Abstract
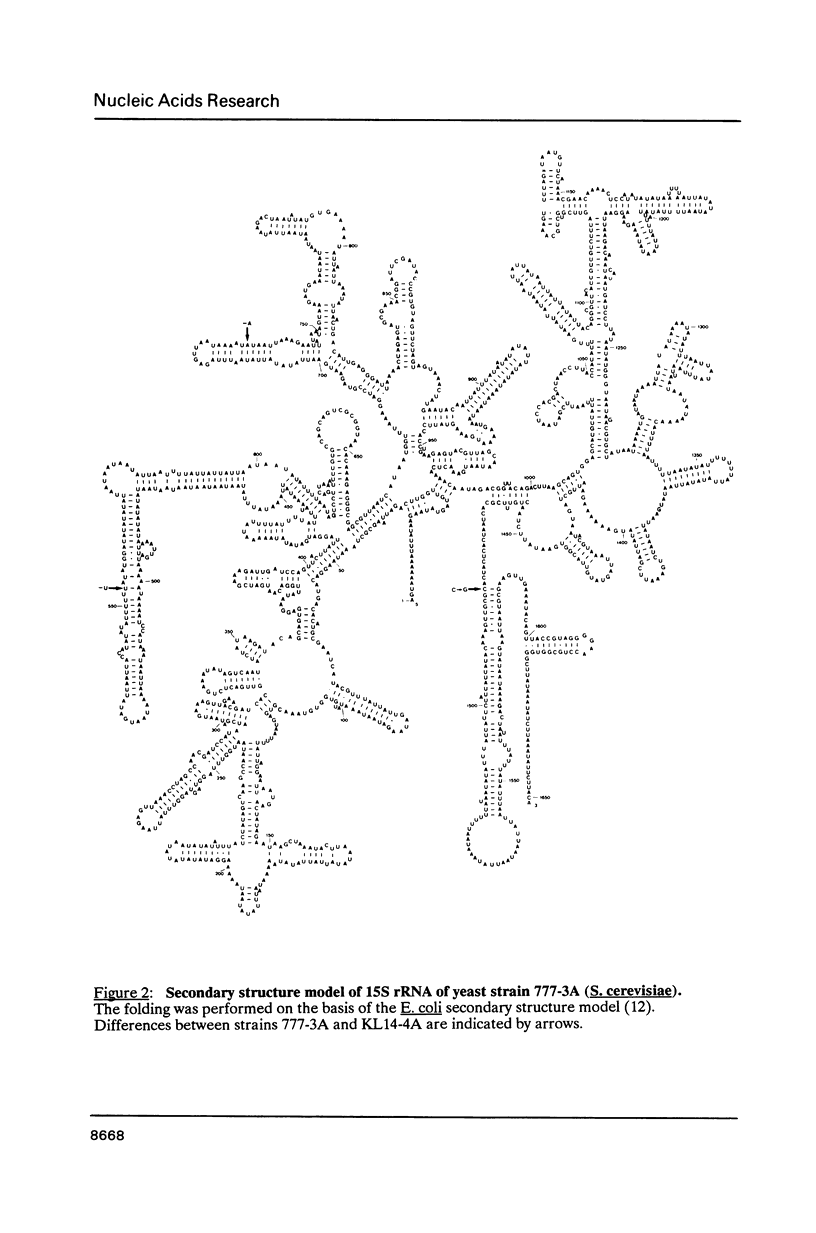
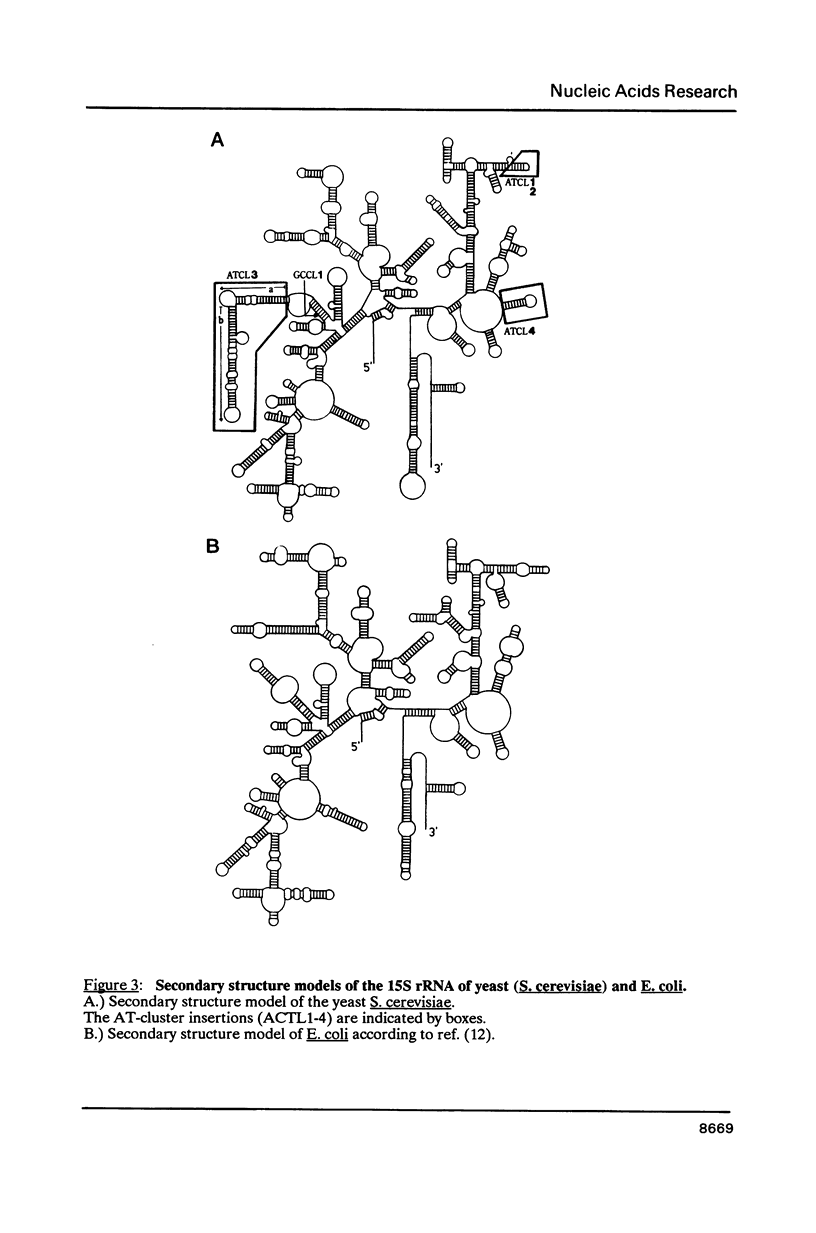
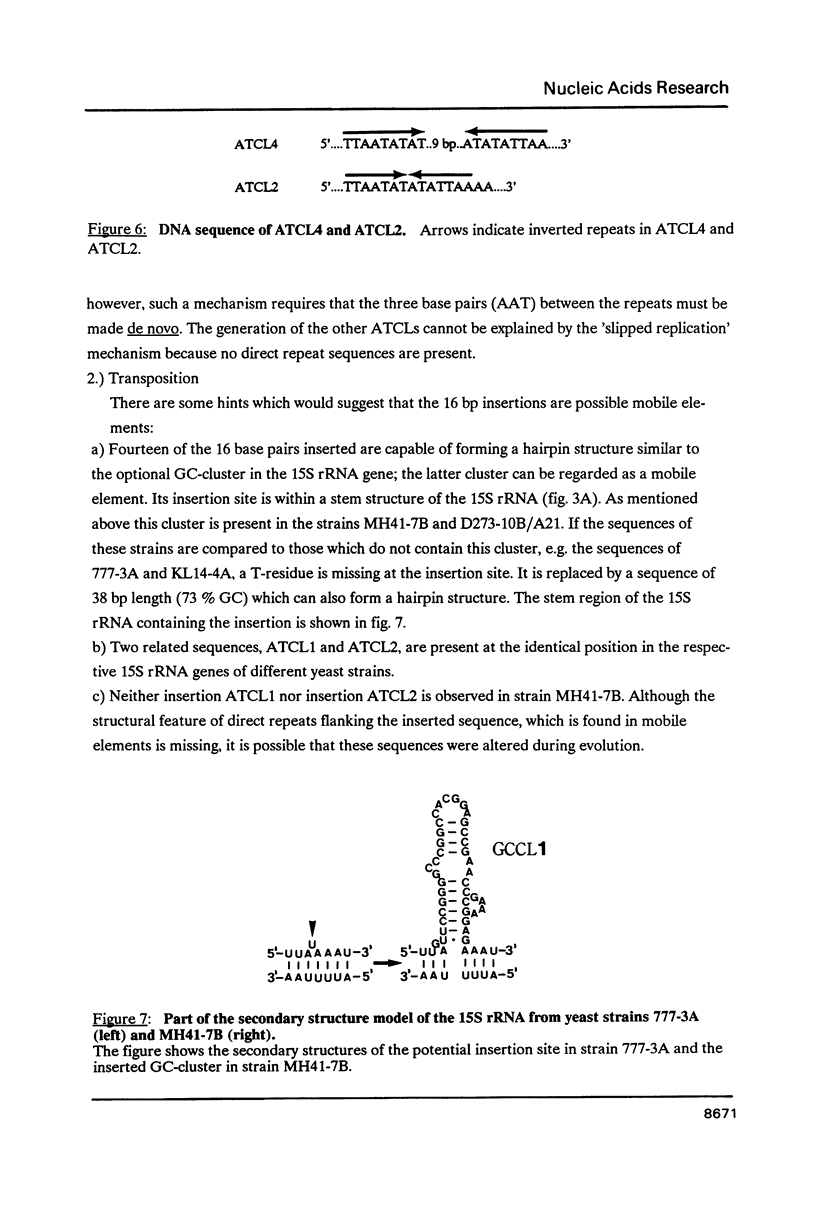
By comparing the mitochondrial 15S rRNA sequences of four wildtype yeast strains together with their respective secondary structures, with those of the 16S-like ribosomal RNA from other organisms we detected two optional and two invariant AT-clusters. The origin of these clusters is discussed with respect to their roles as possible mobile elements.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dobres M., Gerbl-Rieger S., Schmelzer C., Mueller M. W., Schweyen R. J. Deletions in the cob gene of yeast mtDNA and their phenotypic effect. Curr Genet. 1985;10(4):283–290. doi: 10.1007/BF00365624. [DOI] [PubMed] [Google Scholar]
- Fangman W. L., Dujon B. Yeast mitochondrial genomes consisting of only A.T base pairs replicate and exhibit suppressiveness. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7156–7160. doi: 10.1073/pnas.81.22.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans E., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14 (Suppl):r73–118. doi: 10.1093/nar/14.suppl.r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., Underbrink-Lyon K., Martin N. C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982 May 25;257(10):5921–5928. [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Bernardi G. The mitochondrial genome of wild-type yeast cells. IV. Genes and spacers. J Mol Biol. 1974 Jul 15;86(4):825–841. doi: 10.1016/0022-2836(74)90356-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Nature of an inserted sequence in the mitochondrial gene coding for the 15S ribosomal RNA of yeast. Nucleic Acids Res. 1982 Mar 11;10(5):1625–1633. doi: 10.1093/nar/10.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Unequal excision of complementary strands is involved in the generation of palindromic repetitions of rho- mitochondrial DNA in yeast. Cell. 1983 Feb;32(2):391–396. doi: 10.1016/0092-8674(83)90458-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Dujon B., Slonimski P. P. Mitochondrial genetics. V. Multifactorial mitochondrial crosses involving a mutation conferring paromomycin-resistance in Saccharomyces cerevisiae. Mol Gen Genet. 1973 Sep 5;125(1):53–90. doi: 10.1007/BF00292983. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The AT spacers and the var1 genes from the mitochondrial genomes of Saccharomyces cerevisiae and Torulopsis glabrata: evolutionary origin and mechanism of formation. Gene. 1987;54(1):1–22. doi: 10.1016/0378-1119(87)90342-8. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41(1):1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae--a review. Gene. 1986;47(2-3):155–177. doi: 10.1016/0378-1119(86)90060-0. [DOI] [PubMed] [Google Scholar]



