Abstract
Individuals naturally vary in the severity of infectious disease when exposed to a parasite. Dissecting this variation into genetic and environmental components can reveal whether or not this variation depends on the host genotype, parasite genotype or a range of environmental conditions. Complicating this task, however, is that the symptoms of disease result from the combined effect of a series of events, from the initial encounter between a host and parasite, through to the activation of the host immune system and the exploitation of host resources. Here, we use the crustacean Daphnia magna and its parasite Pasteuria ramosa to show how disentangling genetic and environmental factors at different stages of infection improves our understanding of the processes shaping infectious disease. Using compatible host–parasite combinations, we experimentally exclude variation in the ability of a parasite to penetrate the host, from measures of parasite clearance, the reduction in host fecundity and the proliferation of the parasite. We show how parasite resistance consists of two components that vary in environmental sensitivity, how the maternal environment influences all measured aspects of the within-host infection process and how host–parasite interactions following the penetration of the parasite into the host have a distinct temporal component.
Keywords: host–parasite, virulence, genotype-by-environment interaction, genotype-by-genotype interaction, Pasteuria ramosa, food stress
1. Introduction
Infectious disease is inevitably a product of both host and parasite genomes, with studies increasingly highlighting how host and parasite genotypes influence both the onset and severity of infectious disease (GP × GH; [1–5]), as well as how the environment may modify these interactions (GP × GH × E; [6,7]). An important challenge for studies of infectious disease, however, is to not only identify the genetic and environmental basis of the symptoms of infectious disease, but also how these patterns are related to the multiple steps involved in the process of infection. This process begins with behavioural and life-history adaptations that allow animals to reduce the likelihood of infection, followed by the external barriers, such as the cuticle, that physically prevent parasites from entering the host (e.g. pre-infection defences, see table 4.1 and references therein [8]). Much of the complexity of host–parasite interactions is expected in the subsequent within-host phase of the infection process, where immune activation or evasion by the parasite [9], the physiological condition of the host [10,11] and the quality of nutrient intake [12–14] all combine to influence the overall outcome of infection.
In recognition of the series of events that influence the onset and severity of disease, studies of host–parasite interactions have often decomposed infectious disease into estimates of infectivity, parasite growth and host health [15]. Such broad estimates, however, obscure aspects of the multistep infection process, such as the dependence of disease-related traits on events that occur earlier in the infection process [16] and the potential for host genotypes, parasites genotypes and the environment to interact at each underlying step [17]. Variation in infection rates, for example, will depend not only on the effectiveness of the host immune system in preventing parasite colonization, but also on the initial ability of the parasite to encounter and penetrate the host [18,19]. Likewise, subsequent estimates of host mortality or parasite spore loads represent the endpoint of a series of processes, including immune activation and host exploitation [9], each of which is unlikely to be influenced by host genotypes, parasite genotypes and the environment in identical ways.
Instead, by isolating individual steps of the infection process and then dissecting the genetic and environmental basis to disease-related traits, new insights can be generated into the dynamic nature of the within-host infection process. In biomedical studies, for example, variation in the ability of a parasite to encounter and penetrate a host is often circumvented artificially by injecting animals with pathogens of interest. While this has led to a greater understanding of immune function and defence capabilities at the molecular level (e.g. the Drosophila melanogaster immune reactions, reviewed in [20]), such an approach may inadvertently produce a distorted picture of within-host dynamics. By circumventing the natural route of infection, parasite doses may be unrealistic, the initial encounter of the parasite with the host immune system may occur incorrectly (e.g. injection into the wrong tissue) and naturally incompatible host and parasite genotypes may be combined. Studies of naturally occurring infections, by contrast, often rely on the use of parasite samples collected from infected hosts (known as isolates) to generate compatible host–parasite combinations. While the use of isolates mimics natural infections to some degree, the potential presence of multiple parasite genotypes within the host can significantly distort the expression of virulence [21,22].
In this study, we use the water flea Daphnia magna and its bacterial parasite, Pasteuria ramosa, to highlight the importance of the multi-step infection process for understanding the genetic and environmental basis to infectious disease. Although the influence of the environmental [23,24], genotype-by-environment [25,26] and genotype-by-genotype [3,27] interactions has been studied extensively in this system, insights from the stepwise infection process have only recently been incorporated. The recent development of P. ramosa clones, for example, has revealed that parasite susceptibility is highly specific [28] and only certain host–parasite genotype combinations are compatible [29]. Underlying this specificity is the attachment of the spores to the oesophagus of compatible host genotypes, a mechanism that is surprisingly unaffected by environmental variation [19]. Revaluating previous studies, therefore, it is clear that multiple processes are contributing to the onset and severity of disease, from the ability of a parasite to attach to the host oesophagus, to the effectiveness of the host in clearing the parasite and any interactions between different parasite genotypes within the host. Moreover, the environment appears to be able to influence only a subset of these processes, beginning presumably with the penetration of the parasite into the host.
By using compatible host–parasite combinations and manipulating food levels experienced by the mothers of focal Daphnia, our goal was to explore the sources of genetic and environmental variation acting during disease expression. The maternal manipulation is particularly relevant for perturbing the Daphnia–Pasteuria system, as food-stressed mothers have been shown to produce more resistant offspring [23,24]. With this approach, we were able to separate variation in the ability of a parasite to encounter and penetrate a host, from the post-penetration processes (within-host phase) that influence the ability of a parasite to colonize and proliferate within the host. We then measured the probability of a host becoming infected and estimated both parasite (spore load) and host fitness (offspring production and survival) at two time points after the parasite had entered the host: (i) in the middle of the within-host phase of infection as estimated at 28 days post exposure to the parasite, and (ii) at the terminal stage of infection as estimated at host death, approximately 50–60 days post exposure. In doing so, we aimed to find evidence for the ability of Daphnia to resist infectious disease; to explore the role of the maternal environment in the within-host process of infection; and to characterize the consistency of genotype and environmental interactions during the within-host infection process.
2. Material and methods
(a). The study system
The host, D. magna, is a freshwater crustacean that reproduces via cyclical parthenogenesis and is host to a range of bacterial, fungal and microsporidial parasites [30]. The parasite, P. ramosa, is an endospore-forming bacteria of Daphnia, which causes a severe loss of host fitness via castration and reduced survival [31]. After infection, the parasite grows within the host, filling the body cavity with several million endospores and giving an infected host a distinctive reddish-brown coloration. Transmission occurs horizontally with the spores released from the decaying cadaver of a formerly infected host [32].
For this experiment, all hosts were derived from two D. magna clones: HO2, which originates from Hungary; and M10, which originates from Belgium. The P. ramosa material was based on two clones: C1, derived via limited dilution from an isolate (P5) originating from Moscow, Russia; and C19, derived via single-spore infection from an isolate (P1) originating from North Germany. These geographical diverse combinations of host and parasite clones were chosen for two reasons. First, the two host clones and the two parasite clones are perfectly compatible with each other, which is necessary to exclude variation in the early infection steps. Second, we know that C1 and C19 are genetically distinct as they have different patterns of infection across a range of host clones [28]. While the use of such material cannot generate insights into the co-evolutionary processes occurring within populations, our goal was to increase our ability to dissect the genetic and environmental basis of the within-host infection process.
(b). Maternal environment manipulation
We manipulated the maternal environment using food stress following the process outlined in Ben-Ami et al. [24]. We began with 4-day-old Daphnia placed individually in a 100 ml jar filled with 20 ml of artificial medium [33]. On day 5, the high food treatment received 1 × 106 algae cells of Scenedesmus gracilis per Daphnia per day, which was increased to 2 × 106, 2.5 × 106, 3 × 106 and 8 × 106 algal cells per animal per day on days 6, 9, 11 and 13, respectively. For the low food treatment, food levels were initially 0.5 × 106 cells per animal per day on day 5, and increased to 1 × 106 and 2 × 106 cells per animal per day on days 11 and 13, respectively. The artificial medium was replaced weekly beginning on day 12 with 100 ml of fresh medium. The influence of the maternal environment was consistent for both Daphnia clones, as the offspring of low-food mothers produced approximately 10 fewer offspring over this period for both the M10 (low: 102.56 ± 2.11; high: 117.61 ± 3.94) and HO2 clones (low: 73.50 ± 1.77; high: 79.71 ± 2.32).
(c). Experimental trials
We used a factorial experimental design where two parasite clones (C1 and C19) were exposed to two host clones (HO2 and M10), each under two different maternal environmental (high food and low food). For each of the eight host–parasite–environment combinations, 28 individuals were allocated to be sampled 28 days after parasite exposure and 28 individuals to be sampled on the day of death. An additional 28 individuals were allocated to the parasite-free control group for each combination of host genotype and maternal environment (resulting in 560 individuals in total). Animals were collected daily from the food-manipulated mothers and placed individually in 100 ml jars filled with 20 ml of medium on day 4. On day 5, animals received either 10 000 spores of the appropriate exposure group (C1 or C19) or the equivalent volume of a control, uninfected Daphnia suspension. We replaced the medium with 100 ml of fresh medium one week later (day 12). Food levels followed the same feeding schedule as for the high food treatment of the mothers outlined above. All individuals were maintained within a single climate-controlled incubator (16 L : 8 D, 20°C ± 0.5°C) where the position within the incubator was regularly shuffled to reduce positional effects.
After this initial period, we moved individuals to new jars with fresh medium every 3 days and counted the number of offspring produced. Individuals were monitored daily, with deaths recorded and the Daphnia individually frozen (−20°C) for the assessment of infection status and spore production. Animals that died during the experiment before day 16 were excluded from all subsequent analyses as infection cannot be determined before this date. The numbers of mature P. ramosa spores were estimated by crushing the cadavers and counting two independent samples of this suspension in a counting chamber (Neubauer improved). Based on this design we could estimate three sets of informative traits: (i) the ability of the parasite to establish and proliferate within the host (based on the prevalence of disease); (ii) the genetic and environment basis to the growth of the parasite and the reduction in host fecundity at 28 days post exposure to the parasite; and (iii) the terminal characteristics of the infection based on estimates of host and parasite fitness at host death.
(d). Data analysis
We assessed the effects of parasite genotype, host genotype and the maternal environment on the characteristics of infectious disease using three-factor analysis of variance. The prevalence of disease was analysed using a generalized linear model with a binomial error distribution, with individuals included from both sampling points (28 days post infection and host death). All other traits were analysed using a least-squares linear model. However, for the characteristics of the early phase of infection, only individuals that survived until day 28 post parasite exposure were included in the analysis. The appropriate sample sizes for each treatment group are labelled accordingly on the figures. Before analysis we transformed lifetime offspring production using natural logarithms as the data were positively skewed, although the figures here are presented on the original scale to facilitate a comparison with the number of offspring produced before castration. All analyses were conducted in JMP (v. 9: SAS Institute Inc, NC, USA).
3. Results
(a). The ability of a parasite to establish within a host
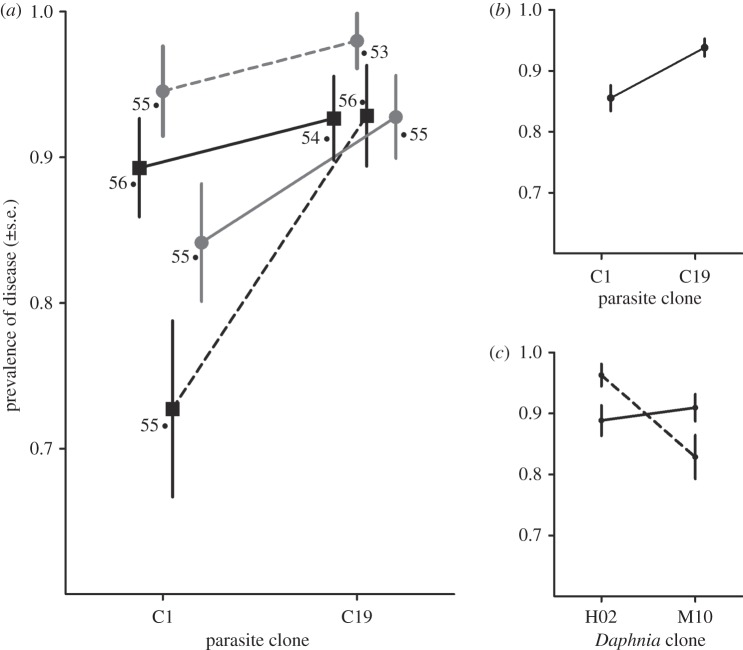
To characterize the ability of the parasite to initially establish within the host, we estimated the proportion of infected animals in each group. Based on visible signs of infection, we found that the ability of the parasites to establish within the host was influenced by the parasite genotype, the host genotype and an interaction between host genotype and the maternal environment (table 1 and figure 1a). The prevalence of disease was higher for individuals infected with the C19 parasite clone than the C1 parasite clone (figure 1b), while the low maternal food stress environment resulted in a decrease in the probability of disease for the HO2 Daphnia clone, but the reverse pattern was observed for the M10 Daphnia clone (figure 1c). These data indicate that following the penetration of P. ramosa into the host, components of the parasite genotype, host genotype and the environment all influence how likely it is that the parasite escapes being cleared by the host.
Table 1.
Effects of parasite genotype, host genotype and the maternal environment on the characteristics of disease at an intermediate stage of the within-host infection process. (Traits measured include the prevalence of disease, the number of offspring produced and the spore load of infected individuals as estimated at 28 days post exposure to the parasite. Asterisks denotes significant effects (α = 0.05).)
| prevalence of disease |
early offspring production |
early spore production |
||||
|---|---|---|---|---|---|---|
| effect tests | 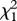 |
p-value | F1,184 | p-value | F1,184 | p-value |
| parasite genotype (GP) | 7.840 | 0.005* | 1.817 | 0.179 | 0.257 | 0.613 |
| Daphnia genotype (GH) | 3.931 | 0.048* | 25.252 | <0.001* | 2.772 | 0.098 |
| maternal environment (E) | 1.022 | 0.312 | 17.368 | <0.001* | 0.007 | 0.663 |
| GP × GH | 0.001 | 0.989 | 0.771 | 0.381 | 6.247 | 0.013* |
| GP × E | 0.880 | 0.348 | 9.780 | 0.002* | 0.796 | 0.374 |
| GH × E | 6.937 | 0.008* | 0.476 | 0.491 | 0.190 | 0.663 |
| GP × GH × E | 0.368 | 0.544 | 1.638 | 0.202 | 0.003 | 0.987 |
Figure 1.
Influence of parasite genotype, host genotype and the maternal environment on the infection status of individuals following the penetration of the parasite into the host. Shown are both the treatment means (a), as well as the significant main effects for parasite genotype (b) and the host genotype by maternal environment interaction (c). Sample sizes are indicated for each treatment group in (a). (a) Filled box denotes M10 Daphnia clone; filled circle denotes H02 Daphnia clone; solid line denotes low-food maternal environment; and dashed line denotes high food maternal environment. (c) Solid line denotes low-food mothers and dashed line denotes high-food mothers.
(b). Characteristics of disease at 28 days after parasite exposure
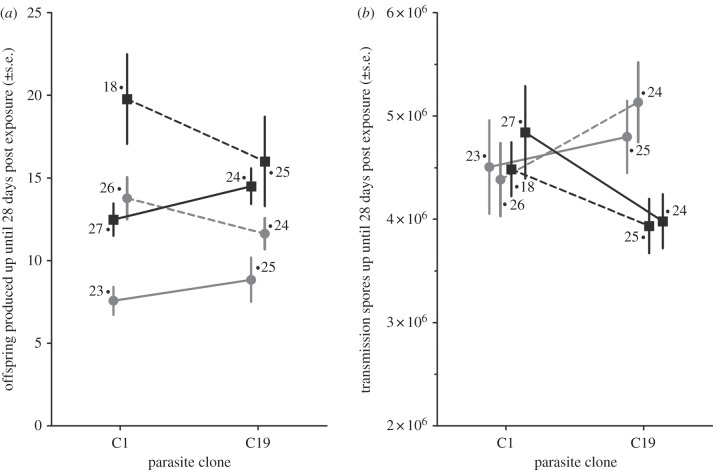
We measured the production of offspring and parasite spore loads at 28 days after parasite exposure in order to characterize the proliferation of the host once infection was successful. Interactions between parasite genotypes, host genotypes and maternal environment were all involved in aspects of disease at this stage. Host genotype, maternal environment and a parasite genotype by maternal environment interaction had a significant effect on the ability of the parasite to castrate the host (table 1 and figure 2a). Host fecundity was lowest for the H02 Daphnia clones in general, while the difference between low- and high-food maternal environments was greater for the C1 parasite clone than for the C19 parasite clone. In contrast to the patterns of fecundity, the number of parasite spores after 28 days depended on the interaction between host and parasite genotypes, but not on the influence of the maternal environment (table 1 and figure 2b). For the C19 parasite genotype, the increase in the number of spores clearly depends on the genotype of the infected host, while there was no difference in the increase in spore numbers between the different hosts for the C1 parasite.
Figure 2.
Influence of parasite genotype, host genotype and the maternal environment on traits describing the severity of disease at 28 days post exposure to the parasite. Shown are the treatment means for the number of offspring produced before host castration (a) and the number of parasite transmissions spores (b). Sample sizes are indicated for each treatment group. Filled square denotes M10 Daphnia clone; filled circle denotes H02 Daphnia clone; solid lines denote low-food maternal environment; dashed line denotes high-food maternal environment.
(c). Characteristics of disease at host death
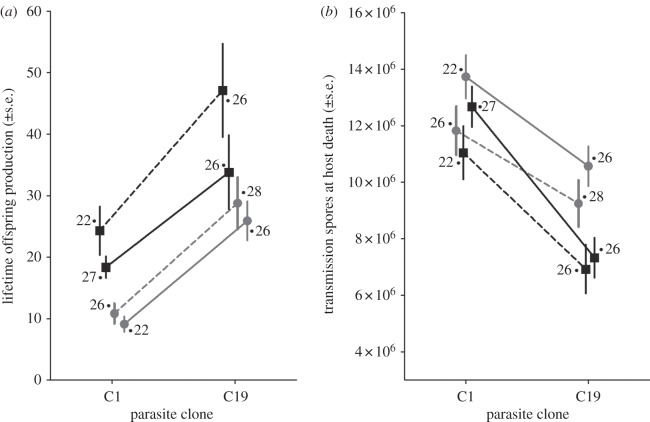
To characterize the genetic and environmental basis to the terminal phase of infection, we monitored infected individuals throughout and measured host survival, lifetime offspring production and final spore counts at host death. We found no significant treatment effects on the survival of Daphnia after infection (table 2; figure not shown). On average, all infected individuals survived for approximately 50 days after being exposed to the parasite (52.23 ± 0.58). This represents a substantial decrease from the survival expected for control individuals, who survived on average 108 days. For the overall severity of disease, however, the lifetime production of offspring by an infected individual was dependent on the host and parasite genotypes, the maternal environment and a marginal interaction between host genotype and maternal environment (table 2). The dominant feature of the treatment effects (figure 3a) is that the overall severity of disease is much less for Daphnia infected by the C19 parasite, irrespective of host genotype and maternal environment.
Table 2.
Results of the analyses describing the effects of parasite genotype, host genotype and the maternal environment on the characteristics of disease at the terminal stage of the within-host infection process. (Traits measured include the survival and lifetime offspring production of an infected host, as well as the lifetime spore production of the parasite, as estimated at time of host death. Asterisks denotes significant effects (α = 0.05).)
| host survival |
lifetime offspring production |
lifetime spore production |
||||
|---|---|---|---|---|---|---|
| effect tests | F1,192 | p-value | F1,195 | p-value | F1,195 | p-value |
| parasite genotype (GP) | 0.675 | 0.412 | 38.576 | <0.001* | 37.279 | <0.001* |
| Daphnia genotype (GH) | 0.265 | 0.607 | 14.269 | <0.001* | 12.038 | <0.001* |
| maternal environment (E) | 0.351 | 0.555 | 5.799 | 0.017* | 6.146 | 0.014* |
| GP × GH | 0.165 | 0.685 | 0.050 | 0.824 | 1.319 | 0.252 |
| GP × E | 0.369 | 0.544 | 0.389 | 0.534 | 0.426 | 0.515 |
| GH × E | 1.350 | 0.267 | 4.070 | 0.045* | 0.890 | 0.347 |
| GP × GH × E | 1.314 | 0.253 | 0.090 | 0.764 | 0.011 | 0.915 |
Figure 3.
Influence of parasite genotype, host genotype and the maternal environment on traits describing the severity of disease at host death. Shown are the treatment means for the lifetime production of offspring by an infected host (a) and the number of parasite transmissions spores produced at host death by the parasite (b). Sample sizes are indicated for each treatment group. Filled square denotes M10 Daphnia clone; filled circle denotes H02 Daphnia clone; solid line denotes low-food maternal environment; and dashed line denotes high-food maternal environment.
This pattern appears to be driven by the ability of the C19-infected Daphnia to produce eggs late after infection (after 28 days), as the proportion of individuals producing at least one clutch after this time was highest for C19 (74 of 106 individuals), whereas almost no individuals infected with C1 were able to reproduce again (only 3 of 94 individuals). Otherwise, the differences between the Daphnia appears to remain consistent between the initial proliferation of the parasite within the host (figure 2a) versus the terminal phases of infection at host death (figure 3a), with M10 clones producing more offspring in both cases. In comparison, we found that the number of spores produced when a host dies depended on the parasite genotype, host genotype and the maternal environment (table 2 and figure 3b). There were no significant interactions among these factors. Parasite spore loads were, on average, highest for infections involving the C1 parasite clone, the H02 Daphnia clone and Daphnia whose mothers had experienced the low-food environment.
4. Discussion
In this study, we build on recent advances in our understanding of the infection process in D. magna and its parasite P. ramosa, to dissect the influence of genetic and environmental factors on the outcome of bacterial infection. Our results reveal how host genotype, parasite genotype and the maternal environment were all involved in the ability of the parasite to colonize the host, the associated reduction in host fecundity and the proliferation of the parasite within the host. While similar influences of genotype and environmental factors on patterns of disease has been examined previously in the Daphnia–Pasteuria system [3,25–27], our findings are based on the use of single-genotype parasite clones and compatible host–parasite combinations. By separating the ability of a parasite to encounter and penetrate a host, from the post-penetration processes that influence the ability of a parasite to colonize and proliferate within the host, our findings generate new insights into the process by which hosts fight infection and the role that the environment has in shaping infectious disease.
(a). Evidence for the effectiveness of the within-host defence cascade
First, we show that even when the parasite is certain to naturally penetrate the host, the host can still completely recover from infection. For many studies of host–parasite interactions, such results should not be surprising. However, the ability of Daphnia to clear the parasite has recently been questioned, as it is now known that the infection process begins with specific parasite clones being able to attach to and penetrate the oesophagus of compatible host genotypes [19,28]. By using compatible host–parasite combinations and experimentally removing variation in this highly specific step, we confirm that the subsequent, within-host phase can also contribute significantly to resistance. It remains unclear whether this within-host resistance is related to the ability of the Daphnia immune system to aid in parasite clearance, as physiological studies of the phenoloxidase cascade, cellular response and nitric oxide pathways have not conclusively related constituent levels of immune activity to patterns of disease resistance [34–36]. Nonetheless, our results, in combination with those of Duneau et al. [19], reinforce how resistance for P. ramosa appears to have two distinct components: one based on host–parasite compatibility for the attachment of the parasite spores to the host oesophagus; and the other based on the ability of the within-host immune response or physiology to prevent the parasite from establishing.
Our findings also highlight how recognition of these two distinct components of resistance is essential for understanding the role of the environment in shaping infectious disease. The attachment and penetration of P. ramosa via the host oesophagus, for example, has previously been shown to depend on strong genetic interactions between hosts and parasites, but not on a range of environmental factors (e.g. food level, temperature and crowding [19]). By contrast, our findings show how food stress and the maternal environment influences multiple aspects of the subsequent within-host infection process, from the ability of the host to clear the infection (figure 1), through to the production of parasite spores and the reduction of host fecundity (figures 2 and 3). Similar conclusions have been made in plant–pathogen systems, where pathogen infectivity and host resistance (presumably under the control of major effect genes) have been shown to remain stable over a range of environmental conditions [37]. In general, however, the role of the environment in shaping the development of disease within compatible host–parasite combinations, and thus potentially overriding patterns of resistance, remains underexplored in animals.
(b). Food stress, the maternal environment and infectious disease
Second, while the influence of the maternal environment on all measured aspects of the within-host infection process adds to the growing awareness of how important maternal effects can be for the onset and severity of infectious disease [23,24,38,39], our results also highlight how challenging it can be to predict the impact of the maternal environment. Based on previous studies [23,24], our prediction was that the low-food maternal environment would enhance the ability of offspring to avoid disease. By contrast, our results indicate that the influence of maternal environment on the resistance to P. ramosa was specific to the genotype of the host (GH × E; figure 1c). The maternal environment also modified the ability of a parasite genotype to reduce host fecundity within the first month after parasite exposure (GP × E; figure 2a). The genotype-specific impact of the maternal environment (G × E), together with the rapid fluctuation in food quantity that commonly occurs in freshwater ponds or lakes, suggests that food-related maternal stress may play an important role in maintaining variation in natural Daphnia populations [15,40].
In combination with the study of Stjernman & Little [41], we can further identify the specific component of the infection process which will be affected. In both studies, only genotype-by-environment interactions describing the ability of the parasite to establish in the host (and hence evidence for parasite clearance) resulted in a change in the rank order of host genotypes (e.g. crossing reaction norms; figure 1c). Furthermore, our findings show how at the time of host death, any maternal effects had equalized between different genotypes, such that the overall severity of disease was greater for the low-food maternal environment (lower fecundity, higher parasite load; figure 3), irrespective of the host or parasite genotype. These findings suggest that the potential for food stress and the maternal environment to maintain genetic variation is related specifically to parasite susceptibility and the initial ability of the host to prevent the parasite from establishing.
An additional aspect of our experimental design was the ability to assess if the maternal environment could modify the outcome of the interaction between host and parasite genomes (GP × GH × E). To date, only a limited number of studies have explored such complex three-way interactions [6,7,27]. In Daphnia, evidence for GP × GH × E interactions has previously been explored by Vale and Little [27] via temperature manipulation. As with this previous study, we also did not find evidence of a GP × GH × E interaction for any of the estimated disease-related traits. Nonetheless, with the ongoing development of new P. ramosa clones and the resulting increase in available genotypes, it is likely that environmental modification of host–parasite genetic interactions will be identified for some component of the infection process. Instead, the challenge will be to identify the underlying physiological mechanisms shaping the interaction between host genomes, parasite genomes and the environment.
(c). The temporal aspect of the within-host infection process
Finally, our study highlights how genetic and environmental interactions within the host have a distinct temporal component. Host–parasite interactions almost one month after parasite exposure were characterized by both genotype-by-environment (G × E) and genotype-by-genotype (G × G) interactions. Host GH × E interactions influenced infection rates (figure 1c), parasite GP × E interactions influenced the reduction in host fecundity (figure 2a) and parasite and host GH × GP interactions were important for the early proliferation of the parasite within the host (figure 2b). By contrast, measures taken at the terminal (lifetime host and parasite reproductive success estimated at host death) stage of the infection process were characterized by the largely independent effects of host genotype, parasite genotype and maternal environment (table 2 and figure 3). Contributing to these differences may be an unrecognized aspect of P. ramosa biology, where individuals infected with certain parasite genotypes (only C19) were able to produce a limited number of clutches again late in life. These late reproducing hosts, however, did not clear the infection, but only temporarily produced more offspring, remaining infected until they died.
Nonetheless, we believe that the absence of strong genetic and environmental interactions late in the infection process may be a common feature of host–parasite interactions. In studies of D. magna and P. ramosa, for example, complex genotype and environmental interactions (G × G and G × E) may be readily expected soon after a host encounters the parasite, owing to the activation of the host immune system [34,36,42,43], and the gradual control of host reproduction and growth [31]. Conversely, at the terminal phase of infection, the lack of strong G × G and G × E interactions may instead reflect how lifetime offspring and spore production ultimately depend on how resources were shared between host and parasites [31]. Other invertebrate studies have also suggested that immune activation and the potential parasite clearance can occur in a matter of hours, while the control of host resources by the parasite is a much more gradual process [18]. For many host–parasite studies, however, this dynamic aspect of the within-host infection process site may be overlooked as disease-related traits are often only measured at a single time point following the exposure of the parasite to a host.
5. Conclusion
In summary, by using compatible host–parasite combinations and focusing exclusively on the within-host infection process, we have demonstrated how different mechanisms of resistance can combine in the fight against infectious disease (for a review on this topic see [44]). Contrasting the resistance based on host–parasite genetic compatibility versus the within-host defence cascade, we show how the maternal environment is involved in all measurable aspects underlying the severity of disease. Reaffirming the importance of the dynamic nature of interactions occurring within the host, however, we suggest that the role of the maternal environment in maintaining genetic variation in disease traits may diminish as the within-host infection process progresses. Extending this work to focus on the direct effects of food stress on the parasite in subsequent infection events would complement our current findings and provide a complete transgenerational understanding of infection for P. ramosa.
Acknowledgements
This study was supported by an EU Marie Curie Incoming International Fellowship (PIIF-GA-2009-252417) to M.D.H. and by the Swiss National Science Foundation. We thank F. Ben-Ami, M. Clerc and two anonymous referees for comments on the manuscipt and J. Hottinger and U. Stiefel for laboratory assistance.
References
- 1.Lazzaro B. P., Sackton T. B., Clark A. G. 2006. Genetic variation in Drosophila melanogaster resistance to infection: a comparison across bacteria. Genetics 174, 1539–1554 10.1534/genetics.105.054593 (doi:10.1534/genetics.105.054593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambrechts L., Halbert J., Durand P., Gouagna L. C., Koella J. C. 2005. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malaria J. 4, 3. 10.1186/1475-2875-4-3 (doi:10.1186/1475-2875-4-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carius H. J., Little T. J., Ebert D. 2001. Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution. 55, 1136–1145 [DOI] [PubMed] [Google Scholar]
- 4.Schulenburg H., Ewbank J. J. 2004. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4, 49. 10.1186/1471-2148-4-49 (doi:10.1186/1471-2148-4-49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvaudon L., Héraudet V., Shykoff J. A. 2005. Parasite–host fitness trade-offs change with parasite identity: genotype-specific interactions in a plant-pathogen system. Evolution 59, 2518–2524 [PubMed] [Google Scholar]
- 6.Bryner S. F., Rigling D. 2010. Temperature dependent genotype by genotype interaction between a pathogenic fungus and its hyperparasitic virus. Am. Nat. 177, 65–74 10.1086/657620 (doi:10.1086/657620) [DOI] [PubMed] [Google Scholar]
- 7.Sadd B. M. 2011. Food-environment mediates the outcome of specific interactions between a bumblebee and its trypanosome parasite. Evolution 65, 2995–3001 10.1111/j.1558-5646.2011.01345.x (doi:10.1111/j.1558-5646.2011.01345.x) [DOI] [PubMed] [Google Scholar]
- 8.Schmid-Hempel P. 2011. Evolutionary parasitology: the integrated study of infections, immunology, ecology, and genetics. Oxford, UK: Oxford University Press [Google Scholar]
- 9.Schmid-Hempel P. 2008. Parasite immune evasion: a momentous molecular war. Trends Ecol. Evol. 23, 318–326 10.1016/j.tree.2008.02.011 (doi:10.1016/j.tree.2008.02.011) [DOI] [PubMed] [Google Scholar]
- 10.Krist A. C., Jokela J., Wiehn J., Lively C. M. 2004. Effects of host condition on susceptibility to infection, parasite developmental rate, and parasite transmission in a snail–trematode interaction. J. Evol. Biol. 17, 33–40 10.1046/j.1420-9101.2003.00661.x (doi:10.1046/j.1420-9101.2003.00661.x) [DOI] [PubMed] [Google Scholar]
- 11.Seppälä O., Liljeroos K., Karvonen A., Jokela J. 2008. Host condition as a constraint for parasite reproduction. Oikos 117, 749–753 10.1111/j.2008.0030-1299.16396.x (doi:10.1111/j.2008.0030-1299.16396.x) [DOI] [Google Scholar]
- 12.Lee K. P., Simpson S. J., Wilson K. 2008. Dietary protein-quality influences melanization and immune function in an insect. Funct. Ecol. 22, 1052–1061 10.1111/j.1365-2435.2008.01459.x (doi:10.1111/j.1365-2435.2008.01459.x) [DOI] [Google Scholar]
- 13.Bedhomme S., Agnew P., Sidobre C., Michalakis Y. 2004. Virulence reaction norms across a food gradient. Proc. R. Soc. Lond. B 271, 739–44 10.1098/rspb.2003.2657 (doi:10.1098/rspb.2003.2657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost P. C., Ebert D., Smith V. H. 2008. Responses of a bacterial pathogen to phosphorus limitation of its aquatic invertebrate host. Ecology 89, 313–318 10.1890/07-0389.1 (doi:10.1890/07-0389.1) [DOI] [PubMed] [Google Scholar]
- 15.Wolinska J., King K. C. 2009. Environment can alter selection in host-parasite interactions. Trends Parasitol. 25, 236–244 10.1016/j.pt.2009.02.004 (doi:10.1016/j.pt.2009.02.004) [DOI] [PubMed] [Google Scholar]
- 16.Schmid-Hempel P., Ebert D. 2003. On the evolutionary ecology of specific immune defence. Trends Ecol. Evol. 18, 27–32 10.1016/S0169-5347(02)00013-7 (doi:10.1016/S0169-5347(02)00013-7) [DOI] [Google Scholar]
- 17.Lambrechts L., Fellous S., Koella J. C. 2006. Coevolutionary interactions between host and parasite genotypes. Trends Parasitol. 22, 12–6 10.1016/j.pt.2005.11.008 (doi:10.1016/j.pt.2005.11.008) [DOI] [PubMed] [Google Scholar]
- 18.Haine E. R., Moret Y., Siva-jothy M. T., Rolff J. 2008. Antimicrobial defense and persistent infection in insects. Science 322, 1257–1259 10.1126/science.1165265 (doi:10.1126/science.1165265) [DOI] [PubMed] [Google Scholar]
- 19.Duneau D., Luijckx P., Ben-Ami F., Laforsch C., Ebert D. 2011. Resolving the infection process reveals striking differences in the contribution of environment, genetics and phylogeny to host–parasite interactions. BMC Biol. 9, 11. 10.1186/1741-7007-9-11 (doi:10.1186/1741-7007-9-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemaitre B., Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 10.1146/annurev.immunol.25.022106.141615 (doi:10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 21.Ben-Ami F., Mouton L., Ebert D. 2008. The effects of multiple infections on the expression and evolution of virulence in a Daphnia-endoparasite system. Evolution 62, 1700–1711 10.1111/j.1558-5646.2008.00391.x (doi:10.1111/j.1558-5646.2008.00391.x) [DOI] [PubMed] [Google Scholar]
- 22.Wegner K. M., Berenos C., Schmid-Hempel P. 2009. Host genetic architecture in single and multiple infections. J. Evol. Biol. 22, 396–404 10.1111/j.1420-9101.2008.01657.x (doi:10.1111/j.1420-9101.2008.01657.x) [DOI] [PubMed] [Google Scholar]
- 23.Mitchell S. E., Read A. F. 2005. Poor maternal environment enhances offspring disease resistance in an invertebrate. Proc. R. Soc. B 272, 2601–2607 10.1098/rspb.2005.3253 (doi:10.1098/rspb.2005.3253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Ami F., Ebert D., Regoes R. R. 2010. Pathogen dose infectivity curves as a method to analyze the distribution of host susceptibility: a quantitative assessment of maternal effects after food stress and pathogen exposure. Am. Nat. 175, 106–115 10.1086/648672 (doi:10.1086/648672) [DOI] [PubMed] [Google Scholar]
- 25.Mitchell S. E., Rogers E. S., Little T. J., Read A. F. 2005. Host-parasite and genotype-by-environment interactions: temperature modifies potential for selection by a sterilizing pathogen. Evolution 59, 70–80 10.1554/04-526 (doi:10.1554/04-526) [DOI] [PubMed] [Google Scholar]
- 26.Vale P. F., Stjernman M., Little T. J. 2008. Temperature-dependent costs of parasitism and maintenance of polymorphism under genotype-by-environment interactions. J. Evol. Biol. 21, 1418–1427 10.1111/j.1420-9101.2008.01555.x (doi:10.1111/j.1420-9101.2008.01555.x) [DOI] [PubMed] [Google Scholar]
- 27.Vale P. F., Little T. J. 2009. Measuring parasite fitness under genetic and thermal variation. Heredity 103, 102–109 10.1038/hdy.2009.54 (doi:10.1038/hdy.2009.54) [DOI] [PubMed] [Google Scholar]
- 28.Luijckx P., Ben-Ami F., Mouton L., Du Pasquier L., Ebert D. 2010. Cloning of the unculturable parasite Pasteuria ramosa and its Daphnia host reveals extreme genotype–genotype interactions. Ecol. Lett. 14, 125–131 10.1111/j.1461-0248.2010.01561.x (doi:10.1111/j.1461-0248.2010.01561.x) [DOI] [PubMed] [Google Scholar]
- 29.Ben-Ami F., Regoes R. R., Ebert D. 2008. A quantitative test of the relationship between parasite dose and infection probability across different host–parasite combinations. Proc. R. Soc. B 275, 853–859 10.1098/rspb.2007.1544 (doi:10.1098/rspb.2007.1544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert D. 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information; See http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books [Google Scholar]
- 31.Ebert D., Carius H. J., Little T., Decaestecker E. 2004. The evolution of virulence when parasites cause host castration and gigantism. Am. Nat. 164(Suppl.), S19–S32 10.1086/424606 (doi:10.1086/424606) [DOI] [PubMed] [Google Scholar]
- 32.Jensen K. H., Little T. J., Skorping A., Ebert D. 2006. Empirical support for optimal virulence in a castrating parasite. PLoS Biol. 4, e197. 10.1371/journal.pbio.0040197 (doi:10.1371/journal.pbio.0040197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebert D., Zschokke-Rohringer C. D., Carius H. J. 1998. Within-and between-population variation for resistance of Daphnia magna to the bacterial endoparasite Pasteuria ramosa. Proc. R. Soc. Lond. B 265, 2127–2134 10.1098/rspb.1998.0549 (doi:10.1098/rspb.1998.0549) [DOI] [Google Scholar]
- 34.Pauwels K., De Meester L., Decaestecker E., Stoks R. 2011. Phenoloxidase but not lytic activity reflects resistance against Pasteuria ramosa in Daphnia magna. Biol. Lett. 7, 156–159 10.1098/rsbl.2010.0634 (doi:10.1098/rsbl.2010.0634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labbe P., Vale P., Little T. J. 2010. Successfully resisting a pathogen is rarely costly in Daphnia magna. BMC Evol. Biol. 10, 355. 10.1186/1471-2148-10-355 (doi:10.1186/1471-2148-10-355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auld S. K. J. R., Scholefield J. A., Little T. J. 2010. Genetic variation in the cellular response of Daphnia magna (Crustacea: Cladocera) to its bacterial parasite. Proc. R. Soc. B 277, 3291–3297 10.1098/rspb.2010.0772 (doi:10.1098/rspb.2010.0772) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laine A.-L. 2007. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant–pathogen association. J. Evol. Biol. 20, 2371–2378 10.1111/j.1420-9101.2007.01406.x (doi:10.1111/j.1420-9101.2007.01406.x) [DOI] [PubMed] [Google Scholar]
- 38.Sadd B. M., Kleinlogel Y., Schmid-Hempel R., Schmid-Hempel P. 2005. Trans-generational immune priming in a social insect. Biol. Lett. 1, 386–388 10.1098/rsbl.2005.0369 (doi:10.1098/rsbl.2005.0369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Little T. J., O'Connor B., Colegrave N., Watt K., Read A. F. 2003. Maternal transfer of strain-specific immunity in an invertebrate. Curr. Biol. 13, 489–492 10.1016/s0960-9822(03)00163-5 (doi:10.1016/s0960-9822(03)00163-5) [DOI] [PubMed] [Google Scholar]
- 40.Gillespie J. H., Turelli M. 1989. Genotype–environment interactions and the maintenance of polygenic variation. Genetics. 121, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stjernman M., Little T. J. 2011. Genetic variation for maternal effects on parasite susceptibility. J. Evol. Biol. 24, 2357–2363 10.1111/j.1420-9101.2011.02363.x (doi:10.1111/j.1420-9101.2011.02363.x) [DOI] [PubMed] [Google Scholar]
- 42.Mucklow P. T., Vizoso D. B., Jensen K. H., Refardt D., Ebert D. 2004. Variation in phenoloxidase activity and its relation to parasite resistance within and between populations of Daphnia magna. Proc. R. Soc. Lond. B 271, 1175–1183 10.1098/rspb.2004.2707 (doi:10.1098/rspb.2004.2707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labbé P., McTaggart S. J., Little T. J. 2009. An ancient immunity gene duplication in Daphnia magna: RNA expression and sequence analysis of two nitric oxide synthase genes. Dev. Comp. Immunol. 33, 1000–1010 10.1016/j.dci.2009.04.006 (doi:10.1016/j.dci.2009.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker B. J., Barribeau S. M., Laughton A. M., de Roode J. C., Gerardo N. M. 2011. Non-immunological defense in an evolutionary framework. Trends. Ecol. Evol. 26, 242–248 10.1016/j.tree.2011.02.005 (doi:10.1016/j.tree.2011.02.005) [DOI] [PubMed] [Google Scholar]