Abstract
Human arylamine N-acetyltransferase 1, (HUMAN)NAT1, is a phase II xenobiotic-metabolizing enzyme that plays an important role in drug and carcinogen biotransformation and cancer development. Its gene expression has been shown to be regulated by environmental factors. The purpose of the current study is to determine the involvement of nuclear receptors in transcriptional regulation of (HUMAN)NAT1 gene. We show that among the nuclear receptors examined, including the glucocorticoid receptor, retinoid acid receptor-related orphan receptor alpha, constitutive androstane receptor, pregnane X receptor, aryl hydrocarbon receptor, and retinoic acid receptor, the glucocorticoid receptor plays a dominant role in regulating (HUMAN)NAT1 gene expression through distal promoter (P3). The involvement of the glucocorticoid receptor in transcription regulation of (HUMAN)NAT1 gene expression was demonstrated by dexamethasone treatment, reporter assay using plasmid-containing 3 kbp of 5′-end region of promoter 3, and treatment of anti-glucocorticoid RU486 in primary culture of human hepatocytes and transfected HepG2 cells. In addition, translation inhibition did not affect dexamethasone-induced gene expression through P3, suggesting that dexamethasone effect is directly mediated by glucocorticoid receptor activation. Furthermore, deletion analysis revealed the presence of multiple responsive elements within the 3 kbp fragment of P3. Transfection assays in mice using hydrodynamics-based procedure and reporter gene assay in a mouse cell line revealed that glucocorticoid-induced NAT gene expression is species dependent. Dexamethasone treatment of transfected mice and mouse cell line decreased (MOUSE)Nat2 gene expression, (HUMAN)NAT1 homologue. These results suggest that glucocorticoids serve as a modulator for (HUMAN)NAT1 gene expression via the P3-containing 5′-flanking region.
Key words: arylamine N-acetyltransferases, glucocorticoids, phase-II enzymes, promoter analysis, regulation of gene expression, transcriptional regulation
INTRODUCTION
The functional roles of arylamine N-acetyltransferases (NAT) 1 and 2 have been well documented in detoxification, promotion of tumor growth and drug metabolism. These enzymes can catalyze the N-acetylation-mediated detoxification of carcinogenic aromatic amines that account for approximately 12% of carcinogens or O-acetylation reaction for enhancing the toxicity of aromatic hydroxylamines (1,2). The balance of these two processes is dependent not only on the chemical structure of the substrate but also on the activity of NAT enzymes and is thought to contribute to the aromatic amine-induced malignancies (1–3).
(HUMAN)NAT1 over-expression has been shown in human breast carcinomas (4–6) and is linked to enhanced growth and resistance to the cytotoxic drug etoposide in normal breast luminal epithelial cells (4). It was also shown that the (HUMAN)NAT1 gene was down-regulated in late-stage breast cancer (6). In addition, DNA hypomethylation in the (HUMAN)NAT1 gene was reported in cancerous breast tissues (7). Clinical studies have shown a longer relapse-free survival of tamoxifen-treated patients with estrogen receptor (ER)+ and NAT1-over-expressing tumors (5). Interestingly, tamoxifen (8) and other anti-cancer drugs such as cisplatin (9) and disulfiram (10) have been shown to inhibit the NAT1 activity. NAT1 involvement in cancer development also pertains to human prostate cancer. In fact, microarray work by Lapointe et al. (11) on 112 human prostate cancer samples identified three separate tumor subtypes, each with a different expression pattern of NAT1. More recent studies have shown that the (HUMAN)NAT1 is induced by androgens in human prostate cancer cells, with possible implication for cancer risk (12).
In addition to genetic polymorphisms, (HUMAN)NAT1 activity also undergoes environmental modulation. Wakefield et al. (13) have shown epigenetic regulation of (MOUSE)Nat2, suggesting folate-mediated regulation of the (HUMAN)NAT1 orthologue. Activity of the (HUMAN)NAT1 promoters 1 (P1) (14), 2 (P2) (15–18), and 3 (P3) (17–19) has been shown to be affected by some xeno- and/or endo-biotics (12,14,20–25), but limited information is currently available. Computer-based analysis of the 3 kbp fragments of (HUMAN)NAT1 promoters (P1, P2, or P3) revealed numerous putative glucocorticoid-responsive elements, suggesting that (HUMAN)NAT1 may be regulated in part by the glucocorticoid receptors. The frequency of the clinical use of glucocorticoids underscores the importance of determining if and how these drugs affect (HUMAN)NAT1 gene expression, as change of (HUMAN)NAT1 gene expression could influence the outcome of cancer treatment. Discerning the dynamics of (HUMAN)NAT1 gene expression can thus provide insights not only into potential interventions in cancer chemotherapy but also molecular mechanisms of cancer biology. Here, we report that glucocorticoids enhance (HUMAN)NAT1 gene expression through P3. We also report that both human and mouse promoters are down-regulated by glucocorticoids in mouse cells, suggesting a species difference. Our results provide new insights into NAT gene regulation by glucocorticoids.
MATERIALS AND METHODS
Materials
The PCR kit and reagents were from Promega (Madison, WI) and New England Biolabs (Ipswich, MA). SYBR Green Real-time PCR master mix was from Applied Biosystems (Carlsbad, CA). The CTD-2547L16 BAC clone containing the full genomic sequence of (HUMAN)NAT1 gene was from Invitrogen (Carlsbad, CA). PCR primers were synthesized by Sigma-Aldrich (St. Louis, MO). TA cloning kit and the pGL3-Basic Vector were from Invitrogen (Carlsbad, CA) and Promega (Madison, WI), respectively. Restriction enzymes, T4 DNA ligase and DH5α competent cells of Escherichia coli were from New England Biolabs (Ipswich, MA). Purification kits for BAC DNA, plasmid, and PCR products were from Qiagen (Valencia, CA). TRIzol reagent was from Invitrogen (Carlsbad, CA). QuantiTect Reverse Transcription Kit was from Qiagen (Valencia, CA). HepG2 cells were from the American Type Culture Collection (ATCC, Rockville, MD), and cell culture medium was from Sigma-Aldrich (St. Louis, MO) while supplements were from Cellgro (Manassas, VA) and Lonza (Walkersville, MD). Dexamethasone (DEX), cycloheximide, mifepristone (RU486), actinomycin D, melatonin, 3-methyl-cholanthrene, 6-(4-chlorophenyl)imidazo(2,1-b)(1, 3)thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime, rifampicin, and 9-cis-retinoic acid were from Sigma-Aldrich (St. Louis, MO). Luciferase assay kits were from Promega (Madison, WI). Protein assay reagent was from Bio-Rad (Hercules, CA). pCMV-hGRα plasmid was kindly provided by Dr. John Cidlowski (National Institute of Environmental Health Sciences, NIH). pCMX-hRORα, pCMV-hCAR, and pCMV-hPXR were kind gifts from Dr. Wen Xie (University of Pittsburgh). pTARGET-hAhR was obtained from Dr. Oliver Hankinson (University of California, Los Angeles, CA), and pCMX-hRAR was from Dr. James Boyer (Yale University). Freshly plated human primary hepatocytes were from Celsis In Vitro Inc. (Baltimore, MD) or the Department of Pathology at the University of Pittsburgh. Maintenance medium for human primary hepatocyte was from Lonza (Walkersville, MD).
Primary Human Hepatocyte Culture
Freshly isolated human hepatocytes were maintained in hepatocyte maintenance medium supplemented with DEX (10−7 M), insulin (10−7 M), and gentamicin (50 μg/ml). They were cultured for 24 h in DEX-free medium and then exposed to DEX, RU486, or cycloheximide for 24 h before RNA isolation.
Animal Treatment
CD1 male mice (18–20 g) were intraperitoneally (ip) injected with 10 μl/g body weight of dimethyl sulfoxide (carrier solution) with or without DEX or RU486 at the indicated dose for the indicated time. Animals were killed, liver removed, and total RNA isolated from the liver. All animal experiments were conducted in full compliance with regulation and approved by the IACUC at the University of Pittsburgh, Pittsburgh, PA.
Real-time RT-PCR
Total RNA was isolated using 1 ml of TRIzol reagent per well of cell culture or 50 mg of mouse liver according to manufacturer’s instruction. Total RNA concentration was determined at 260 nm, and their integrity was checked on 1% agarose gel. Two micrograms of RNA were reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instruction and diluted to a final RNA concentration of 20 (mouse samples) or 40 ng/μl (human samples). Quantitative PCR was performed on the cDNA using the StepOnePlus Real-time PCR System (Applied Biosystem). Specific primers used to amplify the target sequences included: forward 5′-TTGCATGATTCTCCTGCCTA-3′ and reverse 5′-CCCAGAATCCTGTGAGAAATG-3′ to amplify the cDNAs corresponding to RNA product from P1; forward 5′-ACTTCCTCATAGACCTTGGATG-3′ and reverse 5′-AGGTTATTTCAGCCGGCAAC-3′ to amplify the RNA from P2; forward 5′-GCCAAACTGCACAAATCAGA-3′ and reverse 5′-TCACCTGGTTCCTGCTCTCT-3′ to amplify the cDNAs corresponding to P3 activity; forward 5′-GAGTCAACGGATTTGGTCGT3′ and reverse 5′-GACAAGCTTCCCGTTCTCAG-3′ to amplify the (HUMAN)GADPH; forward 5′-ACACTCCAGCCAATAAGTACAGC-3′ and reverse 5′-GGTAGGAACGTCCAAACCCA-3′ to amplify the (MOUSE)Nat2; forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′ to amplify the (MOUSE)Gadph. The identity of each amplicon was checked by DNA sequencing. The 20 μl of the reaction mixture consisted of SYBR Green PCR Master Mix (Applied Biosystem), cDNA (20 ng for mouse samples or 40 ng for human samples), and 125 nM reverse and forward primers. The program consisted of a denaturing step of 10 min at 95°C plus 40 cycles of denaturing at 95°C for 15 s and combined annealing and extension at 60°C for 1 min, followed by dissociation curve analysis. Expression levels were normalized by using the RNA level of human or mouse GAPDH gene, which showed stable expression under xenobiotic treatment and CT range comparable to that of the genes of interest. All the amplification reactions were run in triplicates (variability less than 0.5 CT). All calculations (ΔΔCT method) were conducted as suggested by the StepOnePlus Real-time PCR manual (26,27).
Plasmid Construction
The promoter-containing DNA fragment of the (HUMAN)NAT1 gene was PCR amplified using BAC clone CTD-2547L16 (Invitrogen) as the template and cloned into pGL3-Basic Vector using the Qiagen large construct kit (Qiagen). Deep VentR DNA polymerase (New England Biolabs) and 100 ng of BAC DNA were used to amplify ∼3 kbp of the P1- and P3-containing region with the following primers: reverse 5′-TCCATGATCCCCTAAGCAAG-3′ and forward 5′-GCCTCAACATGCCGAATTAT-3′ for P1-containing fragment (3,070 bp, 0 to −3070); and reverse 5′-GCAGTTTGGCCTAGGCTTTAT-3′ and forward 5′-AAAATCAACACACCAAAATCACT-3′ for P3-containing fragment (3,122 bp, 0 to −3122). Each promoter-containing sequence was amplified with an annealing temperature at 61°C and 28 amplification cycles. The PCR products were separated on an agarose gel and the proper fragment was isolated and incubated for 10 min at 72°C with Taq DNA polymerase and dATP to add 3′-A overhang. The resulting PCR products and the TA vector were then ligated and transformed into DH5α competent cells of E. coli. Each insertion was confirmed by PCR, restriction enzyme digestion, and DNA sequencing; 3 kbp P1- or P3-containing fragments were then excised by digestion with XhoI/MluI and XhoI/Hind, respectively, and ligated into pGL3-Basic Vector, which was linearized with the same corresponding restriction enzymes. The resulting plasmids (pGL3-3kP1-Luc and pGL3-3kP3-Luc) were then transformed into E. coli (DH5α), purified and confirmed by restriction enzyme digestion.
Deletion Constructs
Each deletion insert was amplified using the Deep VentR DNA polymerase (New England Biolabs) and 100 ng of pGL3-3kP3-Luc as template. All the sequences were amplified using the reverse primer 5′-CCCAAGCTTGCAGTTTGGCCTAGGCTTTAT-3′, containing a HindIII site attached to the 5′-end. The forward primers used for the amplification were synthesized with a XhoI site attached to the 5′-end: 5′-CCGCTCGAGAATTAAAGCAACACGGCAAT-3′ (2,050 bp, 0 to −2,050), 5′-CCGCTCGAGGCACCTCACACCTGTCAGAA-3′ (1,027 bp, 0 to −1,027), 5′-CCGCTCGAGTGGAAAATTATATGGAGTTTCCTAAA-3′ (909 bp, 0 to −909), 5′-CCGCTCGAGTTTATCAAAGAGATGTCCACCA-3′ (801 bp, 0 to −801), 5′-CCGCTCGAGTGTGGTATGTAGACACAATGGAA-3′ (690 bp, 0 to −690), 5′-CCGCTCGAGGGATGGTGATCAAAAGATGCT-3′ (521 bp, 0 to −521). Each P3-containing sequence was amplified with an annealing temperature at 56°C and 28 amplification cycles. To clone each fragment into the pGL3-Basic Vector, the PCR products were separated on an agarose gel and the proper fragment was isolated. After digestion with the proper restriction enzymes, PCR fragments and linearized pGL3-Basic Vector were ligated and transformed into E. coli (DH5α). Each sequence insertion was confirmed by PCR, restriction enzyme digestion and DNA sequencing.
Cell Culture and Transfection
HepG2 and Hepa1-6 cells were cultured in a humidified incubator under 5% CO2 at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) and supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were seeded into 48-well plates (8 × 105 cells/well) and allowed to grow for 24 h. Cells were transfected using linear polyethelynimine as transfection reagent. For each well, cells were washed twice with phosphate-buffered saline and transfected with 2 μg of plasmid DNA at a DNA to linear polyethelynimine ratio of 1:2 (in micrograms per microgram). Of the 2 μg of DNA used for the transfection, 1 μg accounts for the reporter plasmid containing P3 sequence while the other microgram accounts for the increasing amount of pCMV-hGRα, pCMX-hRORα, pTARGET-hAhR, pCMV-hCAR, pCMV-hPXR, or pCMX-hRAR (0, 0.2, 0.4, 0.8, or 1 μg) and decreasing amount of empty vector without nuclear receptor gene (1, 0.8, 0.6, 0.2, or 0 μg). The plasmids were mixed in serum free-DMEM, incubated at room temperature for 15 min to allow the DNA/linear polyethelynimine complex formation, and applied to cells at 300 μl/well. The transfection mixture was replaced 12–14 h later with complete DMEM after washing cells with PBS twice. Then, DEX (1 μM), RU486 (0.1, 1 or 10 μM), melatonin (100 μM), 3-methylcholanthrene (5 μM), 6-(4-chlorophenyl)imidazo(2,1-b)(1,3)thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime (1 μM), rifampicin (10 μM), or 9-cis-retinoic acid (1 μM) was added and kept for 24 h. Cells were washed with PBS and lysed with 200 μl of 0.1 M Tris–HCl, 0.1% Triton X-100, and 2 mM EDTA (pH 7.8) at room temperature for 15 min 24 h later. Cell lysates were centrifuged (12,000 rpm, 10 min, 4°C). The supernatant was used for measurement of luciferase activity and determination of protein concentration at 595 nm, using the Bio-Rad Protein Assay Kit (Bio-Rad, CA).
Plasmid DNA Preparation
Plasmid DNA was purified from transformed E. coli using cesium chloride gradient centrifugation. Plasmid DNA was quantified at 260 nm, checked by gel electrophoresis, and stored in saline.
Hydrodynamic Transfection of Animals
Animals were injected via the tail vein in 3–5 s with a volume of DNA-containing saline equal to 10% of body weight as described previously (28). Each mouse received 1 μg DNA/g of body weight, containing 0.5 μg of pGL3-3kP3-Luc and 0.5 μg of pCMV-hGRα or empty vector without GRα gene. After drug treatment, animals were sacrificed at the indicated time and the livers were harvested. Individual tissue samples of approximately 200 mg were homogenized in 1 ml of lysis buffer, followed by centrifugation (16,000×g, 15 min, 4°C). Protein concentration determined at 595 nm using the Bio-Rad Protein Assay Kit (Bio-Rad, CA), and luciferase activity in the supernatant of liver homogenate was determined.
Luciferase Assay
Ten microliters of supernatant from cell lysate or tissue homogenates were added to 100 μl of luciferase substrate (Promega, Madison, WI). Luciferase activity was measured over 10 s in a luminometer (Autolumat LB953, EG&G, Berthhold, Germany). Luciferase activity in each sample was normalized to relative light units per milligram of extracted proteins.
Statistical Analysis
Data are reported as mean ± SD. The Student’s t test or one-way ANOVA test was performed among groups. A p value equal or less than 0.05 or 0.01 was used to indicate statistical significance.
RESULTS
Assessment of (HUMAN)NAT1 Promoters in Response to DEX and Glucocorticoid Receptor
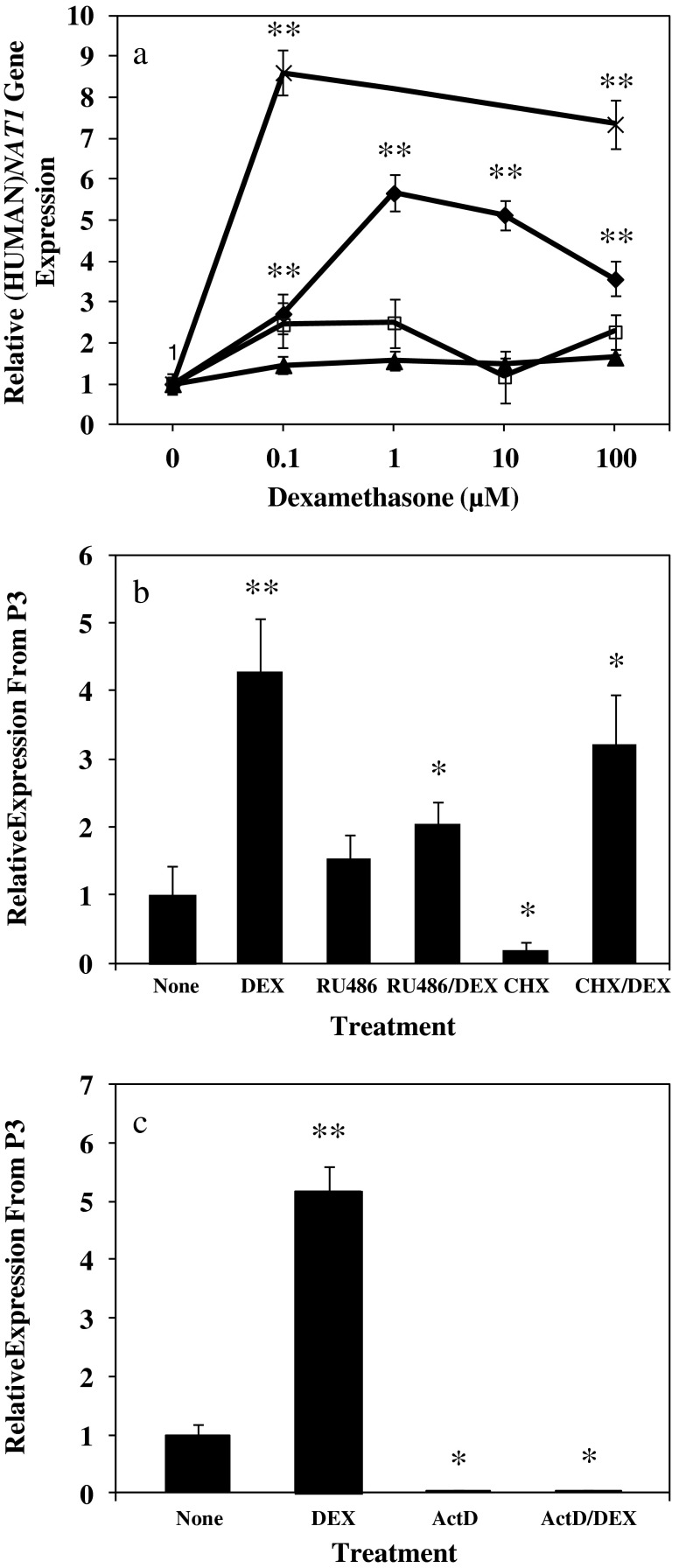
Primary human hepatocytes from a female donor were cultured in hepatocyte maintenance medium without DEX and then treated with DEX for 24 h. (HUMAN)NAT1 transcript levels from P1, P2, and P3 were assessed by real-time RT-PCR using specific primers. Figure 1a shows DEX treatment (0.1, 1, 10, and 100 μM) resulted in no increase in the expression of the (HUMAN)NAT1 gene from P1 and P2 but significant increase in P3-mediated gene expression. DEX treatment resulted in an increase of (HUMAN)NAT1 mRNA level from P3 by ∼3-, 6-, 5-, and 3.5-fold at DEX concentrations of 0.1, 1, 10, and 100 μM, respectively. To confirm that the DEX effect on P3-mediated gene expression is not a gender specific phenomenon, we performed the same set of experiments using primary hepatocytes from a male donor and found even stronger DEX induction (up to ∼8.3- and 7.6-fold) at concentrations of 0.1 and 100 μM (p ≤ 0.01), respectively (Fig. 1a).
Fig. 1.
Effect of dexamethasone (DEX), mifepristone (RU486), cycloheximide (CHX), actinomycin D (ActD) or selected combinations on promoter specific transcription of (HUMAN)NAT1 gene. Primary human hepatocytes were kept in DEX-free medium for 24 h and then treated for 24 h. (HUMAN)NAT1 mRNA levels were determined by real-time RT-PCR and normalized to (HUMAN)GADPH mRNA level. Values represent the mean ± SD of results from two independent experiments of a single culture of human hepatocytes. a Effect of DEX concentration on (HUMAN)NAT1 gene expression driven by P1 (filled triangles), P2 (empty squares), or P3 (diamonds, female donor primary hepatocytes; error marks, male donor primary hepatocytes). b Effect of RU486 (10 μM) and CHX (20 μM) on P3-mediated (HUMAN)NAT1 gene expression with or without DEX (10 μM). c Effect of ActD (5 μM) on promoter 3-mediated transcription of (HUMAN)NAT1 gene with or without DEX (10 μM). *p ≤ 0.01, significantly different from cells treated with DEX; **p ≤ 0.01, significantly different from cells treated with carrier solution (control)
To test whether the activation of the glucocorticoid receptor is directly responsible for DEX-induced (HUMAN)NAT1 gene expression, we examined the effects of treatment with the glucocorticoid receptor antagonist RU486 and protein synthesis inhibitor cycloheximide on DEX-induced (HUMAN)NAT1 gene expression in primary human hepatocytes. Results in Fig. 1b show that DEX alone induced a significant increase (p ≤ 0.01) in P3-mediated (HUMAN)NAT1 gene expression while co-treatment with RU486 blocked DEX-induced expression. It is worth noting that cycloheximide itself decreased (p ≤ 0.01) the level of (HUMAN)NAT1 gene expression from P3. However, inclusion of DEX in the treatment significantly increased the level of (HUMAN)NAT1 RNA level. These results suggest that protein synthesis is not required and activation of pre-existing glucocorticoid receptors is responsible for transcription activation.
The enhancement of P3 activity by DEX was also examined in the presence of the RNA synthesis inhibitor, actinomycin D. As shown in Fig. 1c, actinomycin D treatment caused a significant decrease in (HUMAN)NAT1 mRNA level (p ≤ 0.01), confirming that transcription is required to drive the constitutive and P3-based expression of (HUMAN)NAT1 gene. More importantly, actinomycin D completely abolished the increase of (HUMAN)NAT1 mRNA from P3 in response to DEX (p ≤ 0.01), suggesting that glucocorticoid receptors act through a transcriptional mechanism rather than the improvement of pre-existing mRNA stability. The overall data indicate that DEX-mediated up-regulation of (HUMAN)NAT1 gene expression occurs through P3 and directly involves activation of glucocorticoid receptor.
Glucocorticoid Receptor-Mediated Regulation of (HUMAN)NAT1 Gene Expression in Reporter Assay
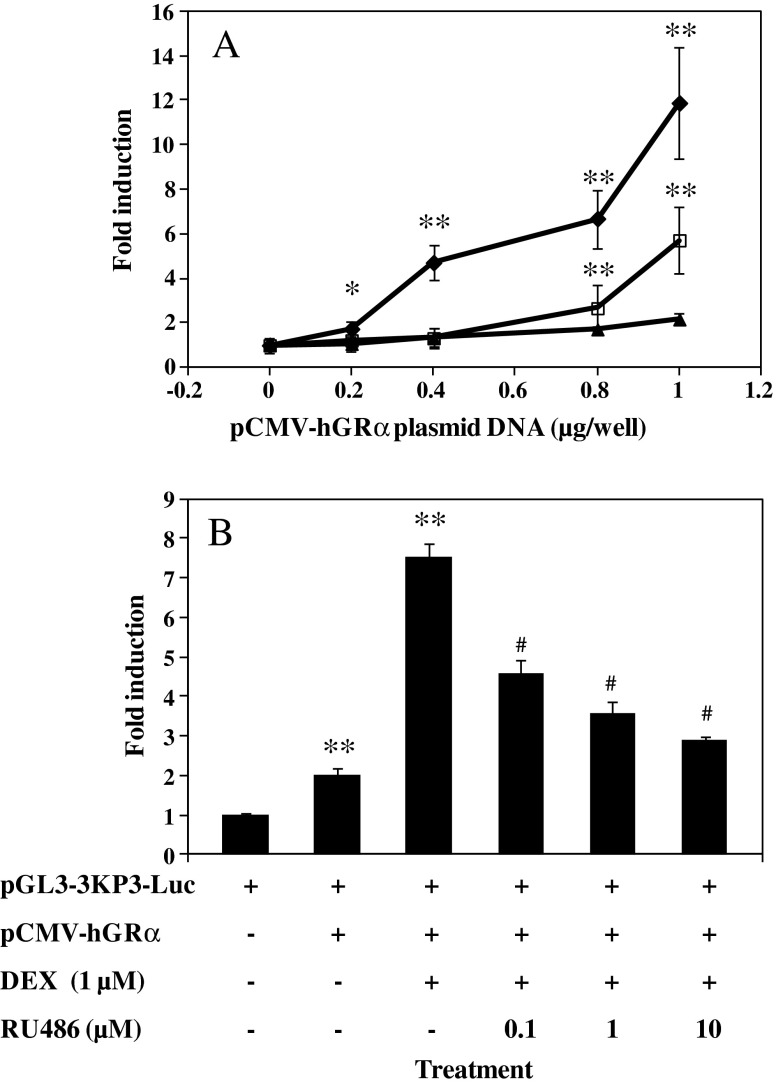
Further evidence for regulation of P3-based (HUMAN)NAT1 gene expression by glucocorticoid receptors was sought. A 3-kbp fragment at the 5′-end of exon 1 including P3 was cloned into a pGL3-Basic Vector containing luciferase reporter gene to generate a new plasmid, pGL3-3kP3-Luc. HepG2 cells were co-transfected with the new construct and the plasmid containing human glucocorticoid receptor α gene under control of a CMV promoter. Plasmid containing the same length of DNA fragment at the 5′-end of exon 9 including P1 (pGL3-3kP1-Luc) was included as a control. All cells were treated with DEX at the same concentration. Results in Fig. 2a show no increase of luciferase activity in cells co-transfected with pGL3-3kP1-Luc and pCMV-hGRα. However, a progressive increase in luciferase activity was seen in cells co-transfected with pGL3-3kP3-Luc and an increasing amount of pCMV-hGRα DNA. In fact, amounts of pCMV-hGRα as low as 0.2 μg already exhibited a significant up-regulation (p ≤ 0.05) of reporter gene expression. Progressively, the induction kept growing up to ∼5-, 7-, and 12-fold (p ≤ 0.01) in the presence of 0.4, 0.8, and 1 μg of pCMV-hGRα, respectively. A significant (p ≤ 0.01) but much lower level of induction was also observed in the absence of DEX. Overall, these data confirm lack of responsiveness in P1 to DEX and over-expression of glucocorticoid receptor α in transfected cells is sufficient to enhance P3-mediated gene expression. Glucocorticoid receptor activation by DEX appears necessary for maximal level of P3-activity.
Fig. 2.
Effect of dexamethasone (DEX) and/or mifepristone (RU486) on P3-mediated reporter gene expression. HepG2 cells (8 × 105 cells/well) were seeded in a 48-well plate 24 h prior to transfection. For each well, cells were incubated for 12 h with 2 μg of plasmid DNA mixed with 4 μg of linear polyethelynimine as transfection reagent and then maintained in serum-containing medium for 24 h. Cells were then treated for additional 24 h with DMSO (carrier solution) with or without DEX or DEX/RU486 in combination. Luciferase activity in each well was determined using luciferase assay. a Effect of co-transfection of HepG2 cells with increasing amount of pCMV-hGRα plasmid DNA on luciferase gene expression from either pGL3-3kP3-Luc or pGL3-3kP1-Luc plasmids. Of the 2 μg of plasmid DNA used for transfection, 1 μg accounts for reporter plasmid while the other accounts for increasing amount of pCMV-hGR (0, 0.2, 0.4, 0.8, or 1 μg) and decreasing amount of empty vector (pGL3-Luc without promoter; 1, 0.8, 0.6, 0.2, or 0 μg). b Blockade of DEX-enhanced luciferase gene expression by RU486. HepG2 cells were transfected with equal amount (1 μg) of pGL3-3kP3-Luc and pCMV-hGRα for 12 h and maintained in serum-containing medium for 24 h. Cells were then treated for another 24 h with DMSO (carrier solution) with or without DEX (1 μM) or increasing amount of RU486 (0.1, 1, or 10 μM) before luciferase assay. Values represent the mean ± SD of three independent transfections. *p ≤ 0.05; **p ≤ 0.01, significantly different from cells transfected without pCMV-hGRα; #(p ≤ 0.01), significantly different from DEX treatment without RU486. Filled diamonds, pGL3-3kP3-Luc/DEX; empty squares, pGL3-3kP3-Luc/DMSO; filled triangles, pGL3-3kP1-Luc/DEX
A dose response curve was established for RU486 to verify direct involvement of glucocorticoid receptors in the P3-mediated induction in reporter assay. As shown in Fig. 2b, DEX treatment increased (p ≤ 0.01) luciferase level by 7-fold compared with an approximate 2-fold increase (p ≤ 0.01) resulting from over-expression of human glucocorticoid receptor α (no DEX included). However, increasing amounts of RU486 progressively abolished (p ≤ 0.01) DEX-induced up-regulation of reporter gene expression. At equal molar concentration (1 μM) of DEX, RU486 reduced the level of DEX-induced luciferase expression by about 60% (p ≤ 0.01), consistent and in agreement with the well-known fact that higher binding affinity of RU486 to human glucocorticoid receptor overrides the activity of DEX in glucocorticoid receptor activation (29).
Identification of the Functional Elements in the 5′-flanking Region of P3 that Are Responsible for Glucocorticoid Receptor-Mediated Activation
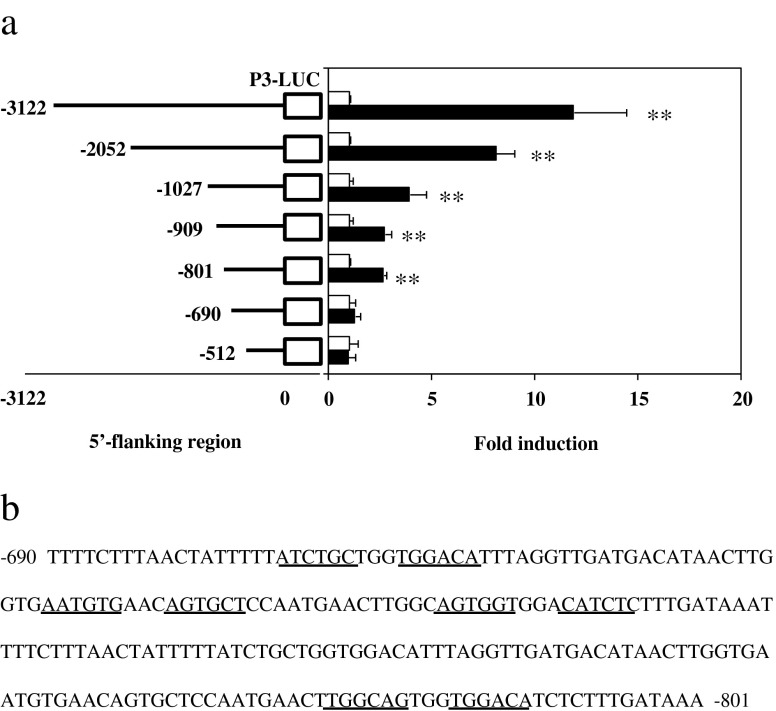
A series of deletion constructs were made in an effort to identify glucocorticoid response elements in the P3 region (Fig. 3a). Glucocorticoid receptor-mediated induction by DEX was determined by the reporter assay as described in Fig. 2. It is apparent that the longer the P3-containing fragment, the higher the DEX- and glucocorticoid receptor-induced enhancement of reporter gene expression. With the full length of the 3 kbp fragment, there is approximately a 12-fold increase (p ≤ 0.01) in luciferase gene expression in the presence of DEX and glucocorticoid receptors, which declined to 8-fold (p ≤ 0.01) and 4-fold (p ≤ 0.01) when 2 and 1 kbp P3-containing DNA fragments were used, respectively. Additional deletion of about 100 to 200 bp from the 1 kbp fragment resulted in further decline to 3-fold (p ≤ 0.01). The DEX- and glucocorticoid receptor-induced response was not seen when the DNA fragment was shorter than 690 bp. Overall, these data suggest that multiple glucocorticoid response elements lie within the 3 kbp fragment located upstream of the exon 1 and control glucocorticoid receptor-mediated induction of (HUMAN)NAT1 gene expression from P3. The most proximal element(s) to the distal promoter P3 seems to be located within the −690 to −801 bp region (Fig. 3b).
Fig. 3.
Identification of the putative GREs in the P3-containing region of the (HUMAN)NAT1 gene. a Effect of the length of 5′-flanking region of P3 on DEX and glucocorticoid receptor mediated induction of luciferase gene expression. A series of deletion reporter constructs were made by inserting a desirable P3-containing fragment into pGL3-basic vector. HepG2 cells were co-transfected with the new plasmid and pCMV-hGRα (solid bars) at 1 μg each per well. The controls were co-transfected with empty plasmid without hGRα (open bars). Cells were transfected for 12 h, maintained in regular medium for 24 h, and treated with DEX (1 μM) for additional 24 h before cell lysis and luciferase assay. Values represent the mean ± SD of three independent transfections. **p ≤ 0.01, significantly different from control transfected with empty vector. b Putative 3′- or 5′-half of GREs (underlined) in the region between −690 and −801 bp upstream of exon 1
Reporter Gene Expression from pGL3-3kP3-Luc Plasmids and Expression of (MOUSE)Nat2 Gene Is Down-regulated in Mouse Liver
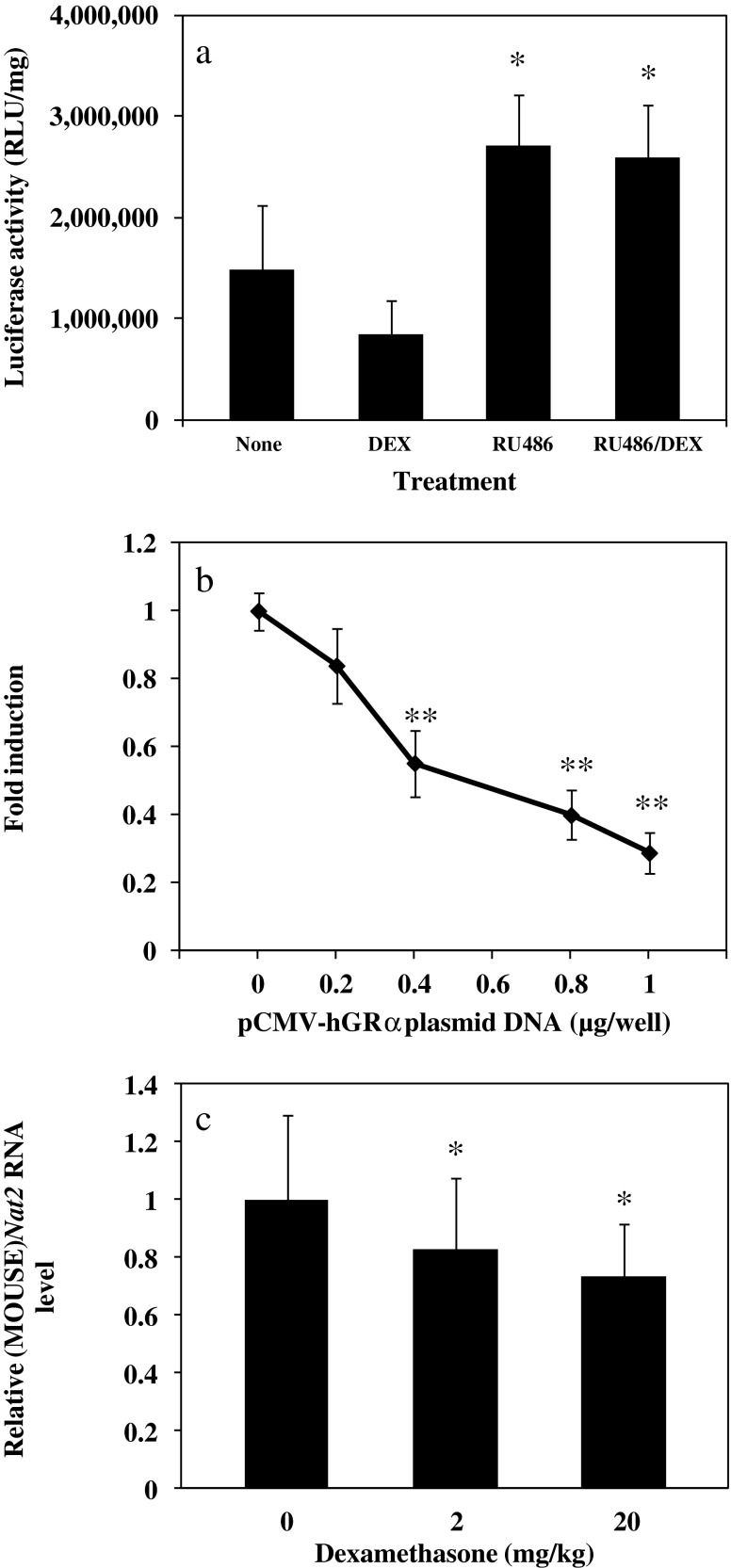
Mice are the most commonly used animal model for cancer research. We thus wondered if the same regulation mechanism seen in (HUMAN)NAT1 gene also applies to that of mice. To explore a species-specific effect of glucocorticoids and/or glucocorticoid receptors on NAT gene expression, we hydrodynamically transfected mice with the reporter construct of pGL3-3kP3-Luc only and administered DEX and/or RU486 (ip) with an intention to assess the effects of DEX and the involvement of the glucocorticoid receptors on (HUMAN)NAT1 gene expression in the mouse liver. As shown in Fig. 4a, DEX induced up to a 50% decrease in luciferase gene expression. Moreover, exposure to RU486—both in the presence or absence of DEX (p ≤ 0.05)—brought the level of luciferase activity above the control level. To explore whether the observed difference in NAT gene expression could be explained by species difference in ligand-dependent activation of the glucocorticoid receptors, we co-transfected pGL3-3kP3-Luc and pCMV-hGRα into the mouse hepatoma cell line Hepa1-6. In this setting (Fig. 4b), we observed a progressive decrease (up to more than 70%, p ≤ 0.01) in luciferase activity, even in the absence of DEX. Similarly, the same results were obtained in hydrodynamically co-transfected mice exposed to various concentrations of DEX (data not shown). Finally, we determined the change in the expression of (MOUSE)Nat2, the homologue of (HUMAN)NAT1, as a consequence of DEX administration. We again found a down-regulation up to about 30% when the highest dose of DEX was employed in vivo (Fig. 4c). These data suggest that DEX and/or glucocorticoid receptor α-mediated regulation of (HUMAN)NAT1 gene expression in mice and human is different and likely dependent on dissimilar signaling rather than the miss- recruitment of mouse co-repressive factors by the human nuclear receptors on the human sequence. The single (MOUSE)Nat2 promoter (30) equivalence to the (HUMAN)NAT1 P2 promoter (31) rather than to the P3 may account for the differencial regulation of the two orthologue genes in mice and humans.
Fig. 4.
DEX and glucocorticoid receptor α-dependent down-regulation of reporter gene expression in pGL3-3kP3-Luc and (MOUSE)Nat2 gene in the mouse liver. a Male CD1 mice were hydrodynamically transfected with pGL3-3kP3-Luc and empty vector without GRα gene. Three h post-transfection mice were injected (ip) with DMSO (carrier solution), DEX (6 mg/kg), RU486 (20 mg/kg), or in combination. Animals were sacrificed 8 h later and luciferase activity was determined in liver homogenate. b Hepa1-6 cells were transfected for 12 h with a total of 2 μg/well of plasmids containing 1 μg of pGL3-3kP3-Luc, increasing amount of pCMV-hGRα (0, 0.2, 0.4, 0.8, or 1 μg) and decreasing amount of empty vector (1, 0.8, 0.6, 0.2, or 0 μg), maintained in serum-containing medium for 24 h, and treated with DEX (1 μM) for another 24 h before cell lysis and luciferase assay. c Male CD1 mice were injected (ip) with DEX (2 or 20 mg/kg) and killed 12 h later. Total RNA was isolated from the liver and (MOUSE)Nat2 mRNA was determined by real-time RT-PCR and normalized to (MOUSE)Gadph mRNA level. Values represent the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01, significantly different from control animals (treated with carrier solution) (a, c) or cells transfected with empty plasmid without hGRα gene (b)
Evaluation on P3-Mediated Regulation by Other Nuclear Receptors
Finally, the inducibility of the P3-based gene expression by other nuclear receptors such as retinoic acid receptor-related orphan receptor α; aryl hydrocarbon receptor; constitutive androstane receptors; pregnane X receptor; and retinoic acid receptor α was examined. Co-transfection of pGL3-3kP3-Luc plasmid with plasmid containing individual nuclear receptor gene was performed and treated with receptor specific ligands. Results (Fig. 5) show modest regulation by some of the nuclear receptors examined (retinoic acid receptor-related orphan receptor α, aryl hydrocarbon receptor, retinoic acid receptor α, p ≤ 0.01), suggesting that other nuclear receptors could also play a role in regulating (HUMAN)NAT1 gene expression. However, compared with the degree of enhancement obtained, DEX induced the highest enhancement in gene expression, suggesting that glucocorticoid receptors are a predominant regulator for P3-mediated (HUMAN)NAT1 gene expression.
Fig. 5.
Transcription activation of P3 by retinoic acid receptor α (RARα), RAR-related orphan receptor α (RORα), constitutive androstane receptor (CAR), pregnane X receptor (PXR), and aryl hydrocarbon receptor (AhR). HepG2 cells were co-transfected for 12 h with a total of 2 μg of plasmid DNA containing 1 μg of pGL3-3kbP3-Luc, an increasing amount of pCMX-hRORα, pTARGET-hAhR, pCMV-hCAR, pCMV-hPXR, or pCMX-hRAR (0, 0.2, 0.4, 0.8, or 1 μg) and a decreasing amount of empty vector (1, 0.8, 0.6, 0.2, or 0 μg). The transfected cells were then cultured in serum-containing medium for additional 24 h followed by 24 h treatment with activators appropriate to individual nuclear receptor at appropriate concentration of melatonin (100 μM), 3-methylcholanthrene, (5 μM), 6-(4-chlorophenyl)imidazole(2,1-b)(1,3)thiazole-5-carbaldehyde O-(3,4-dichloronezyl)oxime (1 μM), refampicin (10 μM), or 9-cis-retinoic acid (1 μM). Cell extracts were prepared and luciferase activity was determined. Values represent the mean ± SD of three independent transfections. *p ≤ 0.05; **p ≤ 0.01, significantly different from cells transfected with empty vector without nuclear receptor gene
DISCUSSION
In this study we demonstrated that the glucocorticoid receptor plays an important role in regulating (HUMAN)NAT1 gene expression. Among the three promoters in (HUMAN)NAT1 gene, we showed that there are putative glucocorticoid responsive elements in the 5′-end sequence containing P3. We also showed that P3-mediated activation in human liver cells requires activation of glucocorticoid receptors by glucocorticoids. In addition, we provided direct evidence in support that glucocorticoid receptor-induced gene expression is animal species dependent. While it enhances (HUMAN)NAT1 gene expression in human hepatocytes, glucocorticoid treatment of mouse cells in culture and of hydrodynamically transfected mouse liver reduced the P3-mediated transcription.
Our study focused initially on the effect of DEX at the genomic level and found that among the 3 promoters, P3 is the one that is highly responsive to glucocorticoids (Fig. 1). We thus restricted our further study on P3. Considering that glucocorticoids exert their biological functions through multiple signaling pathways, we wondered whether glucocorticoid receptors are directly involved in DEX-induced (HUMAN)NAT1 gene expression. The use of the RU486 abrogated the increase in (HUMAN)NAT1 mRNAs originating from the endogenous distal promoter in primary human hepatocytes, implicating the involvement of the glucocorticoid receptors. Also, the use of protein synthesis inhibitor cycloheximide revealed that the glucocorticoid receptor effect is direct and does not require any de novo synthesized protein (Fig. 1b). Further investigation suggested that the glucocorticoid receptors work on (HUMAN)NAT1 gene through P3 activation rather than pre-existing mRNA stabilization, as the transcription inhibitor actinomycin D completely abrogated DEX/glucocorticoid receptor-mediated induction of P3 activity (Fig. 1c).
Further confirmation of the glucocorticoid receptor involvement came from transiently transfected HepG2 cells. We cloned ∼3 kbp of the (HUMAN)NAT1 P3-containing sequence and used the resulting constructs to conduct reporter gene assay. We observed that DEX treatment of transfected HepG2 cells is necessary to boost the glucocorticoid receptor α transactivation of P3, supporting the glucocorticoid receptor involvement (Fig. 2a). In addition, co-treatment with RU486 significantly inhibited the DEX-mediated transactivation of the exogenous P3 in HepG2 (Fig. 2b). In this context, it is noteworthy that the level of induction recorded in our study (∼7–12-fold) is among the highest ever recorded for a glucocorticoid receptor-regulated gene in reporter gene assay (32).
Next, we generated a series of deletion constructs to identify the glucocorticoid responsive elements. Usually, multiple glucocorticoid responsive elements are distributed in the flanking region of target genes and their number and location can vary substantially (33,34). In our analysis (Fig. 3a), we found that the (HUMAN)NAT1 gene does not make exception to this rule, as significant level of reporter gene induction was observed till the original P3-containing fragment shorter than 690 bp. This suggests that multiple glucocorticoid response elements work cooperatively to induce the expression of the (HUMAN)NAT1 gene from P3. We also found that the closest glucocorticoid responsive elements to P3 are located between −690 and −801 bp upstream of exon 1. Inspection of this region revealed the presence of 3′- or 5′-half glucocorticoid responsive elements, which show sequence identity to glucocorticoid responsive elements previously identified in glucocorticoid-regulated genes (Fig. 3b). Additional studies are needed to verify the function of these putative glucocorticoid responsive elements in −690 to −810 bp region. In addition, similar study is needed to identify the glucocorticoid responsive elements located even further upstream in the 5′-flanking region.
We then verified P3 responsiveness to glucocorticoids and glucocorticoid receptors in vivo. Interestingly, we found the NAT expression is regulated differently in humans and the mouse (Fig. 4). In fact, hydrodynamics-based transfection of pGL3-3kP3-Luc plasmid into the mouse liver and subsequent DEX administration revealed a sharp decrease of reporter gene expression. DEX effects are very complex and reported to be opposite in vivo and in vitro (35,36). However, this scenario would appear to neither apply to our data nor seem to depend on a species difference in ligand-dependent activation of the glucocorticoid receptors. In fact, co-transfection of the reporter construct with the plasmid carrying the human glucocorticoid receptors brought about the same outcomes in both Hepa1-6 cells and mouse liver. Consistently, the in vivo DEX-mediated down-regulation of the (MOUSE)Nat2 gene expression, the murine counterpart of (HUMAN)NAT1, completely addressed the possibility of a species-specific difference in regulation of gene expression.
Glucocorticoid-mediated NAT regulation has been previously reported in rats (37) and rabbits (38) while Glowinski and Weber (39), Smolen et al. (40), and Estrada et al. (41) reported androgen-dependent differences in renal NAT2 activity between male and female mice. In our study, we show that steroid regulation may also apply to humans through the glucocorticoid receptor mediated-increase of the (HUMAN)NAT1 gene expression from the distal promoter P3. Considering that the transcriptional regulation is a major determinant for the overall levels of gene expression, we believe that our data may have pharmacological and toxicological implications. Besides significant drug interactions, glucocorticoid exposure could result in enhanced metabolic activation of NAT1 carcinogenic substrates. Also, if our findings can be applied to tissues other than the liver—e.g., breast or prostate, where (HUMAN)NAT1 has been shown to highly contribute to tumor malignancy—concerns should arise about the administration of glucocorticoids to cancer patients. In this context, it should be noted that Wakefield et al. (6) showed that among different ER+ breast cancer cell lines, only those with the highest NAT1 activity (ZR-75-1) had NAT1 transcripts originating from P3, suggesting the inducibility/activation of this promoter and potential pertinence of our findings to tissues are different from the liver. The recurrent prescription of glucocorticoids as cancer co-treatment (42) could thus exacerbate cancer malignancy, due to the increasing evidence of (HUMAN)NAT1 over-expression and its contribution to tumor initiation and progression. Our data, together with the reported (HUMAN)NAT1 gene overexpression in prostate (11) and breast tumors (4–6) as well as inducibility by androgens (12,20) suggest that steroids may regulate the (HUMAN)NAT1 gene expression and thus potentially modify the risk associated with cancers where (HUMAN)NAT1 is implicated. Therefore, it would be relevant to extend the study of the (HUMAN)NAT1 gene responsiveness to glucocorticoids in breast and prostate tissues, as well as to other steroid hormone receptors.
Our results also stress the complexity of glucocorticoid effects. Glucocorticoids can regulate diverse biological processes by different signaling pathways and control the same biological activity in dissimilar ways in vivo and in vitro or in different species. For example, they induce the (RAT)Sult1a1 gene expression (43), do not affect the expression of (HUMAN)SULT1A1 (44), but decrease the mRNA expression of (MOUSE)Sult1a1 in C57BL/6J mice (45). Consistently, we showed that the type of cellular environment plays a fundamental role when the effects of complex drugs and NRs such as glucocorticoids and glucocorticoid receptors are investigated. In fact, sequence identity (as in the case of pGL3-3kP3-Luc transfected in both human cell and mouse) or overall homology, as in the case of (HUMAN)NAT1 and (MOUSE)Nat2 (Fig. 6) (46), is not sufficient to observe the same regulation. Therefore, it is likely that the observed differences depend on the repressive/activating mechanism(s) prevailing in the context of any given experimental model. Hypothetically, in mouse cells, glucocorticoid receptor-mediated signaling pathway on (MOUSE)Nat2 gene expression might depend on a high co-repressor/co-activator ratio while cells of human origin offer to the nuclear receptors a permissive environment, leading to increase of (HUMAN)NAT1 expression. In this context, it is worth noting that Estrada-Rodgers et al. (47) identified a hormone response element upstream of the (MOUSE)Nat2* coding region, which was positively regulated by glucocorticoid hormones and glucocorticoid receptor in reporter gene assays of transiently transfected CV1 (monkey) cell line. The intriguing question of what makes glucocortcoids and glucocorticoid receptor function in species-specific manner still stands and deserves further investigations. Attention should thus be paid to studying cancer-relevant signaling pathways involving the (HUMAN)NAT1 gene in mice in order to apply data obtained from mice to clinical situations.
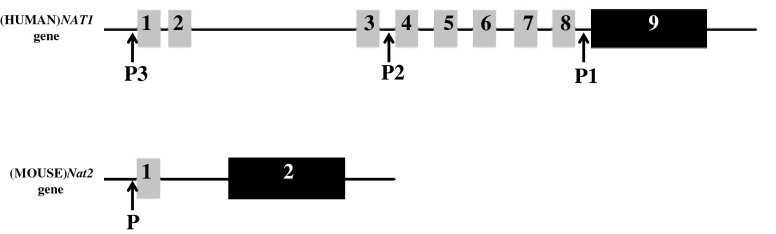
Fig. 6.
Schematic representation of genomic organization of (HUMAN)NAT1 and (MOUSE)Nat2 gene. Both genes contain non-coding exons (grey bars; 1 to 8 for (HUMAN)NAT1, 1 for (MOUSE)Nat2 gene) upstream of the open reading frame. Coding exons are 9 and 2 (black bars) for the (HUMAN)NAT1 and (MOUSE)Nat2 gene, respectively. P promoter
In conclusion, the results of this study demonstrate that a DEX-mediated increase in (HUMAN)NAT1 gene expression is transcriptionally regulated by the direct activity of the glucocorticoid receptors on the P3-containing region via multiple glucocorticoid responsive elements in humans. We also demonstrated the inducibility of the P3-containing fragment by evaluating the possible involvement of other nuclear receptors. Although we found significant regulation for some of the nuclear receptors examined (retinoic acid receptor related orphan receptor α, aryl hydrocarbon receptor and retinoic acid receptor α), none were able to show levels of induction comparable to the glucocorticoid receptor, thus suggesting that glucocorticoid receptors are the primary regulators for P3-mediated NAT gene expression. We also show that DEX effects on NAT expression are species-specific, as reporter gene driven by P3 and endogenous (MOUSE)Nat2 are down-regulated in the mice and in vitro. Overall, these results give new and significant insights into the mechanisms governing (HUMAN)NAT1 gene expression, suggesting caution in the deliberation of clinical treatment of oncological pain, nausea and inflammation with glucocorticoids as well as prudence in extrapolating animal data to human risk when the regulation of cancer-associated xenobiotic-metabolizing enzymes by oncology-relevant drugs is under investigation.
ACKNOWLEDGEMENTS
We thank Drs. John Cidlowski, Wen Xie, Oliver Hankinson, and James Boyer for providing us with the nuclear receptor expression plasmids. This work was supported in part by the grants from the National Institute of Health (RO1EB007357 and RO1HL098295). Barbara Bonamassa was supported by the Rotary Foundation Ambassadorial Scholarship/Research Grant for the academic year 2009–2010.
REFERENCES
- 1.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 2.Hein DW. N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene. 2006;25:1649–1658. doi: 10.1038/sj.onc.1209374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badawi AF, Hirvonen A, Bell DA, Lang NP, Kadlubar FF. Role of aromatic amine acetyltransferases, NAT1 and NAT2, in carcinogen-DNA adduct formation in the human urinary bladder. Cancer Res. 1995;55:5230–5237. [PubMed] [Google Scholar]
- 4.Adam PJ, Berry J, Loader JA, Tyson KL, Craggs G, Smith P, De Belin J, Steers G, Pezzella F, Sachsenmeir KF, Stamps AC, Herath A, Sim E, O’Hare MJ, Harris AL, Terrett JA. Arylamine N-acetyltransferase-1 is highly expressed in breast cancers and conveys enhanced growth and resistance to etoposide in vitro. Mol Cancer Res. 2003;1:826–835. [PubMed] [Google Scholar]
- 5.Bieche I, Girault I, Urbain E, Tozlu S, Lidereau R. Relationship between intratumoral expression of genes coding for xenobiotic-metabolizing enzymes and benefit from adjuvant tamoxifen in estrogen receptor alpha-positive postmenopausal breast carcinoma. Breast Cancer Res. 2004;6:R252–R263. doi: 10.1186/bcr784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakefield L, Robinson J, Long H, Ibbitt JC, Cooke S, Hurst HC, Sim E. Arylamine N-acetyltransferase 1 expression in breast cancer cell lines: a potential marker in estrogen receptor-positive tumors. Genes Chromosomes Cancer. 2008;47:118–126. doi: 10.1002/gcc.20512. [DOI] [PubMed] [Google Scholar]
- 7.Kim SJ, Kang HS, Chang HL, Jung YC, Sim HB, Lee KS, Ro J, Lee ES. Promoter hypomethylation of the N-acetyltransferase 1 gene in breast cancer. Oncol Rep. 2008;19:663–668. [PubMed] [Google Scholar]
- 8.Lee JH, Lu HF, Wang DY, Chen DR, Su CC, Chen YS, Yang JH, Chung JG. Effects of tamoxifen on DNA adduct formation and arylamines N-acetyltransferase activity in human breast cancer cells. Res Commun Mol Pathol Pharmacol. 2004;115–116:217–233. [PubMed] [Google Scholar]
- 9.Ragunathan N, Dairou J, Pluvinage B, Martins M, Petit E, Janel N, Dupret JM, Rodrigues-Lima F. Identification of the xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 as a new target of cisplatin in breast cancer cells: molecular and cellular mechanisms of inhibition. Mol Pharmacol. 2008;73:1761–1768. doi: 10.1124/mol.108.045328. [DOI] [PubMed] [Google Scholar]
- 10.Malka F, Dairou J, Ragunathan N, Dupret JM, Rodrigues-Lima F. Mechanisms and kinetics of human arylamine N-acetyltransferase 1 inhibition by disulfiram. FEBS J. 2009;276:4900–4908. doi: 10.1111/j.1742-4658.2009.07189.x. [DOI] [PubMed] [Google Scholar]
- 11.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butcher NJ, Minchin RF. Arylamine N-acetyltransferase 1 gene regulation by androgens requires a conserved heat shock element for heat shock factor-1. Carcinogenesis. 2010;31:820–826. doi: 10.1093/carcin/bgq042. [DOI] [PubMed] [Google Scholar]
- 13.Wakefield L, Boukouvala S, Sim E. Characterisation of CpG methylation in the upstream control region of mouse Nat2: evidence for a gene-environment interaction in a polymorphic gene implicated in folate metabolism. Gene. 2010;452:16–21. doi: 10.1016/j.gene.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Butcher NJ, Arulpragasam A, Pope C, Minchin RF. Identification of a minimal promoter sequence for the human N-acetyltransferase type I gene that binds AP-1 (activator protein 1) and YY-1 (Yin and Yang 1) Biochem J. 2003;376:441–448. doi: 10.1042/BJ20030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husain A, Zhang X, Doll MA, States JC, Barker DF, Hein DW. Functional analysis of the human N-acetyltransferase 1 major promoter: quantitation of tissue expression and identification of critical sequence elements. Drug Metab Dispos. 2007;35:1649–1656. doi: 10.1124/dmd.107.016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Husain A, Barker DF, States JC, Doll MA, Hein DW. Identification of the major promoter and non-coding exons of the human arylamine N-acetyltransferase 1 gene (NAT1) Pharmacogenetics. 2004;14:397–406. doi: 10.1097/01.fpc.0000114755.08559.6e. [DOI] [PubMed] [Google Scholar]
- 17.Boukouvala S, Sim E. Structural analysis of the genes for human arylamine N-acetyltransferases and characterisation of alternative transcripts. Basic Clin Pharmacol Toxicol. 2005;96:343–351. doi: 10.1111/j.1742-7843.2005.pto_02.x. [DOI] [PubMed] [Google Scholar]
- 18.Butcher NJ, Arulpragasam A, Goh HL, Davey T, Minchin RF. Genomic organization of human arylamine N-acetyltransferase type I reveals alternative promoters that generate different 5′-UTR splice variants with altered translational activities. Biochem J. 2005;387:119–127. doi: 10.1042/BJ20040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker DF, Husain A, Neale JR, Martini BD, Zhang X, Doll MA, States JC, Hein DW. Functional properties of an alternative, tissue-specific promoter for human arylamine N-acetyltransferase 1. Pharmacogenet Genomics. 2006;16:515–525. doi: 10.1097/01.fpc.0000215066.29342.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butcher NJ, Tetlow NL, Cheung C, Broadhurst GM, Minchin RF. Induction of human arylamine N-acetyltransferase type I by androgens in human prostate cancer cells. Cancer Res. 2007;67:85–92. doi: 10.1158/0008-5472.CAN-06-2635. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell KR, Warshawsky D. Xenobiotic inducible regions of the human arylamine N-acetyltransferase 1 and 2 genes. Toxicol Lett. 2003;139:11–23. doi: 10.1016/S0378-4274(02)00437-X. [DOI] [PubMed] [Google Scholar]
- 22.Hsia TC, Yang JH, Lin HJ, Yu CS, Yu FS, Chung JG. Paclitaxel inhibits N-acetyltransferase activity and gene expression in human stomach tumor cells (SC-M1) Res Commun Mol Pathol Pharmacol. 2004;115–116:21–38. [PubMed] [Google Scholar]
- 23.Yu FS, Yu CS, Lin JP, Chen SC, Lai WW, Chung JG. Diallyl disulfide inhibits N-acetyltransferase activity and gene expression in human esophagus epidermoid carcinoma CE 81T/VGH cells. Food Chem Toxicol. 2005;43:1029–1036. doi: 10.1016/j.fct.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Cheng KC, Li YC, Yu CS, Yu FS, Lee JH, Lin ML, Yang JS, Chung JG. Ketoprofen-inhibited N-acetyltransferase activity and gene expression in human colon tumor cells. Anticancer Res. 2006;26:1105–1111. [PubMed] [Google Scholar]
- 25.Westerink WM, Schoonen WG. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol In Vitro. 2007;21:1592–1602. doi: 10.1016/j.tiv.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 26.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 27.Bustin SA, Beaulieu JF, Huggett J, Jaggi R, Kibenge FS, Olsvik PA, Penning LC, Toegel S. MIQE precis: practical implementation of minimum standard guidelines for fluorescence-based quantitative real-time PCR experiments. BMC Mol Biol. 2010;11:74. doi: 10.1186/1471-2199-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 29.Heikinheimo O, Kontula K, Croxatto H, Spitz I, Luukkainen T, Lahteenmaki P. Plasma concentrations and receptor binding of RU 486 and its metabolites in humans. J Steroid Biochem. 1987;26:279–284. doi: 10.1016/0022-4731(87)90083-5. [DOI] [PubMed] [Google Scholar]
- 30.Fakis G, Boukouvala S, Buckle V, Payton M, Denning C, Sim E. Chromosome mapping of the genes for murine arylamine N-acetyltransferases (NATs), enzymes involved in the metabolism of carcinogens: identification of a novel upstream noncoding exon for murine Nat2. Cytogenet Cell Genet. 2000;90:134–138. doi: 10.1159/000015648. [DOI] [PubMed] [Google Scholar]
- 31.Boukouvala S, Price N, Plant KE, Sim E. Structure and transcriptional regulation of the Nat2 gene encoding for the drug-metabolizing enzyme arylamine N-acetyltransferase type 2 in mice. Biochem J. 2003;375:593–602. doi: 10.1042/BJ20030812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Wieland S, Schatt MD, Rusconi S. Role of TATA-element in transcription from glucocorticoid receptor-responsive model promoters. Nucleic Acids Res. 1990;18:5113–5118. doi: 10.1093/nar/18.17.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jantzen HM, Strahle U, Gloss B, Stewart F, Schmid W, Boshart M, Miksicek R, Schutz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987;49:29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- 35.Luo LG, Bruhn T, Jackson IM. Glucocorticoids stimulate thyrotropin-releasing hormone gene expression in cultured hypothalamic neurons. Endocrinology. 1995;136:4945–4950. doi: 10.1210/en.136.11.4945. [DOI] [PubMed] [Google Scholar]
- 36.Estupina C, Belmar J, Tapia-Arancibia L, Astier H, Arancibia S. Rapid and opposite effects of dexamethasone on in vivo and in vitro hypothalamic somatostatin release. Exp Brain Res. 1997;113:337–342. doi: 10.1007/BF02450331. [DOI] [PubMed] [Google Scholar]
- 37.Zaher H, Svensson CK. Glucocorticoid induction of hepatic acetyl CoA:arylamine N-acetyltransferase activity in the rat. Res Commun Chem Pathol Pharmacol. 1994;83:195–208. [PubMed] [Google Scholar]
- 38.Reeves PT, Minchin RF, Ilett KF. Induction of sulfamethazine acetylation by hydrocortisone in the rabbit. Drug Metab Dispos. 1988;16:110–115. [PubMed] [Google Scholar]
- 39.Glowinski IB, Weber WW. Biochemical characterization of genetically variant aromatic amine N-acetyltransferases in A/J and C57BL/6J mice. J Biol Chem. 1982;257:1431–1437. [PubMed] [Google Scholar]
- 40.Smolen TN, Brewer JA, Weber WW. Testosterone modulation of N-acetylation in mouse kidney. J Pharmacol Exp Ther. 1993;264:854–858. [PubMed] [Google Scholar]
- 41.Estrada L, Kanelakis KC, Levy GN, Weber WW. Tissue- and gender-specific expression of N-acetyltransferase 2 (Nat2*) during development of the outbred mouse strain CD-1. Drug Metab Dispos. 2000;28:139–146. [PubMed] [Google Scholar]
- 42.Evans-Storms RB, Cidlowski JA. Delineation of an antiapoptotic action of glucocorticoids in hepatoma cells: the role of nuclear factor-kappaB. Endocrinology. 2000;141:1854–1862. doi: 10.1210/en.141.5.1854. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, Klaassen CD. Regulation of hepatic sulfotransferases by steroidal chemicals in rats. Drug Metab Dispos. 1996;24:854–858. [PubMed] [Google Scholar]
- 44.Duanmu Z, Locke D, Smigelski J, Wu W, Dahn MS, Falany CN, Kocarek TA, Runge-Morris M. Effects of dexamethasone on aryl (SULT1A1)- and hydroxysteroid (SULT2A1)-sulfotransferase gene expression in primary cultured human hepatocytes. Drug Metab Dispos. 2002;30:997–1004. doi: 10.1124/dmd.30.9.997. [DOI] [PubMed] [Google Scholar]
- 45.Gong H, Jarzynka MJ, Cole TJ, Lee JH, Wada T, Zhang B, Gao J, Song WC, DeFranco DB, Cheng SY, Xie W. Glucocorticoids antagonize estrogens by glucocorticoid receptor-mediated activation of estrogen sulfotransferase. Cancer Res. 2008;68:7386–7393. doi: 10.1158/0008-5472.CAN-08-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boukouvala S, Fakis G. Arylamine N-acetyltransferases: what we learn from genes and genomes. Drug Metab Rev. 2005;37:511–564. doi: 10.1080/03602530500251204. [DOI] [PubMed] [Google Scholar]
- 47.Estrada-Rodgers L, Levy GN, Weber WW. Characterization of a hormone response element in the mouse N-acetyltransferase 2 (Nat2*) promoter. Gene Expr. 1998;7:13–24. [PMC free article] [PubMed] [Google Scholar]