Abstract
The purpose of this study was to characterize and evaluate differences of protamine sulfate, a highly basic peptide drug, obtained from five different sources, using orthogonal thermal and spectroscopic analytical methods. Thermogravimetric analysis and modulated differential scanning calorimetry showed that all five protamine sulfate samples had different moisture contents and glass transition and melting temperatures when temperature was modulated from 25 to 270°C. Protamine sulfate from source III had the highest residual moisture content (4.7 ± 0.2%) at 105°C, resulting in the lowest glass transition (109.7°C) and melting (184.2°C) temperatures compared with the other four sources. By Fourier-transform infrared (FTIR) spectroscopy, the five sources of protamine sulfate had indistinguishable spectra, and the spectra were consistent with a predominantly random coil conformation in solution and a minor population in a β-sheet conformation (~12%). Circular dichroism spectropolarimetry confirmed the FTIR results with prominent minima at 206 nm observed for all five sources. Finally, proton (1H) nuclear magnetic resonance spectroscopy showed that all five protamine sulfate sources had identical spectra with backbone amide chemical shifts between 8.20 and 8.80 ppm, consistent with proteins with predominantly random coil conformation. In conclusion, thermal analyses showed differences in the thermal behavior of the five sources of protamine sulfate, while spectroscopic analyses showed the samples had a predominantly random coil conformation with a small amount of β-sheet present.
KEY WORDS: circular dichroism, differential scanning calorimetry, Fourier-transform infrared spectroscopy, nuclear magnetic resonance, peptide, protamine sulfate, thermogravimetric analysis
INTRODUCTION
Therapeutic proteins are usually derived from exogenous sources such as microbial cells (e.g. E. coli.), plants, or animals, and they play a significant role in pharmacotherapy. Unlike small-molecule drugs, therapeutic proteins can have higher-order structure, resulting from folding of their primary amino acid sequence through numerous relatively weak noncovalent interactions (1). Their manufacturing also involves many steps of extraction, isolation, and purification, which can affect the protein structure. Because the manufacturing processes of proteins are often proprietary, differences may exist between alternate manufacturers for the same therapeutic protein (2). Even small variations during steps of the manufacturing process may result in chemical (such as glycosylation, oxidation, reduction, and hydrolysis) and/or physical (differences in folding, aggregation, precipitation, and denaturation) instability, which could have significant impact on the protein activity (3,4). As a result, heterogeneity may exist among the same proteins from different manufacturers and even between batches from the same manufacturer (4). This heterogeneity could arise from differences in amino acid sequences as a result of one or more amino acid substitutions, deletions, or additions or due to posttranslational modifications such as glycosylation and pegylation (5).
For protamine sulfate, a highly cationic protein drug that is derived from the sperm nuclei of chum salmon fish, heterogeneity in the amino acid sequence has been reported in different samples from different geographical locations. These reported variations in amino acid sequence have been attributed to differences among individual fish from the separate geographical populations, the use of mixtures of ripe and unripe sperm cells, and/or differences in methods of preparation (6). For example, considerable differences in amino acid sequences were reported to exist in protamine obtained from sperm of fish belonging to the Salmonidae family, which includes salmon and rainbow trout fish from Denmark, Japan, and Canada (6–8). Moreover, its commercial extraction and purification from fish milt are very complicated, consisting of at least 18 steps involving the use of organic solvents alcohol, acetone, or acetonitrile and chemicals such as picric acid (9). There is, therefore, the need to effectively characterize protamine sulfate drug substances obtained from different sources to determine any differences since small differences in their structures or properties may affect their clinical effects.
The purpose of this research was to identify any structural similarities or differences in a model protein drug, protamine sulfate, obtained from three different suppliers using thermal and spectroscopic analytical methods.
MATERIALS AND METHODS
Materials
Five protamine sulfate drug substances were obtained from three different suppliers and were labeled as source I through source V. Source I was protamine sulfate (lot no. 010M1273), USP grade, obtained from Sigma-Aldrich (St. Louis, MO). Source II was protamine sulfate (lot no. R24334) obtained from MP Biomedical (Solon, OH). Source III was protamine sulfate (lot no. D00075119) obtained from Calbiochem (Japan). Source IV was protamine sulfate (lot no. 019K1227) grade II obtained from Sigma-Aldrich (St. Louis, MO), and source V was protamine sulfate (lot no. 079K1504) obtained from Sigma-Aldrich (St. Louis, MO). All the drug substances were used as received. All aqueous solutions were prepared with HPLC-ready 18-MΩ water, obtained in-house, from a Milli-Q Gradient A-10 water purification system (Millipore Corp., Bedford, MA).
Thermogravimetric Analysis
Thermogravimetric analysis (TGA) of protamine sulfate samples was carried out on a TA Q5000 instrument (TA Instruments, New Castle, DE). Approximately 5 mg of powdered samples was accurately weighed and placed in aluminum hermetic pans. The pans were sealed to prevent the effect of moisture on the experiments, since protamine sulfate is hygroscopic. The instrument is equipped with a puncher which punched the sealed pan before it was placed in the furnace. The percentage weight loss of the samples was then monitored at the linear heating rate of 10°C/min from 25 to 280°C. Results were analyzed using Universal Analysis 2000 software (TA Instruments, New Castle, DE).
Loss on Drying Experiment
The loss on drying (LOD) of protamine sulfate samples was carried out according to the USP/NF monograph on protamine sulfate drug substance (USP 34/NF 29). Briefly, 500 mg each of the five protamine sulfate drug substances was accurately weighed and evenly spread in a petri dish and placed in an oven. The samples were heated at a temperature of 105°C for 3 h and weighed. The difference in weight before and after heating was determined as the percent moisture loss.
Modulated Differential Scanning Calorimetry
Thermal transitions in the five protamine sulfate samples were monitored using a DSC Q2000 (TA Instruments, New Castle, DE). Temperature calibration of the instrument was performed using indium (melting point = 156.6°C) as a standard reference. To eliminate the effect of moisture on the analysis, aluminum hermetic pans were used. Before modulated differential scanning calorimetry (MDSC) experiments were performed, conventional differential scanning calorimetry was performed to determine the temperature for the various thermal transitions by applying a linear heating temperature of 10°C/min from 25 to 270°C. Once the transitions were observed, MDSC experiments were performed by accurately weighing 10 mg of protamine sulfate drug substance from the five sources and placing in aluminum pans. The pans were hermetically sealed, equilibrated at 25°C for 5 min, and then heated from 25 to 270°C at an underlying heating rate of 4°C/min and a modulation amplitude of +0.636°C every 60 s. A hermetically sealed empty pan was used as a reference. Data obtained were analyzed using TA Universal Analysis 2000 software (TA Instruments, New Castle, DE).
Fourier-Transform Infrared Spectroscopy
All five protamine sulfate samples were dissolved in deionized water. About 100 mg of each sample was accurately weighed and dissolved in 10 mL of deionized water and slowly stirred to yield a final concentration of 10 mg/mL.
All spectral measurements were conducted with a CONFOCHECK™ Tensor 37 Fourier-transform infrared (FTIR) spectrometer (Bruker Optics, Billerica, MA) equipped with OPUS™ software for data analysis. About 25 μL of the aqueous sample was placed in an AquaAspec™ flow through transmission cell equipped with CaF2 windows of 4 mm thickness and an optical path length of 7 μm. All measurements of 25 scans per measurement in the range of 3,000 and 1,000 cm−1 were carried out at a constant temperature of 25°C at a resolution of 4 cm−1 and an acquisition time of 30 s per sample. Quantification of the secondary structure was performed by comparison of the native transmission protein spectra with a protein transmission spectra library using partial least squares algorithm. Bovine serum albumin (BSA) was used as a reference protein in all the analyses.
Circular Dichroism Spectroscopy
Stock solutions of protamine sulfate from the five sources were prepared by dissolving 20 mg of protamine sulfate in 10 mL deionized water by gentle stirring to yield a final concentration of 2 mg/mL. The solutions were then filtered using 0.22-μm low protein-binding filters. Circular dichroism (CD) spectra were collected using an AVIV Model 410 Circular Dichroism Spectrometer (Lakewood, NJ). Protamine sulfate solutions were diluted to a concentration of 1 mg/mL with 10 mM phosphate buffer, pH 7.4, and spectra were recorded at room temperature from 200 to 260 nm in 1-mm quartz cells with 1-nm steps with a 1-s averaging time. Samples were maintained at 25°C. CD spectra were corrected for solvent contributions and expressed in terms of observed ellipticity versus wavelength.
Nuclear Magnetic Resonance Spectroscopy
All the nuclear magnetic resonance (NMR) experiments were performed on an Agilent 500-MHz Direct Drive spectrometer with a cryoprobe. Five-millimeter tubes were used. A 3-mg aliquot of protamine (~0.8 mM in protamine) was dissolved in 630 μL of 100 mM phosphate buffer with a pH of 4.0 ± 0.1. Seventy microliters of D2O was added for the field-frequency lock with 0.6 mM sodium 2,2-dimethylsilapentane-5-sulphonate (DSS) as an internal chemical shift standard. The Agilent “presat” pulse sequence was used to reduce the water signal present in the 90% H2O samples. An acquisition time of 1 s was used with a sweep width of 7,530 Hz and a relaxation delay of 2 s, and 16 scans were co-added for each sample. A 90° pulse width was used with the probe air temperature regulated at 25°C.
RESULTS AND DISCUSSION
Thermal Analyses
Thermal analysis provides insights into the physical behavior and stability of proteins and can be a guide in the development of an optimized formulation. For example, the presence of residual moisture in pharmaceutical protein or peptide drugs could result in water–protein interactions, which could affect the potency of the drug or cause excessive increase in the level of decomposition (10). Residual moisture can act as a reactant or product in the degradation pathways of the peptide, serve as a medium in which degradation occurs, or act as a plasticizer by reducing the glass transition temperature (Tg) and enhancing the mobility of reactants in the solid matrix (11). Residual moisture refers to the low level of surface water, ranging from less than 1 to 5%, remaining in a lyophilized biological product after the bulk of the aqueous solvent has been removed (12).
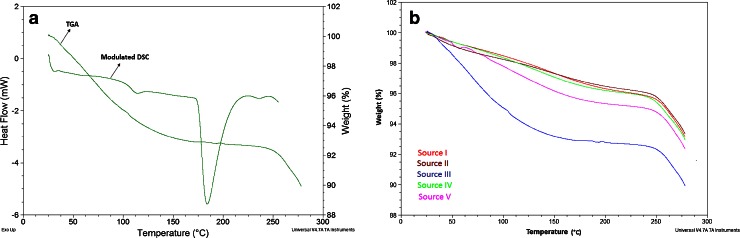
For protamine sulfate, a hygroscopic drug, the USP stipulates that its residual moisture loss upon the application of heat to a temperature of 105°C for 3 h should not be more than 5%. Figure 1a shows an overlay of the TGA and MDSC thermograms for protamine sulfate from source III. All the five samples showed various levels of weight loss between 25 and 270°C (Fig. 1b). Protamine sulfate from source III had the highest moisture loss of 4.95% at 105°C and 7.32 ± 0.24% at 175°C (Table I). At those two temperatures, the weight loss for the other four protamine sulfate samples was less than 4%. At temperatures between 25 and 105°C, the weight loss was attributed to loosely bound water from the hydration region. Above 105°C, the weight loss was attributed to moisture loss from tightly bound water associated with the charged or highly polar groups of the peptide (10), while beyond 240°C, the weight loss was attributed to decomposition of the drug substance (Fig. 1). The moisture loss due to the LOD at 105° was higher than the moisture loss from the TGA experiment at the same temperature (Table I). We attribute this difference to the shorter duration of exposure to the heat of the TGA experiment.
Fig. 1.
a Overlay of TGA and modulated DSC thermograms of protamine sulfate from source III showing weight loss, glass transition, and melting endotherms and b overlay of TGA thermograms for all five protamine sulfate samples showing various levels of weight loss with temperature increase
Table I.
Comparison of Percent Weight Loss of Protamine Sulfate Using TGA and LOD Experiment in an Oven at 105°C
| TGA (105°C) | TGA (175°C) | LOD (105°C) | |
|---|---|---|---|
| Source I | 2.01 ± 0.25 | 3.26 ± 0.14 | 4.15 ± 0.30 |
| Source II | 1.97 ± 0.09 | 3.17 ± 0.10 | 2.51 ± 0.13 |
| Source III | 4.70 ± 0.16 | 7.32 ± 0.24 | 4.89 ± 0.23 |
| Source IV | 1.99 ± 0.25 | 3.47 ± 0.15 | 2.51 ± 0.19 |
| Source V | 2.31 ± 0.15 | 4.38 ± 0.32 | 3.21 ± 0.22 |
Data are representative of three independent experiments
TGA thermogravimetric analysis, LOD loss on drying
MDSC is an extension of conventional DSC, in which a sinusoidal wave modulation is superimposed on a linear temperature program to improve the quality and quantity of information that may be obtained by conventional DSC (13). Generally, the total heat flow from a sample experiencing a linear heating program comprises a heat capacity (reversing) component and contribution from any kinetically hindered (non-reversing) thermal event (14). The ability of MDSC to separate the total heat flow into the reversing and non-reversing components is the result of the different response of the two heat flow signals to the underlying and modulated temperature programs. The application of large modulation amplitudes and low underlying heating rates results in better resolution needed to separate two simultaneous events such as aggregation (exothermic event) and protein unfolding (endothermic event) (15).
When conventional DSC was applied to the protamine sulfate samples at a heating rate of 10°C/min, a broad endothermic peak was observed between 130 and 217°C for all the protamine sulfate samples. The linear heating program of conventional DSC was not able to separate overlapping events that occurred during the heating. However, by using a low underlying heating rate of 4°C/min and modulation amplitude of 0.636°C every 60 s, MDSC was able to separate the overlapping events such as glass transition temperature and melting of the proteins.
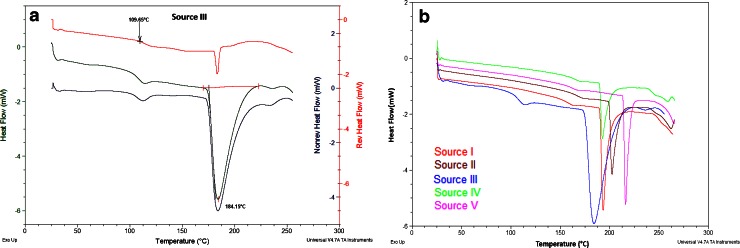
MDSC was able to separate the total heat flow into the reversing and non-reversing flows so that both the heat capacity events and kinetic events could be easily observed (Fig. 2a). A look at the total heat flow for the five protamine sulfate samples (Fig. 2b) revealed a small endothermic event with onset at about 109°C for source III and between 152 and 161°C for the other four samples (Table II). This endothermic event was split into the reversing and non-reversing signals. The endothermic event in the reversing signal could be attributed to the glass transition (Tg), a reversible transition in which an amorphous drug changes its behavior from the glassy state where there is limited molecular mobility to a state with greater molecular mobility as a result of increase in heat. The same endothermic event in the non-reversing signal was due to enthalpic relaxation, a process which is dependent on temperature and time scale of the measurement and is a kinetic event (16). The Tg and enthalpic relaxation for the five protamine sulfate samples are shown in Table II. Whereas protamine sulfate from sources I and II had similar Tg with onset around 161°C, source III had the lowest Tg with onset at about 109°C, followed by source V at 152°C and source IV with Tg at about 158°C. These differences were attributed to differences in the amount of residual moisture present in the samples. For example, the TGA analysis showed that source III had the highest percent weight loss (Fig. 1), and this sample had the lowest Tg and Tm values (Table II). Residual moisture has a plasticizing effect on proteins and acts by reducing the Tg of the protein sample and enhancing molecular mobility in the solid matrix (11). A second prominent endothermic peak was observed for the five samples, which could be attributed to melting (Tm). All the protamine sulfate samples had different melting temperatures with protamine sulfate from source V having the highest melting temperature at 216°C while source III had the lowest melting temperature at 184°C. These differences in thermal properties could be a characteristic of the biological source of the protamine or the process used to manufacture the drug. The range of values observed here may be the normal variation in the currently marketed product.
Fig. 2.
a Deconvoluted heat flow signals for MDSC of protamine sulfate from source III showing the reversing heat signal (top, red), total heat flow (middle, green), and non-reversing heat signal and b total heat flow thermograms for all five protamine sulfate samples showing the differences in their glass transition (T g) and melting (T m) endotherms
Table II.
Comparison of Glass Transition and Melting Temperatures from MDSC Experiments for Protamine Sulfate from Five Different Sources
| T g (°C) | T m (°C) | |
|---|---|---|
| Source I | 161.63 | 193.37 |
| Source II | 161.63 | 202.30 |
| Source III | 109.65 | 184.15 |
| Source IV | 158.58 | 192.66 |
| Source V | 152.05 | 216.11 |
T g glass transition temperature, T m melting temperature
Spectroscopic Analyses
Proteins in solution are frequently composed of a mixture of secondary structural components such as α-helix, β-sheet, and random coil which make up the folded segments of the overall tertiary structure. These secondary structures have characteristic dihedral angles (ϕ, φ) which define the spatial orientation of the peptide backbone and the presence of specific hydrogen bonds (17). For proteins and peptides, of the nine characteristic IR absorption bands, the amide I band is the most widely used in the studies of protein secondary structure. This is because in the amide I region (1,700–1,600 cm−1), which is predominantly (approximately 80%) due to the C=O stretching vibrations coupled with in-plane NH bending (approximately 20%), there is a high sensitivity to small variations in molecular geometry and hydrogen bonding patterns, and the result is that each type of secondary structure gives rise to a somewhat different C=O stretching frequency (18).
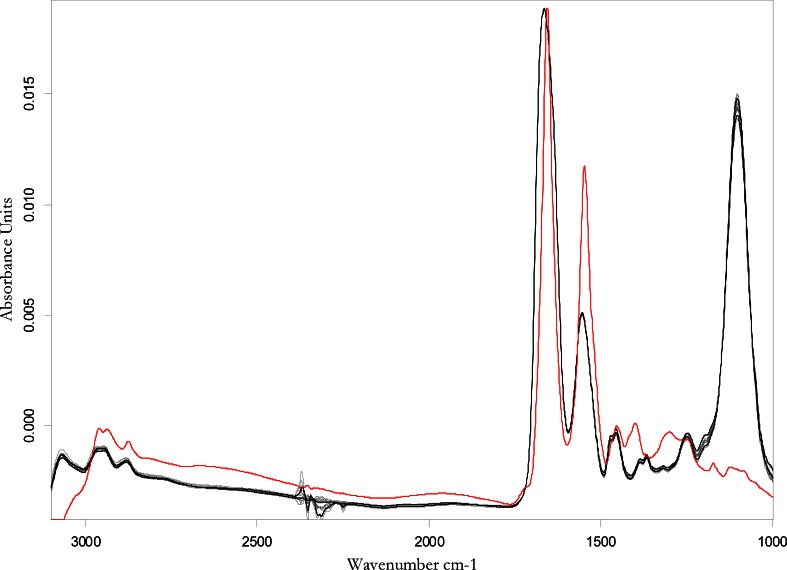
Figure 3 shows the overlaid FTIR spectra of all the protamine sulfate samples, together with the spectrum of the reference protein standard, BSA. Both protamine sulfate and BSA spectra showed prominent bands at 1,600 and 1,550 cm−1 corresponding to amide I and amide II bands, respectively. The only difference between the protamine sulfate spectra and that of BSA is the intense band around 1,100 cm−1 for protamine sulfate which was attributed to arginine that comprises ~67% of the total amino acid content of protamine.
Fig. 3.
Overlaid FTIR spectra of five protamine sulfate samples (black line) and reference standard, BSA (red line)
However, the observed raw spectra cannot be resolved by simple visual inspection because the observed amide I band consists of many overlapping component bands that represent different populations of structural elements such as α-helices, β-sheets, turns, and non-ordered or irregular structures. Thus, identification and assignment of discrete bands to the particular substructures can be difficult (17). Second derivative transformation was therefore applied to the raw spectra to enhance the resolution of the amide I band. For the protamine sulfate samples, two distinct bands were observed at 1,680 and 1,634 cm−1 consistent with the canonical values for a protein with some β-sheet conformation in water (Fig. 4) (18). By contrast, for the reference standard, only one band was observed at 1,656 cm−1 (Fig. 4). Generally, in water, bands between 1,654 and 1,658 cm−1 are assigned to α-helices, as observed for the reference standard, BSA, which has a predominantly α-helix structure (19).
Fig. 4.
Second derivative FTIR spectra of protamine sulfate amide I band show two bands at 1,634 and 1,680 cm−1 while the reference standard BSA (red) shows only one band at 1,656 cm−1
Quantitative analysis of the secondary structure was performed by comparison of the native transmission protein spectra with a protein transmission spectral library using partial least squares algorithms. The β-sheet composition for the five protamine sulfate samples ranged between 11.9 ± 0.1% for source I to 12.3 ± 0.3% for source III (Table III). No α-helical structure was apparent, and the rest of the peptide structure was assigned as random coil. Analysis of variance showed no statistical significance in the percent β-sheet composition of the protamine sulfate samples (p > 0.05). This observation of a predominantly random coil conformation of protamine sulfate in solution was consistent with the observations of other laboratories (20). An alternate metric, the Malhalanobis distance, is a measure of the closeness of the observed value of the percent β-sheet composition to the predicted value from the spectral library. A Malhalanobis distance value of less than the limit of 0.12 for each source of protamine sulfate clearly indicates the presence of a population of β-sheet structures in protamine sulfate solutions.
Table III.
Deconvoluted Amide I Band Assignments of β-Sheet Composition for Protamine Sulfate Samples in Water
| Sample | α-Helix (%) | β-Sheet (%) | Malhalanobis distance | Limit |
|---|---|---|---|---|
| BSA | 59.0 | 1.9 | 0.069 | 0.12 |
| Source I | 0 | 11.85 ± 0.06 | 0.013 ± 0.00 | 0.12 |
| Source II | 0 | 12.20 ± 0.46 | 0.012 ± 0.001 | 0.12 |
| Source III | 0 | 12.28 ± 0.33 | 0.012 ± 0.001 | 0.12 |
| Source IV | 0 | 12.15 ± 0.30 | 0.012 ± 0.001 | 0.12 |
| Source V | 0 | 12.10 ± 0.41 | 0.012 ± 0.001 | 0.12 |
BSA was used as a reference standard. Data are representative of three independent experiments for all protamine sulfate samples (n = 3)
BSA bovine serum albumin
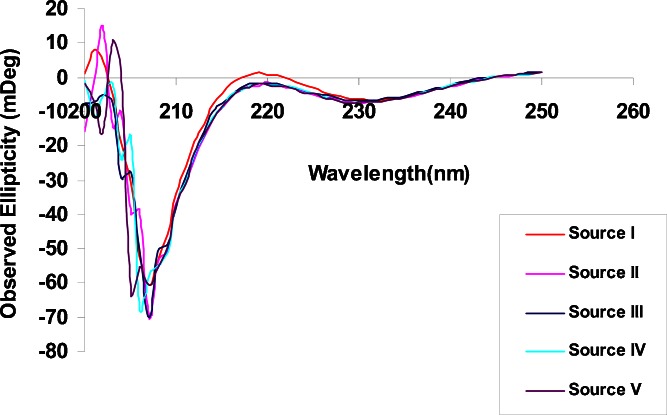
The secondary structure of the protamine samples was further examined by CD. CD measures the differential absorption of left-handed and right-handed circularly polarized light. In a typical CD spectrum, a negative signal or band means greater absorption of left circularly polarized light while a positive band denotes greater absorption of right circularly polarized light (21). Because protamine sulfate has no aromatic amino acids in its sequence, all measurements were carried out in the range of 200 to 250 nm, where the amide chromophores of the peptide bonds predominate (22). Figure 5 shows the overlaid CD spectra of the protamine sulfate samples. For all five samples, the most predominant peak was a strong negative band observed around 206 nm. The peak at 206 nm is consistent with a mixture of predominantly random coil peptides with a smaller population of peptides with a β-sheet conformation in solution. Proteins with substantial α-helical structure show negative bands at 222 and 208 nm while proteins with substantial β-sheets display a negative band near 216 nm and a positive band between 195 and 200 nm. Proteins with random coil conformation also show a strong negative band at 200 nm (22,23). The fact that there was no negative band at or close to 220 nm effectively rules out the presence of α-helix in the secondary structure of protamine sulfate under the solution conditions used in this study. These data are consistent with our FTIR results where no α-helical bands in protamine sulfate spectra were observed. The CD measurements were scanned in the 200- to 250-nm range because of noise in the region at 200 nm and below where the detector became saturated due to salts in the sample.
Fig. 5.
Overlaid CD spectra for the five protamine sulfate samples. The overlaid spectra show a negative band at 206 nm which is in agreement with a predominantly random coil structure in solution with a minor contribution from a population in a β-sheet conformation
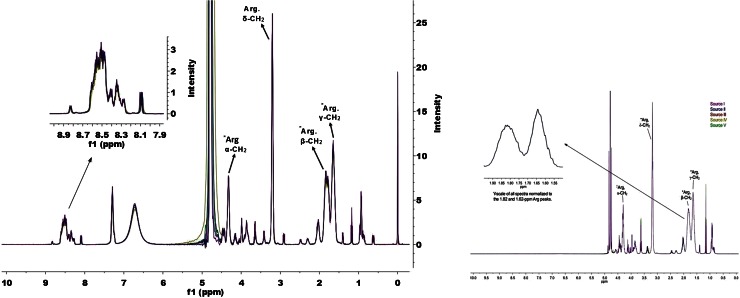
One-dimensional proton (1H) NMR spectroscopy was also used to study the structure of the five protamine sulfate samples. Overlaid spectra of the five protamine sulfate samples showed identical patterns of NMR signal over the entire chemical shift range (Fig. 6). The sharp singlet at 0 ppm is the chemical shift reference standard, DSS, and the signal at ~4.77 ppm is the residual water signal after presaturation was applied. Prominent signals from the arginine amino acids are observed (denoted in Fig. 6).
Fig. 6.
Overlaid spectra of the 0- to 10-ppm portion of the 500-MHz 1H-NMR of the five protamine sulfate samples. The region between 8.1 and 8.8 ppm shows bands for the backbone amide bonds which are typical for proteins with a predominantly random coil conformation. Peak assignment of arginine residues is based on previously reported values (29)
The range of amide or CαH backbone proton chemical shifts is very sensitive to differences in amino acid sequence, secondary or tertiary structure, and solution conditions (24,25). Figure 6 shows an indistinguishable pattern of backbone amide proton signals between 8.10 and 8.80 ppm for all five protamine sulfate samples. This chemical shift range observed for the backbone amide protons (see insert in Fig. 6) is consistent with a predominately random coil conformation for protamine at pH 4 in solution (24,26). Thus, NMR spectroscopy further confirmed the results obtained in FTIR and CD spectroscopy that all the five protamine sulfate lots have very similar amino acid composition and are predominantly random coil conformation in solution.
Peptide Analysis
Protamine sulfate from all the different vendors were analyzed by a validated HPLC procedure (27), and there were no observable differences in the peptide abundance for any of the sources. Also, there were no significant differences in the impurity profiles of all the sources of the drug substance studied. Also, another study performed by Hoffman et al. (28) showed the amino acid sequence for four peptides of protamine sulfate from chum salmon source. The protamine sulfate in our study was obtained from the same biological source, and the profile of the four peptide peaks from that study (28) matched with our peak profiles (27). Therefore, further amino acid sequencing was not conducted in this study.
CONCLUSION
Thermal and spectroscopic analytical techniques were successfully applied to determine differences in protamine sulfate obtained from different sources. The five protamine sulfate samples showed some differences in their thermal properties such as moisture contents, glass transition, and melting temperatures. Protamine sulfate from source III had the highest moisture content among the five protamine sulfate sources. This high moisture content resulted in low glass transition and melting temperatures for source III compared with the other sources. Spectroscopic analytical techniques such as FTIR, CD spectroscopy, and NMR all showed that the five protamine sulfate samples existed in solution as predominantly random coil conformation with no alpha-helical structures and small amount of β-sheet conformation.
For a protein drug such as protamine sulfate, it is not known how such differences in their thermal characteristics could affect properties of the drug such as shelf-life stability, clinical efficacy, and product safety, including immunogenicity. To the best of our knowledge, this is the first study to systematically characterize the solution conformation of protamine sulfate from different manufacturers. These findings could be important in establishing guidelines for assessing similar protein drugs from different manufacturers.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Matthew Weinstock from the University of Utah School of Medicine for help with CD spectroscopy studies, Dr. Cruz Hinojos from Bruker Optics, Texas, for help with FTIR spectroscopy studies, and Dr. Robert Berendt from DPQR, FDA, for his helpful insight into NMR.
Disclaimer
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
REFERENCES
- 1.Manning MC, Patel K, Borchardt RT. Stability of protein pharmaceuticals. Pharm Res. 1989;6:903–918. doi: 10.1023/A:1015929109894. [DOI] [PubMed] [Google Scholar]
- 2.Hincal F. An introduction to safety issues in biosimilars/follow-on biopharmaceuticals. J Med CBR Def. 2009;7.
- 3.Crommelin DJA, Storm G, Verrijk R, de Leede L, Jiskoot W, Hennink WE. Shifting paradigms: biopharmaceuticals versus low molecular weight drugs. Int J Pharm. 2003;266:3–16. doi: 10.1016/S0378-5173(03)00376-4. [DOI] [PubMed] [Google Scholar]
- 4.Chirino AJ, Mire-Sluis A. Characterizing biological products and assessing comparability following manufacturing changes. Nat Biotechnol. 2004;22:1383–1391. doi: 10.1038/nbt1030. [DOI] [PubMed] [Google Scholar]
- 5.Ward WW, Swiatek G. Protein purification. Curr Anal Chem. 2009;5(2):85–105. doi: 10.2174/157341109787846171. [DOI] [Google Scholar]
- 6.McKay DJ, Renaux BS, Dixon GH. Rainbow trout protamines. Eur J Biochem. 1986;158:361–366. doi: 10.1111/j.1432-1033.1986.tb09759.x. [DOI] [PubMed] [Google Scholar]
- 7.Sakai M, Fujii-Kuriyama Y, Saito T, Muramatsu M. Closely related mRNA sequences of protamines in rainbow trout testis. J Biochem. 1981;89:1863–1868. doi: 10.1093/oxfordjournals.jbchem.a133388. [DOI] [PubMed] [Google Scholar]
- 8.States JC, Connor W, Wosnick MA, Aiken JM, Gedamu L, Dixon GH. Nucleotide sequence of a protamine component CII gene of Salmo gairdnerii. Nucleic Acids Res. 1982;10:4551–4563. doi: 10.1093/nar/10.15.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill TA, Singer DS, Thompson JW. Purification and analysis of protamine. Process Biochem. 2006;41:1875–1882. doi: 10.1016/j.procbio.2006.04.001. [DOI] [Google Scholar]
- 10.Towns JK. Moisture content in proteins: its effects and measurement. J Chromatogr A. 1995;705:115–127. doi: 10.1016/0021-9673(94)01257-F. [DOI] [PubMed] [Google Scholar]
- 11.Shalaev EY, Zografi G. How does residual water affect the solid-state degradation of drugs in the amorphous state? J Pharm Sci. 1996;85:1137–1141. doi: 10.1021/js960257o. [DOI] [PubMed] [Google Scholar]
- 12.Center for Biologics Evaluation and Research (CBER). Guideline for the determination of residual moisture in dried biological products. Food and Drug Administration, 1990. At: http://www.fda.gov/ohrms/dockets/dockets/05d0047/05d-0047-bkg0001-Tab-11.pdf. Accessed September 10, 2011.
- 13.Coleman NJ, Craig DQM. Modulated temperature differential scanning calorimetry: a novel approach to pharmaceutical thermal analysis. Int J Pharm. 1996;135:13–29. doi: 10.1016/0378-5173(95)04463-9. [DOI] [Google Scholar]
- 14.Hill VL, Craig DQM, Feely LC. Characterisation of spray-dried lactose using modulated differential scanning calorimetry. Int J Pharm. 1998;161:95–107. doi: 10.1016/S0378-5173(97)00334-7. [DOI] [Google Scholar]
- 15.Badkar A, Yohannes P, Banga A. Application of TZERO calibrated modulated temperature differential scanning calorimetry to characterize model protein formulations. Int J Pharm. 2006;309:146–156. doi: 10.1016/j.ijpharm.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Royall PG, Craig DQM, Doherty C. Characterisation of the glass transition of an amorphous drug using modulated DSC. Pharm Res. 1998;15:1117–1121. doi: 10.1023/A:1011902816175. [DOI] [PubMed] [Google Scholar]
- 17.Pelton JT, McLean RL. Spectroscopic methods for analysis of protein secondary structure. Anal Biochem. 2000;277:167–176. doi: 10.1006/abio.1999.4320. [DOI] [PubMed] [Google Scholar]
- 18.Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin. 2007;39:549–559. doi: 10.1111/j.1745-7270.2007.00320.x. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama T, Katoh S, Nakajima M, Nabetani H, Abbott TP, Shono A, et al. FT-IR analysis of BSA fouled on ultrafiltration and microfiltration membranes. J Membr Sci. 2001;192:201–207. doi: 10.1016/S0376-7388(01)00502-6. [DOI] [Google Scholar]
- 20.Balhorn R. The protamine family of nuclear proteins. Genome Biol. 2007;8(9):227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correa DHA, Ramos CHI. The use of circular dichroism spectroscopy to study protein folding, form and function. Afr J Biochem Res. 2009;3(5):164–173. [Google Scholar]
- 22.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta Protein Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2007;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wuthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. p. 127. [Google Scholar]
- 25.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 26.Mielke SP, Krishnan VV. Characterization of protein secondary structure from NMR chemical shifts. Prog Nucl Magn Reson Spectrosc. 2009;54(3–4):141–165. doi: 10.1016/j.pnmrs.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awotwe-Otoo D, Agarabi C, Faustino PJ, Habib MJ, Lee S, Khan MA, et al. Application of quality by design elements for the development and optimization of an analytical method for protamine sulfate. J Pharm Biomed Anal. 2012;62:61–67. doi: 10.1016/j.jpba.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann JA, Chance RE, Johnson MG. Purification and analysis of the major components of chum salmon protamine contained in insulin formulations using high-performance liquid chromatography. Protein Expr Purif. 1990;1(2):127–133. doi: 10.1016/1046-5928(90)90005-J. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes SD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]