W. 2001. Evolution of the angiosperms: calibrating the family tree. Proc. R. Soc. Lond. B 1482, 2211–2220 10.1098/rspb.2001.1782 (doi:10.1098/rspb.2001.1782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Magallon S., Castillo A. 2009. Angiosperm diversification through time. Am. J. Bot. 96, 349–365 10.3732/ajb.0800060 (doi:10.3732/ajb.0800060) [DOI] [PubMed] [Google Scholar]
- 51.Ehleringer J. R., Cerling T. E., Helliker B. R. 1997. C4 photosynthesis, atmospheric CO2, and climate. Oecologia 112, 285–299 10.1007/s004420050311 (doi:10.1007/s004420050311) [DOI] [PubMed] [Google Scholar]
- 52.Christin P.-A., Besnard G., Samartitani E., Duvall M. R., Hodkinson T. R., Savolainen V., Salamin N. 2008. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43 10.1016/j.cub.2007.11.058 (doi:10.1016/j.cub.2007.11.058) [DOI] [PubMed] [Google Scholar]
- 53.Eldrett J. S., Greenwood D. R., Harding I. C., Huber M. 2009. Increased seasonality through the Eocene to Oligocene transition in northern high latitudes. Nature 459, 969–973 10.1038/nature08069 (doi:10.1038/nature08069) [DOI] [PubMed] [Google Scholar]
- 54.Willis K. J., McElwain J. C. 2002. The evolution of plants. Oxford, UK: Oxford University Press [Google Scholar]
- 55.Munns R., Tester M. 2008. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 10.1146/annurev.arplant.59.032607.092911 (doi:10.1146/annurev.arplant.59.032607.092911) [DOI] [PubMed] [Google Scholar]
- 56.Flowers T. J., Colmer T. D. 2008. Salinity tolerance in halophytes. New Phytol. 179, 945–963 10.1111/j.1469-8137.2008.02531.x (doi:10.1111/j.1469-8137.2008.02531.x) [DOI] [PubMed] [Google Scholar]
- 57.Colmer T. D., Voesenek L. A. C. J. 2009. Flooding tolerance: suites of plant traits in variable environments. Funct. Plant Biol. 36, 665–681 10.1071/FP09144 (doi:10.1071/FP09144) [DOI] [PubMed] [Google Scholar]
- 58.Osborne C. P., Sack L. 2012. Evolution of C4 plants: a new hypothesis for an interaction of CO2 and water relations mediated by plant hydraulics. Phil. Trans. R. Soc. B 367, 583–600 10.1098/rstb.2011.0261 (doi:10.1098/rstb.2011.0261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan M. A., Ungar I. A., Showalter A. M. 2005. Salt stimulation and tolerance in an intertidal stem-succulent halophyte. J. Plant Nutr. 28, 1365–1374 10.1081/PLN-200067462 (doi:10.1081/PLN-200067462) [DOI] [Google Scholar]
- 60.Christin P. A., Sage T. L., Edwards E. J., Ogburn R. M., Khoshravesh R., Sage R. F. 2011. Complex evolutionary transitions and the significance of C3-C4 intermediate forms of photosynthesis in Molluginaceae. Evolution 65, 643–660 10.1111/j.1558-5646.2010.01168.x (doi:10.1111/j.1558-5646.2010.01168.x) [DOI] [PubMed] [Google Scholar]
- 61.Yang Y., Berry P. E. 2011. Phylogenetics of the Chamaesyce clade (Euphorbia, Euphorbiaceae): reticulate evolution and long-distance dispersal in a prominent C4 lineage. Am. J. Bot. 98, 1486–1503 10.3732/ajb.1000496 (doi:10.3732/ajb.1000496) [DOI] [PubMed] [Google Scholar]
- 62.Ocampo G., Columbus J. T. 2010. Molecular phylogenetics of suborder Cactineae (Caryophyllales), including insights into photosynthetic diversification and historical biogeography. Am. J. Bot. 97, 1827–1847 10.3732/ajb.1000227 (doi:10.3732/ajb.1000227) [DOI] [PubMed] [Google Scholar]