Abstract
Metabolomics is a method for investigation of changes in the global metabolite profile of cells. This paper discusses the technical application of the approach, considering metabolite extraction, separation, mass spectrometry and data interpretation. A particular focus is on the application of metabolomics to the study of stem cell physiology in the context of biomaterials and regenerative medicine. Case studies are used to illustrate key points, focusing on the use of metabolomics in the examination of mesenchymal stem cell responses to titania-nanopillared substrata designed for orthopaedic applications.
Keywords: metabolomics, stem cells, biomaterials, mass spectrometry, regenerative medicine, titanium
1. Introduction
In the context of regenerative medicine, metabolomics has a considerable advantage with respect to proteomics, the other major global functional-level analysis tool for biomaterials analysis, in that a large number of metabolites can be detected with very small quantities of sample material. In addition, typical extraction and sample processing protocols are less technically demanding than for high-sensitivity saturation labelled two-dimensional difference gel electrophoresis (saturation DiGE; discussed in greater detail in McNamara et al. [1] and in relation to biomaterials research in McNamara et al. [2]), making the technique more accessible for researchers with access to high-sensitivity mass spectrometers. The techniques are complementary, however, and the combination of protein abundance data generated by a proteomic approach, such as saturation DiGE, and the metabolic data from a metabolomic study should both facilitate the global comprehension of changes in stem cell profiles during differentiation. In this paper, technical application of metabolomics is discussed, with a particular focus on stem cell biology in regenerative medicine. A case study is used to illustrate the application of metabolomics to the study of mesenchymal stem cell (MSC) responses to titania-nanopillared substrata. The results highlighted the involvement of extracellular signal-regulated kinase (ERK) 1/2 as a likely central regulator in the differentiation of MSCs on 15 nm high nanopillars, and implicated l-ornithine as a key metabolite that is significantly modulated in abundance between different heights of nanopillared titania.
2. Metabolomics: current applications in stem cell research
Metabolomics has been used to demonstrate that the redox status of embryonic stem (ES) cells is modulated during differentiation [3]. Greater understanding of the ‘normal’ changes in metabolomic profile according to cell maturation and differentiation will prove useful when more attention is given to the response of these cells to biomaterials and implanted devices. Recent research has also begun to apply metabolomics to the investigation of stem cell responses to topographically structured substrates with the ability to retain stem cell plasticity [4] and promote osteogenesis [5]. On an osteogenic titania surface of 15 nm nanopillar-like structures, MSCs displayed a more metabolically ‘active’ phenotype, with upregulation of metabolites related to differentiation and osteogenesis [5]. MSCs cultured on an ordered array of polymer nanopits that promoted retention of stem cell characteristics, in contrast, had a greater proportion of unsaturated metabolites (containing C=C bonds). These unsaturated metabolites are predicted to provide a capacity for redox reactions during differentiation [4]. Metabolomics has also been used to highlight distinctions in the metabolic profile of classes of stem cells. Haematopoietic stem cells, for example, were shown to use glycolysis rather than aerobic respiration, owing to their hypoxic niche [6]. Interestingly, induced pluripotent stem (iPS) cells were metabolically distinct from ES cells and the somatic cell types from which the iPS cells were derived [7]. iPS cells also contained lower levels of a number of key unsaturated metabolites than ES cells, and the authors suggested that changes in metabolite abundance could be involved in the epigenetic reprogramming. Following reprogramming from mature cell types, iPS cells transitioned from oxidative to glycolytic metabolism [7]. Metabolomic analysis of culture medium conditioned by fibroblasts (to be used in ES cell culture) illustrated that some metabolites (such as lactate and alanine) had become enriched in the conditioned medium, while the abundance of others (such as tryptophan) were reduced in the presence of fibroblasts [8]. Another study investigated ES cell metabolism under normal culture conditions, and in response to the teratogenic drug valproate [9]. It was demonstrated that stem cells respond differently to topographical stimuli depending on the conditions, with low oxygen tension promoting enhanced alignment of MSCs to a nanogrooved surface [10]. Given the differential responses of stem cells to topography under normoxic and hypoxic conditions, it would seem appropriate and timely to characterize the metabolic profile of stem cells under a variety of conditions, including hypoxia, to facilitate understanding of the response to biomaterial surfaces and implanted devices.
3. Challenges and future potential for metabolomics in regenerative medicine
3.1. Maintenance of ‘stemness’
In order for the ‘stemness’ of stem cell cultures to be retained, a number of important factors have to be considered, including the density of the cell cultures, the culture medium used and the culture conditions. The culture environment affects this, for example, certain topographical cues can enhance the retention of MSC phenotype [4] or promote differentiation [4,5], substrate stiffness affects lineage commitment [11], and the structure of scaffolds (two-dimensional versus three-dimensional constructs) modulates gene expression in differentiating stem cells [12]. More prolonged culture for the expansion of stem cells can deplete the number of stem cells in a population: MSCs, for example, often undergo spontaneous differentiation in culture, so the use of early passage cells is usually advisable. In the case of iPS cells, however, the use of later passage cells is advantageous. The early passage iPS cells appear to retain more characteristics of the parent cell types from which the iPS cells were derived, but these are gradually lost with increasing passage number, probably as a result of epigenetic changes [13]. The chosen seeding density can also affect cell fate [14]. In addition, variation between different ES and iPS cell lines could also introduce undue biological ‘noise’ into the analysis. Panopoulous [7] used the same iPS cell line to study the progression of the metabolic profile from early to late passage number in chemically defined culture media to avoid this issue and minimize variation in culture conditions. Therefore, it is important to establish culture conditions that retain the stemness of the cells of interest prior to commencement of the study, which can be checked using techniques such as fluorescence-activated cell sorting (FACS) to confirm the presence of appropriate stem cell markers.
3.2. Solvent extraction
Metabolomics offers great potential for improving our understanding of the metabolic processes accompanying and underpinning the changes that occur during differentiation. The technique can be used for the in vitro characterization of cell responses to biomaterials with different surface chemistries and topographies, with attention paid to stress responses, which may give information about the tolerance of the implant or device in the body. When the metabolic profile of differentiating stem cells is better characterized in vitro, both under inducing conditions and in the absence of induction media, this should provide a useful benchmark profile for the metabolic changes during differentiation, at least in vitro. With such sensitive global approaches, it is likely that there will appear to be some disparities between datasets owing to small variations in cell state. This would become an insurmountable issue if excess technical variation is introduced; this must be minimized by efficient experimental technique and planning. Technical rigour is required to maximize the reproducibility of the results within and between experiments, and careful experimental planning is necessary to capture the metabolites in their most ‘lifelike’ state.
Rapid quenching of metabolic reactions is required to minimize the occurrence of unwanted metabolic stress and ‘run-on’ reactions. In our experience, direct precipitation of adherent stem cells from biomaterial surfaces, using chilled extraction solvent (1 : 3 : 1 chloroform:methanol:double-distilled water) with agitation of the culture dishes on a shaking platform, is a useful and rapid means of isolating metabolites (described in McNamara et al. [5]). This approach also negates the requirement for harvest of the cells from the surfaces (for example, by trypsinization) prior to extraction, and should greatly reduce the development of artefactual changes in the stem cell metabolome. In addition, the volume of extraction solvent is minimized, which carries the additional benefit that sample concentration (e.g. using a vacuum concentrator with heat, which has the potential to affect labile metabolites) should not be required and thus the metabolites should be more readily detected by the mass spectrometric workflow. A related direct quenching approach is described in Teng et al. [15], which uses methanol as the quenching agent, followed by cell scraping to harvest the extracted material. The authors report that their approach gives a sample recovery yield that is 50 times greater than the equivalent protocols incorporating centrifugation steps prior to quenching (for example, requiring trypsinization to harvest the cells). The choice of solvent, together with the chromatographic separation method (as discussed in greater detail below), affects the balance of metabolites that can be extracted and resolved. If a particular subset of metabolites, such as lipids, is of particular interest, the extraction solvent should be chosen to maximize the extraction and resolution of these metabolites. The quenching solution that is commonly used in our laboratory (1 : 3 : 1 chloroform:methanol:double-distilled water) is useful as a ‘general’ quenching agent, intended to extract a relatively broad range of metabolites.
The use of a ‘shake test’ prior to the main experiment is a helpful way of determining the minimal volume of solvent required to precipitate the metabolites from the particular biomaterial surfaces of interest without excessive evaporation during the extraction period. In the shake test, the extraction solvent is added to unseeded biomaterial samples at a range of different (minimal) volumes and agitated using a shaking platform under chilled conditions (4°C, for example, in a cold room), for the time (typically 1 h) to be used in the main experiment. The speed of platform rotation should be chosen to ensure the liquid is optimally agitated to facilitate cell disruption, and the samples remain covered with solution for the duration of the extraction. If the biomaterial substrates are scarce or difficult to fabricate, an alternative material of comparable size and thickness could be used for the trial, but use of the actual material in the shake test is helpful in order to assess how the solvent interacts with the material surface (as some surfaces can be degraded), and whether this is likely to adversely affect the experiment. It may also be necessary to test how many samples are required to generate sufficient material to ensure that the cell number (i.e. metabolite abundance) is not a limiting factor to metabolite detection. This could comprise, for example, testing metabolic extracts from stem cells cultured in different numbers of tissue culture plastic (TCP) culture wells (if material conservation is necessary, and culture on TCP represents a comparable proliferation rate to that when the cells are cultured on the material of interest), with cells seeded at the same density and for the length of culture period that will be used in the main experiment. We have previously highlighted the importance of using controls with the same surface chemistry as the topographically structured ‘test’ materials in metabolomics [5] and discussed the possibility of pooling substrates to generate more material in the context of proteomics [2]. For metabolomics studies, it is important that the samples remain sufficiently concentrated to detect the metabolites—this will be affected by the cell seeding density and the amount of extraction solvent used per sample, and should be tested prior to the main experiment. A volume of extraction solvent in the region of 200 μl per structure (for samples that fit within a well of a 24-well culture plate) is often a useful starting point for optimization experiments.
If the stem cells of interest are not adherent (for example, haematopoietic stem cells), we suggest the following modification as a potential alternative protocol. The cells could first be pelleted by centrifugation and then all culture medium removed. The cells could be washed twice in chilled sterile phosphate-buffered saline (PBS) and lysed in a small volume of extraction solvent in compatible tubes.
The choice of extraction solvent will affect the panel of metabolites that can be analysed, and the type of liquid chromatography (LC) will modulate the chromatographic separation and resolution of the metabolites. The appropriate stationary phase should generally be chosen to give the widest range of metabolites when there is no particular class of metabolites of interest. For a summary of typical metabolomics columns and their applications, see table 1.
Table 1.
Applications of metabolomic column types.
| column type | applications | limitations | reference |
|---|---|---|---|
| reversed phase | hydrophobic metabolites, lipids | organic extracts require drying and resuspending in aqueous solvent/dilution. | [16] |
| HILIC | polar metabolites, amino acids, sugars | poor retention of most lipids, salt elution during gradient. | [17] |
| ion exchange | di- and tri-phosphosugars, organic acids, cations or anions | only cations or anions per run. Overall retention not as good as HILIC. | [18,19] |
Hydrophilic interaction chromatography (HILIC) columns are increasing in popularity owing to their improved selectivity for polar metabolites, especially those that are critical in the tricarboxylic acid (TCA) cycle and glycolysis, although their retention of lipid and hydrophobic molecules is poor [20]. This gives broad retention characteristics for small molecules [17]. Another benefit is that their selectivity allows organic solvents to be used for metabolite extraction without further processing prior to loading on the column. Reverse phase is still used as a standard separation column for metabolomics, mostly owing to the multitude of high-quality columns available for the purpose [16]. The largest limitation to the use of reversed phase columns is the requirement to load samples in an aqueous solvent, which is a non-ideal extraction solvent, and that many of the key metabolites are too polar to be retained and therefore elute in the wash-through. They are commonly applied to lipids and non-polar metabolites. Finally, ion chromatography shows promise for many metabolites, including organic acids and multiply phosphorylated species, although it has a more limited selectivity than the previously described separation methods [18]. A major benefit of ion chromatography columns is that as long as the ionic concentration of the sample is low, the sample can be in any extraction solvent without deleterious effects.
3.3. Control extractions and metabolomic ‘compartments’
The use of additional control extractions is also advisable during the main experiment. LC-mass spectrometry (LC-MS) analysis of a portion of the extraction solvent alone gives an indication of the ‘background’ compounds (which are predominantly traces of plasticizers, polysiloxanes from the atmosphere and detergents) that are present within it. This is even more informative if the solvent is incubated under the same conditions as the other samples. If the samples are material discs with cultured cells being extracted from cell culture plates, for example, the same volume of solvent should be incubated in an empty well of the plate. This would then include the chemical contribution from any degradation of the tissue culture plastic. Similarly, a control with solvent in the presence of the biomaterial also accounts for the release of constituents of the unseeded biomaterial that are detectable by LC-MS. This additional information affords greater consideration (and potentially subtraction) of these metabolites in later analyses.
‘Fresh medium’ control samples (complete cell culture medium, containing all the same components as the medium to be incubated with the samples, including any supplements or inhibitors to be added) prepared with the extraction solvent would also indicate the background metabolic signature of the culture medium. If the same volume of culture medium is harvested from the seeded cell samples and extracted separately, this can give an indication of modulations in the exometabolome (the extracellular metabolome), for comparison with the endometabolome (metabolites within the cell). This also gives an indication of the success of the wash steps in removing the culture medium from the cells prior to extraction, as the metabolites that are highly enriched in the medium should generally be present at lower levels in the cells and intracellular metabolites enriched in the converse manner. Owing to the high concentration of certain metabolites in culture medium (such as l-glutamine and other enrichments), a much smaller quantity of medium is required for extraction, compared with the cell extracts, but this volume should be kept consistent between the fresh and spent medium samples, to ensure comparability between the analytes.
In addition to the blank medium and extraction solvent samples, quality control samples are also important in a metabolomic study. These usually consist of two different forms: standards and pooled samples. Standard compounds are used to calibrate retention time parameters and ensure that instrument sensitivity is appropriate. A pooled sample consists of a mixture of samples to be run, and an aliquot from the pool should be analysed periodically during the batch. This allows tracking of non-enzymatic degradation during sample analysis, as well as any loss in sensitivity or retention time drift that occurs as the mass spectrometer analyses the samples.
4. Mass spectrometry in metabolomics: the process of metabolite identification and analysis
4.1. Mass spectrometry
MS has replaced nuclear magnetic resonance spectroscopy (NMR) as the most common method of performing metabolomic analysis. MS is essentially a means of detecting and separating molecules based on their mass (actually, mass-to-charge ratio, as all commercially available MSs detect ions). This is an extremely powerful tool for metabolomics, as it allows the analysis of complex mixtures of molecules. When hyphenated to a separation system, e.g. liquid or gas chromatography, analyses approaching (but, it is important to bear in mind, not reaching) the metabolomic scale of complexity are available. A plethora of different MS instruments are available, but we have restricted our discussion to those most commonly employed for metabolomics research.
4.2. Gas chromatography mass spectrometry
Gas chromatography MS (GC-MS) consists of a gas chromatograph usually coupled with a single quadrupole or time of flight (ToF) MS. GC-MS is still commonly used in metabolomics: the very high resolution of GC separations matches well with the complexity of metabolomic samples, and the reproducible fragment patterns available from electron impact (EI) ionization, which are enhanced by the accurate mass available from GC-ToF instruments. Limitations are the derivitization step of the analysis, which can add complexity to the analysis, and the commonly absent molecular ion, which can result in issues with metabolite identification.
4.3. Liquid chromatography-mass spectrometry instruments
4.3.1. Triple quad
Triple quadrupole instruments are usually used for targeted metabolomic analyses. The main reason for this is the multiple reaction monitoring technique available with this type of instrument, where the first quadrupole may be locked to a specific ion of interest. Fragments are then generated in the second quadrupole (a collision cell) and the third quadrupole selects one or more characteristic fragments that can be used to confirm the initial ion of interest. Although only unit resolution is available for the selection of the precursor and detection of the fragment ions, the specificity gained is unparalleled, since identification is based on intact mass and the internal structure of the molecule [19].
Of course, the major limitation of such a targeted analysis is that only expected metabolites will ever be observed. It is for this reason that accurate mass MS has been crucial in the development of untargeted metabolomics. Accurate mass (to within five parts per million of the true mass) is essential to detect the small chemical shifts that distinguish compounds with the same nominal mass. In some cases, the accurate mass alone can be sufficient to derive the empirical formula of a compound. The most commonly used accurate mass instruments are quadrupole ToF instruments (Q-ToFs) and Fourier transform (FT) deconvolution-based instruments such as the Orbitrap.
4.3.2. Quadrupole time of flight
The Q-ToF has been a workhorse instrument for proteomics for many years, but it has only recently achieved the level of mass accuracy and resolution required for metabolomic analysis. It consists of a mass selecting quadrupole coupled with a ToF apparatus, and thus offers ion selection and fragmentation, as well as accurate mass [21]. The major benefit of the Q-ToF in metabolomics is speed. Resolution of a ToF is decoupled from scan rate, and therefore high-quality modern instruments with a resolution of 30 000 and scan rate of 20 Hz are available. This is especially important in high-throughput studies where ultra high-performance LC (UHPLC) separations with peak widths of 1 s are common.
4.3.3. Orbitrap
The Orbitrap is a relatively new type of mass spectrometer, first commercially available in 2006 [22]. It couples the ultra-high mass accuracy with high sensitivity and ease of use. Resolutions of 100 000 and mass accuracies of less than 1 ppm are routine on this type of instrument. As each scan consists of a waveform detected from the movement of ion packets in the trap itself, however, the resolution is proportional to the scan time, and a 100 000 resolution scan requires roughly 2 s to perform, thus increasing the duty cycle and limiting the power of the Orbitrap for high-throughput analysis.
4.3.4. Comments on data analysis and interpretation
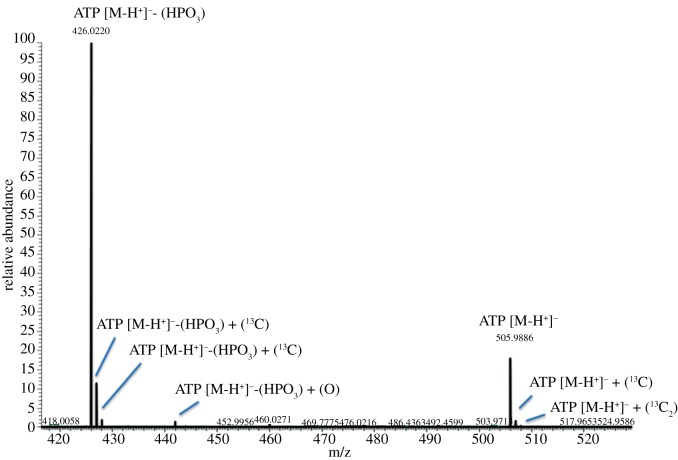
During data analysis, the dataset must be simplified to remove artefacts and noise. While the key benefit of electrospray ionization (as commonly used in LC-MS) is the formation of molecular ions from the majority of compounds, fragmentation events and unusual adducts can and will occur (figure 1). Fragments especially may have the same empirical chemical formula and consequently exactly the same mass as a ‘real’ metabolite yet be at an inappropriate retention time for that species. Therefore, any metabolomics software must be able to both effectively pick peaks (as distinguishable from background noise, i.e. a feature's intensity must rise and fall over a specified period), and take fragments and adducts into account. MzMine [23] supports both functions, and our in-house software incorporates the peak-picking functions of XCMS [24] and the ‘related peaks’ annotation from MzMatch [25].
Figure 1.
Common adducts and fragments of the molecule ATP. Note the isotope peaks (+ 1 13C). Phosphate loss in ionization is very common with ATP, such that the ADP peak is considerably more intense. Additionally, evidence of oxidization of ADP is present. These features considerably complicate the identification of individual metabolites.
During metabolite identification, sometimes multiple identities are assigned to the same peak, since the peak masses are similar. This ambiguity decreases in inverse proportion to the mass accuracy of an instrument. The use of retention time data from known metabolite standards can provide a valuable means of validating the metabolite identities, allowing the exclusion of possible identities that do not closely match the retention time of the observed metabolite. In addition, retention time prediction for compounds for which standards are not available shows promise as a method for dramatically reducing false identifications [26]. One disadvantage of metabolomic analysis is that mass spectrometry alone cannot readily distinguish between isomers (which share the same molecular formula, but have a different structural arrangement of atoms), which can complicate the analysis of certain types of metabolites, such as sugars. It should be noted that the nature of metabolomics is such that any small molecule in a sample may be detected, which necessitates manual annotation to distinguish physiologically relevant metabolites from contaminants. An example of this is the presence of streptomycin, added as an antibiotic to the cell culture medium, or the omnipresent phthalates, resulting from plasticizers in sample containers. For this reason, although it is possible to produce a shortlist of candidate compounds from accurate mass alone, it is customary to apply an authentic standard to confidently identify a metabolite, based on accurate mass/fragment pattern and retention time [27]. Failing this, a database of potential identifications can be used to draw attention to putative biological interpretations of a peak.
In addition, metabolomics is almost exclusively performed in a quantitative manner, to ascertain differences between samples or states. For this reason, visualization of quantitative information is critical for metabolomic analysis. Conditional formatting using newer versions of Microsoft Excel permits the colour coding of metabolite intensities to facilitate the rapid visual assessment of trends in abundance: for consistency between biological and technical replicates, and differential abundance between test samples. Graphical representation of individual metabolites can be useful to focus on metabolites of interest, particularly when combined with larger scope pathway analysis (e.g. see McMurray et al. [4] and McNamara et al. [5]).
In our laboratory, Ingenuity pathway analysis (IPA) and Pathos [28] (software that overlays metabolite identities (IDs) onto Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways) are used to analyse the contextual functional significance of the metabolites. The facilities of these two approaches are compared with each other, and with databases of metabolite IDs, in table 2. IPA can be used to categorize the differential metabolite abundance graphically and also as networks of interacting molecules. Such combinations of focused studies and global interactions are useful in order to evaluate the modulation in individual molecules and at the level of inter-molecular interactions. Owing to the complexity and conditionality of metabolomes, however, metabolites are frequently detected that are currently poorly characterized in terms of biological functionality, and some may even be completely novel. Only metabolites with functional annotation can be analysed using IPA, and only those that can be classified and mapped to a KEGG map of known metabolites can be analysed with Pathos.
Table 2.
Comparison of the functionality of some of the data analysis and representation approaches available.
| function | Pathos (KEGG-map-based) analysis | IPA analysis | metabolite databases (e.g. HMDB) |
|---|---|---|---|
| description of function of individual characterized metabolites | yes | yes | yes |
| generation of novel interaction networks based on up-loaded data | not at present | yes | not at present |
| graphical representation of involvement in known metabolic pathways | yes | yes | not at present |
| colour-coded maps indicating differential metabolite abundance | yes | yes | not at present |
| classification of groups of metabolites into known functional classes with graphical representation | yes | yes | not at present |
| facility for comparative analysis with proteomic or microarray data | not at present | yes | not at present |
| illustration of structural formulae and other metabolite annotation | yes (via links to external metabolite databases) | yes (via links to external metabolite databases) | yes |
5. Case studies
This section will describe three brief biomaterials case studies performed in our laboratories using nanostructured titania and MSCs (isolated from patients undergoing routine hip arthroplasty in accordance with the previously described protocol [29]). The nanostructured titania discs and polished controls were fabricated as described in McNamara et al. [5], with cell culture and metabolomic analysis performed as previously described [5]. In order to use an IPA approach to evaluate the relationships between the metabolites using network analysis, and other ‘invited’ nodes (molecules that become incorporated into the network via their associations with metabolites that were examined directly), the ratios of peak intensities on the 15 nm: planar substrates were calculated and up-loaded to the IPA server for analysis.
5.1. Case study I: depletion of extracellular guanosine by mesenchymal stem cells on nanofeatured titania
Guanosine is required for a number of important processes including DNA and protein synthesis, the latter of which would be particularly important during differentiation. Previous research from our group has shown that this metabolite is enriched in MSCs cultured on titania, and particularly on the 15 nm nanopillar-like surface. Interestingly, MSCs cultured on nanostructured titania substrates depleted this metabolite from the extracellular environment, but the effect was not observed with MSCs cultured on TCP (figure 2). This suggests that greater levels of guanosine are required by MSCs on titania surfaces, in particular by MSCs cultured on the osteogenic 15 nm pillars. In addition, these data reiterate the importance of selecting appropriate controls, as there were distinctions in cell response owing to surface chemistry.
Figure 2.
Enrichment of (a) an endometabolite (sphingosine) in MSCs cultured on the osteogenic 15 nm substrate and (b) a metabolite (guanosine) that has been depleted from the culture medium by MSCs cultured on the Ti surfaces. Note (a) the enrichment of sphingosine in the cell extracts relative to the extracts from the culture medium and (b) the depletion of guanosine from the Ti surfaces (Spent PL, 15, 55 and 90 nm Ti) but not the TCP substrate. ‘Fresh medium’ denotes extracts from medium that has not been exposed to cells, ‘spent medium’ denotes extracts from the medium exposed to cells, and ‘cell extracts’ were prepared from MSCs cultured on the substrates. Adapted from McNamara et al. [5].
5.2. Case study II: representation of metabolomic changes in tabular format for MSCs on nanofeatured titania
In addition to the representation of individual metabolites graphically, as highlighted in figure 2, it can also be helpful to note the metabolites with the highest magnitude of change in abundance, a process that can be expedited using software such as IPA to gain a measure of fold change in metabolite ‘abundance’ (the intensity of the spectral peaks) between a test sample and a reference control sample. This is a less visual approach, but allows direct comparison between different samples. Using a network-based IPA approach, by contrast, only permits comparison between two samples at a time. Pathos pathways analysis compares pairs of samples, but also has the facility to represent multiple samples graphically. Use of the magnitude of fold change as a mechanism for the evaluation of tabulated data is shown in table 3. Most interestingly, using this approach, the metabolite l-ornithine emerged as highly differentially abundant between the surfaces. It was one of the most highly upregulated metabolites on both the TCP and 15 nm surfaces relative to the planar control Ti, but was one of the most highly downregulated metabolites on the 55 and 90 nm substrata. The difference in abundance of this metabolite in MSCs cultured on the 15, 55 and 90 nm surfaces is consistent with our previous results indicating that cells on the 15 nm substrate are phenotypically distinct, with a more metabolically ‘active’ physiology [5]. It is interesting to note that this non-essential amino acid was upregulated on both the TCP and 15 nm substrata relative to the planar control. Given the physiological distinction in MSC responses to these substrata, it highlights the importance of considering the overall metabolic picture, in addition to changes at the level of individual metabolites.
Table 3.
(a) List of most upregulated metabolites, (b) most downregulated on the TCP, 15, 55 and 90 nm substrates. Note that l-ornithine (highlighted) was one of the most upregulated metabolites on PL and 15 nm substrates, but one of the most downregulated metabolites on the 55 nm and 90 nm Ti surfaces.
| (a) | most upregulated metabolites | functional significance |
|---|---|---|
| TCP:PL | ||
| isovaleric acid | +6.534 | fatty acid |
| methyl-4-tyramine | +5.630 | monoamine derived from tyrosine. Promotes release of catecholamine from storage vesicles |
| urocanic acid | +4.365 | intermediate in the metabolic degradation of l-histidine |
| hexanoic acid | +3.875 | mammalian fatty acid |
| l-ornithine | +3.824 | non-essential amino acid involved in the urea cycle |
| tyramine | +3.664 | neurotransmitter derived from tyrosine |
| 15 nm:PL | ||
| N-acetylserotonin | +127.4335 | intermediate in production of melatonin from serotonin |
| 2′-deoxyadenosine | +11.756 | deoxyribonucleoside, DNA nucleoside A |
| l-methionine sulphoxide | +9.268 | results from the oxidation of methionine by reactive oxygen species |
| l-ornithine | +6.708 | non-essential amino acid involved in the urea cycle |
| 55 nm:PL | ||
| l-histidine | +4.853 | amino acid. Required for protein synthesis |
| indole-3-lactic acid | +3.812 | tryptophan metabolite |
| l-methionine sulphoxide | +3.760 | results from the oxidation of methionine by reactive oxygen species |
| 90 nm:PL | ||
| N-acetylserotonin | +29.437 | intermediate in production of melatonin from serotonin |
| delta-1-piperidine-6-carboxylic acid | +6.303 | intermediate of l-lysine metabolism in the brain |
| l-methionine sulphoxide | +5.412 | results from the oxidation of methionine by reactive oxygen species |
| (b) | most downregulated metabolites | functional significance |
| TCP:PL | ||
| phosphate | −28.158 | energy generation (AMP/ADP/ATP balance), component of nucleic acids, post-translational modification (phosphorylation) of proteins |
| l-kynurenine | −21.922 | metabolite of l-tryptophan that is involved in production of vitamin B3 (niacin). Vitamin B3 alters the balance of low- and high-density lipoproteins in blood |
| pyridoxine | −21.657 | vitamin B6. Involved in maintenance of sodium/potassium balance and haematopoiesis. |
| homoarginine | −13.399 | low levels are associated with cardiac difficulties. |
| folic acid | −12.632 | required for DNA synthesis and repair and is a cofactor in biological processes |
| delta-1-piperidine-6-carboxylic acid | −19.867 | an intermediate in l-lysine metabolism in the brain |
| d-sphingosine | −9.996 | can be phosphorylated to sphingosine-1-phosphate which promotes recirculation of osteoclasts back into tissues and away from bone |
| 15 nm:PL | ||
| 4-aminobutanal | −1.732 | metabolite of putrescine, which are produced by breakdown of amino acids |
| thymidine | −1.727 | nucleoside required for DNA synthesis |
| dexpanthenol | −1.725 | biologically active form of the alcohol analogue of pantothenic acid (vitamin B6). Vitamin B6 is required for synthesis and turnover of proteins, carbohydrates and lipids |
| kyurenic acid | −1.563 | metabolite of l-tryptophan with neuroactive activity |
| 55 nm:PL | ||
| l-ornithine | −122.119 | non-essential amino acid involved in the urea cycle |
| spermidine | −114.439 | polyamine that can stimulate autophagy |
| trimethlenediamine | −38.532 | involved in arginine/proline metabolic pathways and beta-alanine synthesis |
| methylguanidine | −30.524 | guanidine compound resulting from the metabolic degradation of protein |
| l-2-aminoadipate 6-semialdehyde | −21.155 | substrate for an oxidoreductase involved in turnover of the amino acid lysine |
| creatine | −21.155 | promotes regeneration of ATP |
| anthranilic acid | −19.272 | precursor of the amino acid l-tryptophan |
| 5-aminopentanamide | −15.123 | involved in lysine degradation |
| 90 nm:PL | ||
| l-ornithine | −83.821 | non-essential amino acid involved in the urea cycle |
| formiminoglutamic acid | −8.062 | intermediate metabolite in histidine metabolism |
| phosphate | −5.810 | energy generation (AMP/ADP/ATP balance), component of nucleic acids, post-translational modification (phosphorylation) of proteins |
| delta-1-piperideine-2-carboxylic acid | −4.683 | an intermediate in l-lysine metabolism in the brain |
| l-2-aminoadipate 6-semialdehyde | −4.140 | substrate for an oxidoreductase involved in turnover of the amino acid lysine |
| N2-acetyl-l-ornithine | −4.107 | protein and amino acid synthesis |
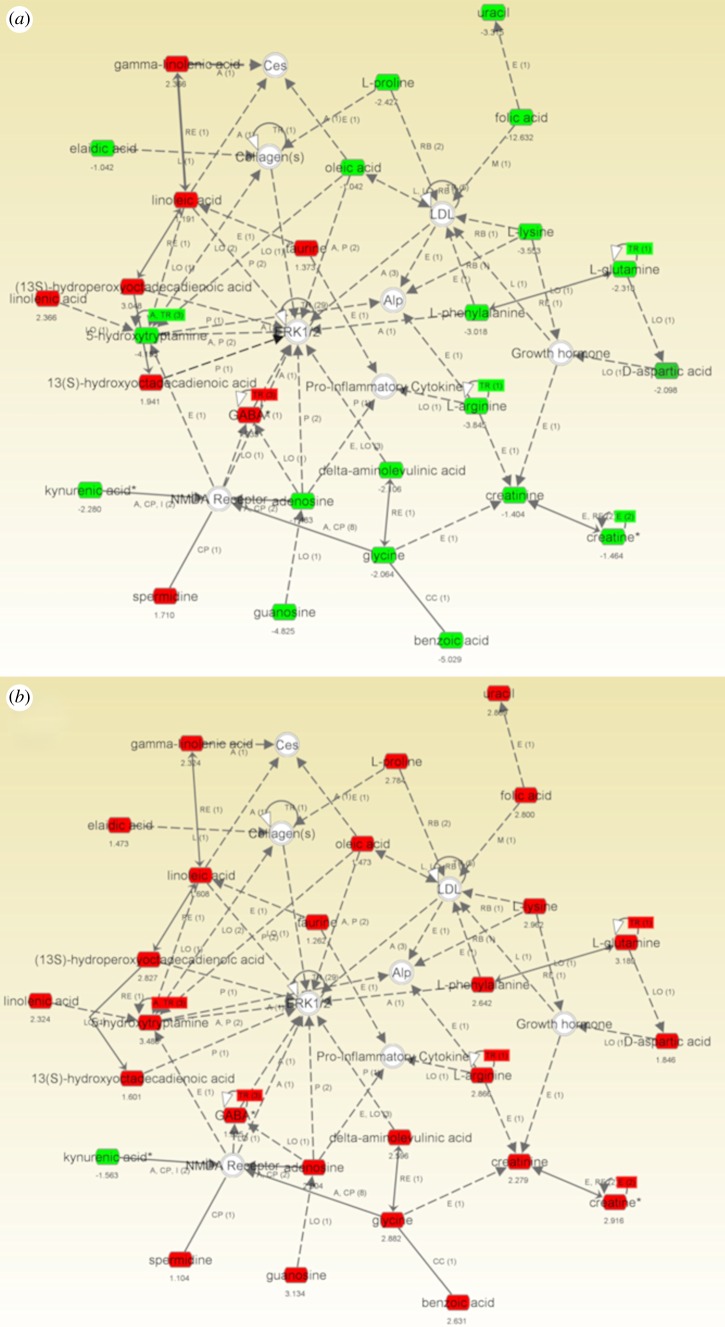
5.3. Case study III: representation of metabolomic changes by network analysis: the importance of extracellular signal-regulated kinase 1/2 and collagen regulation is highlighted in mesenchymal stem cells on 15 nm titania nanopillar-like features
An example of the use of IPA network analysis in the visualization of a complex metabolite dataset via the generation of molecular interaction networks, in this case for MSCs cultured on TCP or the osteogenic 15 nm titania substrates, compared with planar titania [5]. This network illustrated that most metabolites that were increased in abundance on the 15 nm pillar-like features were decreased in abundance on the TCP surface (figure 3). Most interestingly, central invited nodes in this network included ERK 1/2, which is a key protein involved in cell proliferation and differentiation [30], and collagen, which is required for endochondral ossification during bone formation [31].
Figure 3.
Highest scoring IPA network showing the upregulated (red) and downregulated (green) metabolites in MSCs cultured for 3 days on (a) TCP versus planar Ti or (b) 15 nm Ti pillar-like features versus planar Ti. Note that most of the metabolites in the network that were upregulated on 15 nm were downregulated on TCP, and the prominence of the invited nodes ERK 1/2 and collagen in the network.
A number of metabolites that were downregulated on the TCP surface relative to the planar Ti were upregulated on the Ti nanopillared surfaces. This was particularly apparent on the 15 nm substrate, where all the metabolites in the network were upregulated excepting kynurenic acid. Consistent with our previous metabolomic study [5], this analysis suggests that the pillars were promoting activation of metabolic processes including protein synthesis (various amino acids were upregulated) and energy generation (upregulation of guanosine, adenosine and creatine). Although linoleic acid and gamma-linoleic acid can be adipogenic mediators, these metabolites were upregulated on both the TCP surface (non-osteogenic) and the 15 nm substrate (osteogenic), and thus it seems more likely that these antioxidants were acting to modulate the redox status of the cells, and thus affect their differentiation.
6. Future applications: potential value of exometabolome screening in clinical applications
Increased understanding of the metabolites that are depleted or secreted into the medium during the modulation of stem cell fate, including different stem cell populations, should inform our knowledge of cell differentiation, and how to monitor stem cell status in a non-invasive manner in vitro using medium sampled from culture wells. In addition, improved knowledge of the exometabolome (particularly in vivo and human patients) could provide a minimally invasive means of assessing the extent of cell differentiation at the site of an implant, or changes in host response towards an implanted device or scaffold. Greater understanding of the metabolites that are key to these processes will inform the extent to which such diagnostic processes would need to be invasive.
Metabolic changes that require the study of the immediate local tissue environment or the tissue itself would be less favourable, owing to their potential requirement for tissue biopsies. Sampling fluid from the tissue could be a somewhat less damaging alternative, but particularly attractive options that may be feasible in future using high-sensitivity metabolomics include the collection of blood, stool, urine or saliva samples, if the metabolic changes become sufficiently well characterized, and useful marker metabolites can be detected at the required distance from the device, and in the harvested sample type. This would most probably require optimization for individual studies, but further research to improve our knowledge of the changing stem cell metabolome, and the detectability and fluctuations of associated metabolic markers in sample types that can be obtained by non-invasive means, should help to determine the feasibility of different approaches. In a metabolomic study of acetaminophen-induced hepatic toxicity, opthalmate was identified as a metabolite that was associated with liver injury, and could be detected in serum as a marker of oxidative stress [32]. Goldsmith et al. [33] reviewed the literature on metabolic biomarkers that were identified following exposure to toxins and in rejection of tissue grafts, and discussed the advantages of using a metabolomic approach to discover biomarkers of pathology.
With additional research, it is hoped that the markers indicative of processes such as scaffold/graft tolerance versus rejection, stem cell differentiation and early-stage osteolysis around implanted orthopaedic devices, would be detectable in ‘accessible’ bodily fluids. In future, perhaps the supplementation of metabolic deficiencies or promotion of particular processes by tailored addition of metabolites will have potential to improve the clinical outcome of stem cell-based therapies or interactions between the patient's cells and implanted devices. Modulation of sphingosine-1-phosphate levels, for example, could potentially promote recirculation of osteoclasts back into tissues and away from orthopaedic implants, as it has been shown to promote recirculation [34]. Interestingly, the precursor form was enriched in MSCs cultured on the osteogenic 15 nm substrate discussed in the case studies (figure 2a; [5]). Enhanced understanding of the metabolic changes during cell–material interactions could facilitate improvements in the performance and lifetime of medical devices by modulating cell responses with suitable physical and/or chemical cues to induce therapeutically favourable metabolic features.
7. Conclusions
Metabolomics offers insight into the global metabolic profile of stem cells and has great potential for research into stem cell fate and physiology, as well as in vivo and patients as a diagnostic monitoring tool for regenerative medicine. In addition, this technique offers the tantalizing possibility of non-invasive analysis of bodily fluids for marker metabolites, which would be a great advantage in follow-up studies of the clinical efficacy of implanted grafts and devices. Greater characterization of the stem cell metabolome under different conditions, and enhanced annotation of metabolites would be highly beneficial for these applications.
8. Resources
Ingenuity pathways analysis: http://www.ingenuity.com (subscription required)
Pathos: http://motif.gla.ac.uk/Pathos/index.html
Human metabolome database: http://www.hmdb.ca/
KEGG (Kyoto Encyclopaedia of Genes and Genomes): http://www.genome.jp/kegg/kegg1a.html
MZmine: http://mzmine.sourceforge.net/
MZmatch: http://mzmatch.sourceforge.net/.
Acknowledgements
This work was funded by EPSRC grant EP/G048703/1. We thank Kate Murawski (Bone and Joint Research Group) for skeletal stem cell isolations, David Leader and Darren Creek (University of Glasgow) for use of their metabolomics software, and all in CCE for advice and helpful discussion.
References
- 1.McNamara L. E., Kantawong F. A., Dalby M. J., Riehle M. O., Burchmore R. 2011. Preventing and troubleshooting artefacts in saturation labelled fluorescence 2-D difference gel electrophoresis (saturation DiGE). Proteomics 11, 4610–4621 10.1002/pmic.201100135 (doi:10.1002/pmic.201100135) [DOI] [PubMed] [Google Scholar]
- 2.McNamara L. E., Dalby M. J., Riehle M. O., Burchmore R. 2010. Fluorescence two-dimensional difference gel electrophoresis for biomaterial applications. J. R. Soc. Interface 7, S107–S118 10.1098/rsif.2009.0177.focus (doi:10.1098/rsif.2009.0177.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanes O., et al. 2010. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411–417 10.1146/annurev.es.01.110170.000245 (doi:10.1146/annurev.es.01.110170.000245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMurray R. J., et al. 2011. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat. Mater. 10, 637–644 10.1038/nmat3058 (doi:10.1038/nmat3058) [DOI] [PubMed] [Google Scholar]
- 5.McNamara L. E., et al. 2011. Skeletal stem cell physiology on functionally distinct titania nanotopographies. Biomaterials 32, 7403–7410 10.1037/h0040373 (doi:10.1037/h0040373) [DOI] [PubMed] [Google Scholar]
- 6.Simsek T., Kocabas F., Zheng J., DeBerardinis R. J., Mahmoud A. I., Olson E. N., Schneider J. W., Zheng C. C., Sadek H. A. 2010. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 10.1016/j.stem.2010.07.011 (doi:10.1016/j.stem.2010.07.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panopoulous A. D. 2012. The metabolome of induce pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 22, 168–177 10.1038/cr.2011.177 (doi:10.1038/cr.2011.177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacIntyre D. A., Melguizo Sanchís D., Jiménez B., Moreno R., Stojkovic M., Pineda-Lucena A. 2011. Characterisation of human embryonic stem cells conditioning media by 3H-nuclear magnetic resonance spectroscopy. PLoS ONE 6, e16732. 10.1371/journal.pone.0016732 (doi:10.1371/journal.pone.0016732) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cezar G. G., Quam J. A., Smith A. M., Rosa G. J. M., Piekarczyk M. S., Brown J. F., Gage F. H., Muotri A. R. 2007. Identification of small molecules from human embryonic stem cells using metabolomics. Stem Cells Dev. 16, 869–882 10.1089/scd.2007.0022 (doi:10.1089/scd.2007.0022) [DOI] [PubMed] [Google Scholar]
- 10.Zhao F., Veldhuis J., Duan Y., Yang Y., Christoforou N., Ma T., Leong K. W. 2010. Low oxygen tension and synthetic nanogratings improve the uniformity and stemness of human mesenchymal stem cell layer. Mol. Ther. 18, 1010–1018 10.1038/mt.2010.21 (doi:10.1038/mt.2010.21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engler A. J., Sen S., Sweeney H. L., Discher D. E. 2006. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 10.1007/s10539-008-9147-5 (doi:10.1007/s10539-008-9147-5) [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Lin J., Roy K. 2006. Effect of 3D scaffold and dynamic culture condition on the global gene expression profile of mouse embryonic stem cells. Biomaterials 27, 5978–5989 10.1016/j.biomaterials.2006.05.053 (doi:10.1016/j.biomaterials.2006.05.053) [DOI] [PubMed] [Google Scholar]
- 13.Polo J. M. 2010. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855 10.1038/nbt.1667 (doi:10.1038/nbt.1667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 10.1016/S1534-5807(04)00075-9 (doi:10.1016/S1534-5807(04)00075-9) [DOI] [PubMed] [Google Scholar]
- 15.Teng Q., Huang W., Collette T. W., Ekman D. R., Tan C. 2009. A direct cell quenching method for cell-culture based metabolomics. Metabolomics 5, 199–208 10.1007/s11306-008-0137-z (doi:10.1007/s11306-008-0137-z) [DOI] [Google Scholar]
- 16.Dunn W. B. 2009. Changes in the metabolic footprint of placental explant-conditioned culture medium identifies metabolic disturbances related to hypoxia and pre-eclampsia. Placenta 30, 974–980 10.1016/j.placenta.2009.08.008 (doi:10.1016/j.placenta.2009.08.008) [DOI] [PubMed] [Google Scholar]
- 17.Kamleh A., Barrett M. P., Wildridge D., Burchmore R. J. S., Scheltema R. A., Watson D. G. 2008. Metabolomic profiling using Orbitrap Fourier transform mass spectrometry with hydrophilic interaction chromatography: a method with wide applicability to analysis of biomolecules. Rapid Commun. Mass Spectrom. 22, 1912–1918 10.1002/rcm.3564 (doi:10.1002/rcm.3564) [DOI] [PubMed] [Google Scholar]
- 18.Burgess K. E. V., Creek D. J., Dewsbury P., Cook K., Barrett M. P. 2011. Semi-targeted analysis of metabolites using capillary-flow ion chromatography coupled to high-resolution mass spectrometry. Rapid Commun. Mass Spectrom. 22, 3447–3452 10.1002/rcm.5247 (doi:10.1002/rcm.5247) [DOI] [PubMed] [Google Scholar]
- 19.Yost R. A., Enke C. G. 1978. Selected ion fragmentation with a tandem quadrupole mass spectrometer. J. Am. Chem. Soc. 100, 2274–2275 10.1021/ja00475a072 (doi:10.1021/ja00475a072) [DOI] [Google Scholar]
- 20.Creek D. J., Janevics A., Breitling R., Watson D. G., Barrett M. P., Burgess K. E. V. 2011. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal. Chem. 83, 8703–8710 10.1021/ac2021823 (doi:10.1021/ac2021823) [DOI] [PubMed] [Google Scholar]
- 21.Morris H. R., Paxton T., Dell A., Langhorne J., Berg M., Bordoli R. S., Hoyes J., Bateman R. H. 1996. High sensitivity collisionally-activated decomposition tandem mass spectrometry on a novel quadrupole/orthogonal-acceleration time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 10, 889–896 (doi:10.1002/(SICI)1097-0231(19960610)10:8<889::AID-RCM615>3.0.CO;2-F) [DOI] [PubMed] [Google Scholar]
- 22.Makarov A., Denisov E., Kholomeev A., Balschun W., Lange O., Strupat K., Hornin S. 2006. Performance evaluation of a novel linear ion trap/orbitrap mass spectrometer. Anal. Chem. 78, 2113–2120 10.1021/ac0518811 (doi:10.1021/ac0518811) [DOI] [PubMed] [Google Scholar]
- 23.Pluskal T., Castillo S., Villar-Briones A., Oresic M. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 11, 52–78 10.1111/0018-2656.00104 (doi:10.1111/0018-2656.00104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith C. A., Want E. J., O'Maille G., Abagyan R., Siuzdak G. 2006. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching and identification. Anal. Chem. 78, 779–787 10.1021/ac051437y (doi:10.1021/ac051437y) [DOI] [PubMed] [Google Scholar]
- 25.Scheltema R. A., Janevics A., Jansen R. C., Swertz M. A., Breitling R. 2011. PeakML/mzMatch: a file format, Java library, R library and tool-chain for mass spectrometry data analysis. Anal. Chem. 83, 2786–2793 10.1093/bjps/axi157 (doi:10.1093/bjps/axi157) [DOI] [PubMed] [Google Scholar]
- 26.Creek D. J., Jankevics A., Breitling R., Watson D. G., Barrett M. P., Burgess K. E. V. 2011. Towards global metabolomics analysis with liquid chromatography mass spectrometry: improved metabolite identification by retention time prediction. Anal. Chem. 83, 8703–8710 10.1016/0025-5564(78)90077-9 (doi:10.1016/0025-5564(78)90077-9) [DOI] [PubMed] [Google Scholar]
- 27.Sumner L. 2007. Proposed minimum reporting standards for chemical analysis. Metabolomics 3, 211–221 10.1007/s11306-007-0082-2 (doi:10.1007/s11306-007-0082-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leader D. P., Burgess K., Creek D., Barrett M. P. 2011. Pathos: a web facility that uses metabolic maps to display experimental changes in metabolites identified by mass spectrometry. Rapid Commun. Mass Spectrom. 25, 3422–3426 10.1002/rcm.5245 (doi:10.1002/rcm.5245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirmalek-Sani S.-H., Tare R. S., Morgan S. M., Roach H. I., Wilson D. I., Hanley N. A., Oreffo R. O. 2006. Characterization and multipotentiality of human fetal femur-derived cells: implications for skeletal tissue regeneration. Stem Cells 24, 1042–1053 10.1634/stemcells.2005-0368 (doi:10.1634/stemcells.2005-0368) [DOI] [PubMed] [Google Scholar]
- 30.Lai C-F., Chaudhary L., Fausto A., Halstead L. R., Ory D. S., Avioli L. V., Cheng Su-Li. 2001. Erk is essential for growth, differentiation, integrin expression, and cell function in human osteoblastic cells. J. Biol. Chem. 276, 14 443–14 450 10.1074/jbc.M010021200 (doi:10.1074/jbc.M010021200) [DOI] [PubMed] [Google Scholar]
- 31.von der Mark K., von der Mark H. 1977. The role of three genetically distinct collagen types in endochondral ossification and calcification of cartilage. J Bone Joint Surg. Br. 59-B, 458–464 [DOI] [PubMed] [Google Scholar]
- 32.Soga T., et al. 2006. Differential metabolomics reveals opthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 281, 16 768–16 776 10.1074/jbc.M601876200 (doi:10.1074/jbc.M601876200) [DOI] [PubMed] [Google Scholar]
- 33.Goldsmith P., Fenton H., Morris-Stiff G., Ahmad N., Fisher J., Prasad K. R. 2010. Metabonomics: a useful tool for the future surgeon. J. Surg. Res. 160, 122–132 10.1023/A:1005083323183 (doi:10.1023/A:1005083323183) [DOI] [PubMed] [Google Scholar]
- 34.Ishii M., Egen J., Klauschen F., Meier-Schellersheim M., Saeki Y., Vacher J., Proia R. L., Germain R. N. 2009. Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature 458, 524–528 10.1037/0735-7036.110.1.3 (doi:10.1037/0735-7036.110.1.3) [DOI] [PMC free article] [PubMed] [Google Scholar]