Abstract
Malignant brain tumors, including high-grade gliomas, are among the most lethal of all cancers. Despite considerable advances, including multi-modal treatments with surgery, radiotherapy, and chemotherapy, the overall prognosis remains dismal for patients diagnosed with these tumors. With the discovery of RNA interference (RNAi) for target-specific gene silencing via small interfering RNA (siRNA), a novel method to target malignant gliomas has been exposed, an endeavor that is aggressively being carried out in numerous laboratories. However, practical difficulties in tissue- or organ-specific targeting of therapeutic quantities of siRNA still preclude its applicability in a clinical setting. MicroRNA (miRNA), an endogenously expressed form of siRNA, not only presents an alternate method to induce RNAi in a given diseased tissue or organ, but also exposes a unique set of diagnostic markers that can be used to identify, and then differentiate between tumor grades. Thus, miRNA can be considered the cells' answer to siRNA. Discovered over a decade ago, miRNA is fast becoming recognized as crucial in regulating gene expression in cancers. Therein lies the therapeutic potential of miRNA, as it may now be possible to induce or inhibit RNAi in a given diseased cell population by controlling the cells' miRNA expression profile. This review outlines the potential of miRNA as a therapeutic strategy against high-grade gliomas, and also the technological hurdles that need to be addressed before this promising technique can be administered in a clinical setting.
INTRODUCTION
Tumors that originate in the brain are classified as primary brain tumors, which can be either benign or malignant. Gliomas are tumors of neuro-epithelial origin and account for almost half of all primary brain tumors. The World Health Organization categorizes gliomas as either low-grade (WHO grades I and II: pilocytic astrocytoma and diffuse astrocytoma, respectively) or high-grade (WHO grades III and IV: anaplastic astrocytoma and glioblastoma multiforme [GBMs], respectively) (Kleihues and Cavanee, 2000). Gliomas are the most common of all solid tumors, and the second most frequent neoplasm in children (American Brain Tumor Association (ABTA), 2006). In fact, brain tumors are the second leading cause of cancer-related death among children under age 20, the leading cause of cancer-related death among males aged 20–39, and the fifth leading cause of cancer-related death among females in the same age group (ABTA, 2006).
Nearly 50,000 new cases of primary brain and central nervous system (CNS) tumors were expected to be diagnosed in the United States in 2005 (Central Brain Tumor Registry of the United States, 2006), with an incidence rate of 7.4 cases of malignant brain tumors per 100,000 person-years. The global incidence rate is 3.7 and 2.6 cases per 100,000 person-years for males and females, respectively (International Agency for Research on Cancer, 2006).
Metastatic brain tumors are malignant neoplasms that spread to the brain from elsewhere in the body, and represent the most common neurologic manifestation of cancer, occurring in up to 15% of cancer patients. In fact, brain metastases are the most common intracranial tumor in adults, accounting for approximately 40% of intracranial neoplasms. With improved survival of cancer patients, the incidence of brain metastases has been rising. Studies involving multiple, large autopsy series suggest that, in order of decreasing frequency, lung, breast, melanoma, renal, and colon cancers are the most common primary tumors to metastasize to the brain. Altogether 190,000 patients will develop brain metastases in the United States annually (National Brain Tumor Foundation, 2006). Typically, the prognosis of cancer patients with metastatic spread to the brain is also poor.
BRAIN TUMORS
The mammalian brain is largely composed of two principal cell types: neurons and glial cells. While neurons are involved in signal transmission, glial cells form the major constituents of the CNS, where they outnumber neurons by almost 10-fold (Williams and Herrup, 1988). Glial cells play a supportive role that helps delineate synaptic contacts and maintain the signaling ability of neurons. Subtypes of glial cells, commonly referred to as neuroglia or glia, include astrocytes, oligodendrocytes, and microglia. Astrocytes provide metabolites, structural support, and insulation for nerve cells. The primary function of oligodendrocytes is the myelination of neuronal axons. Microglial cells are specialized macrophages capable of phagocytosis. Ependymal cells, also named ependymocytes, line the cavities (ventricles) of the CNS and help circulate the cerebrospinal fluid by ciliary action.
Approximately half of all primary brain tumors arise from glial cells, which are collectively named as gliomas. Gliomas are named according to the specific type of cell they most resemble. Thus, astrocytomas are derived from astrocytes, oligodendrogliomas develop from oligodendrocytes, and ependymomas originate from ependymal cells. Mixed gliomas, such as oligoastrocytomas, contain a mixture of cells derived from both oligodendrocytes and astrocytes. Benign intracranial tumors arise mainly from the meninges (meningiomas), pituitary gland (pituitary adenomas), and the myelin sheath of cranial nerves (neuromas or Schwannomas).
Other types of brain tumors include choroids plexus tumors, primitive neuroectodermal tumors (PNET, e.g., medulloblastoma, neuroblastoma, retinoblastoma, pineoblastoma), tumors originating from neuronal cells (gangliocytoma, central neurocytoma), mixed glio-neuronal tumors (tumors displaying a neuronal as well as a glial component, such as ganglioglioma), and dysembryoplastic neuroepithelial tumors (DNET).
It is the astrocytic tumors that develop from astrocytes or astrocytic progenitors (Ignatova et al., 2002; Hemmati et al., 2003; Singh et al., 2003) which comprise a wide range of neoplasms that differ in their location within the CNS, age and gender distribution, growth potential, extent of invasiveness, morphological features, tendency for progression, and clinical course. There is increasing evidence that these differences reflect the type and sequence of genetic alterations acquired during the process of neoplastic transformation. Inherently, diffuse astrocytomas have a tendency for malignant progression, with the GBMs as the most aggressive phenotypic endpoint. Glioblastoma may manifest at any age, but preferentially affect adults, with a peak incidence between 45 and 70 years. GBMs are among the most malignant of human neoplasms with an average survival of less than 12 months from the time of diagnosis. Despite considerable advances in brain cancer therapy, including surgical intervention followed by adjuvant radiotherapy and/or chemotherapy, prognosis has not significantly improved for these patients. Thus, new strategies need to be tested and evaluated to complement the current standard therapies in order to improve the clinical outcome. It is with this goal that RNA interference (RNAi) is being actively studied in a preclinical setting.
FROM “JUNK-RNA” TO miRNA
Isolation and fractionation of RNA from any tissue invariably results in a minute fraction of very low molecular weight (<200 nucleotides) RNA. Long discarded as an unfortunate by-product due to the inherent instability of RNA during the purification process, this “RNA-waste” has now been shown to harbor microRNA (miRNA), a crucial component of posttranscriptional gene regulation.
Equally unexpected was the fact that these small noncoding RNAs were generated in vivo within chromosome regions which, until recently, had been relegated to the position of “junk-DNA,” that is, the introns within protein-coding genes and the so-called “intergenic” regions. However, it should be noted that miRNAs are themselves encoded by distinct transcription units (i.e., genes) (Nelson et al., 2003; Kim and Nam, 2006), despite being noncoding in nature.
The present review will first address the latest preclinical studies on miRNA and then discuss the potential clinical benefits of using miRNA to improve the diagnostic/prognostic and therapeutic strategies against malignant brain tumors, with emphasis on GBMs, one of the most malignant and lethal of all cancers.
BACKGROUND
The first hint of the presence of endogenously transcribed and processed RNA that regulates global gene expression in an organism came through studies involving the nematode Caenorhabditis elegans (Lee et al., 1993; Nelson et al., 2003). Named lin-4 (abnormal cell lineage-4), this very first miRNA moiety [then denoted as stRNA or small temporally expressed RNA (Reinhart et al., 2000)] was identified due to its ability to base-pair with mRNA for lin-14, a protein important in post-embryonic development. In C. elegans, base pairing between lin-4 and lin-14 resulted in translational repression, thus identifying a mode of posttranscriptional gene silencing that differed from the mechanism for small interfering RNA (siRNA), which activates a cascade of events that result in cleavage of target mRNA.
Thus, miRNAs belong to a family of small noncoding RNA molecules (~21 nucleotides long) that mediate expression of target genes by base pairing with complementary regions within target mRNA, which then either results in inhibition of translation or causes direct destruction of the mRNA template. The latter is thought to closely parallel the mechanism for RNAi via siRNA. The fate of mRNA template is currently thought to be decided based on the pairing between miRNA and the target mRNA template, with a perfect match targeting the mRNA toward “siRNA-type” targeted destruction, while mismatches create conditions for inhibition of translation (Nelson et al., 2003; Krol and Krzyzosiak, 2004; Meltzer, 2005).
Therefore, miRNAs differ from siRNAs in several important aspects. While the size of each moiety is approximately similar, miRNAs have an endogenous origin, such that they are derived from within distinct transcription units in the genome itself. Studies to date indicate that miRNAs can be transcribed from (a) intergenic regions (where, in the classical sense, a gene refers to a transcription unit that encodes a protein), (b) introns, or (c) from the 5′ or 3′ untranslated regions of a given coding gene (Fig. 1).
FIG. 1.
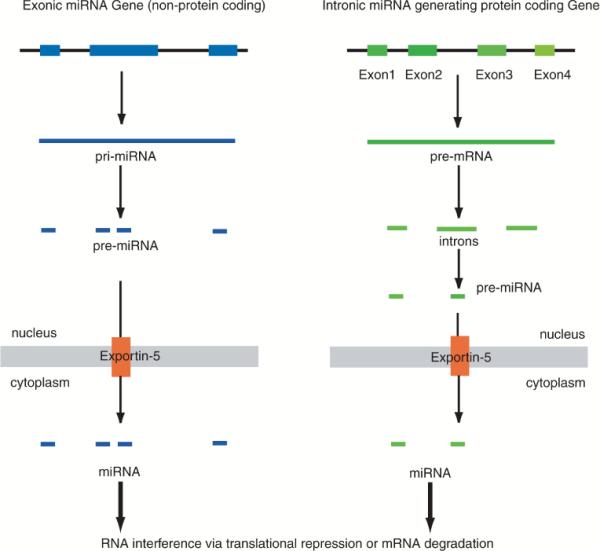
Biogenesis of miRNA. Current research indicates two schemes for biosynthesis of miRNA: via nonprotein-coding “transcription units” (termed exonic-miRNA genes), and those encoded within introns of protein-coding genes (termed intronic miRNA). Each exonic miRNA gene, or a given intron spliced from pre-mRNA, can give rise to multiple miRNAs, each targeting a different set of mRNA transcripts.
In addition, a microarray profiling survey on the expression patterns of 175 human miRNAs across 24 different human organs has shown that proximal pairs of miRNAs are in general co-expressed, with miRNAs that are separated by < 50 kb in the human genome typically being derived from a common transcript (Baskerville and Bartel, 2005). miRNAs that were expressed within introns were usually coordinately expressed with their host gene mRNA, indicating that they also were derived from introns within the same precursor mRNA (pre-mRNA) transcript.
In contrast, siRNAs of therapeutic utility have exogenous origins (although they are also believed to be synthesized in vivo upon viral infection), where (a) they are directly introduced into cells as ~21-bp dsRNA molecules, or (b) they are generated in vivo from introduced expression vector systems, which utilize RNA polymerase III–type promoters (H1 promoter or U6 promoter is commonly used) to synthesize the siRNAs in vivo. In the latter case, siRNAs transcribed from expression vectors are denoted as shRNAs (short-hairpin RNAs), where both sense and anti-sense templates of siRNAs are transcribed as one continuous RNA strand, which then folds in vivo into the double-stranded “active” form.
The readers are also directed to several excellent reviews on the cellular machinery and the cascade of events that are involved in processing of siRNAs, their targeting to specific mRNA species, and the degradation of target mRNA (Mittal, 2004; Dykxhoorn and Lieberman, 2005; Filipowicz, 2005; Filipowicz et al., 2005).
miRNA NOMENCLATURE
Since their initial discovery in C. elegans, several hundred miRNA moieties have been putatively identified among vertebrates, invertebrates, and plants by in silico screening methods (Ambros et al., 2003; Berezikov et al., 2006a; Griffiths-Jones et al., 2006). Even, viral genomes have yielded putative miRNA species. However, most remain to be verified as functional genes that are capable of generating miRNA in vivo.
With the rapid expansion in the number of miRNAs that have been mined both in silico and in vivo, a uniform miRNA nomenclature scheme has been adopted with particular emphasis on functional verification prior to enumeration (Ambros et al., 2003; Griffiths-Jones, 2006; Griffiths-Jones et al., 2006). Thus, experimentally proven novel miRNA moieties are assigned numerical identifiers along with a three- or four-letter abbreviated prefix for species designation. The precursor miRNAs (pre-miRNAs) are given the designation mir, while the mature miRNAs are denoted as miR. Thus, a fully processed human (Homo sapien) miRNA sequence will be designated hsa-miR. Mature miRNAs where the sequence differs at one or two positions are given lower-case-letter suffixes (e.g., miR-1a and miR-1b). Pre-miRNAs from different genomic loci that give rise to identical mature miRNAs are given numerical suffixes (e.g., miR-1-1 and miR-1-2). More complex nomenclature patterns also exist based on the organism and the mode of processing the pre-miRNA (Griffiths-Jones et al., 2006). In order to accommodate the growing populations of miRNAs from different species, a database registry of miRNAs has been formulated (miRBase), which is described in detail elsewhere and online (Griffiths-Jones, 2006).
ALGORITHMS FOR miRNA IDENTIFICATION IN SILICO
To circumvent the difficulties in experimental verification of miRNAs or construction and screening of miRNA libraries, numerous in silico methods for “miRNA mining” off organismal genomes have been put forward (Rajewsky, 2006). However, such prediction methods are of limited utility until detailed experimental verification can be accomplished. One avenue of investigation to improve the “target hit-rate” has been to correlate mRNA levels with the corresponding miRNA expression patterns (Legendre et al., 2005; Wang et al., 2005). However, due to the propensity of miRNAs to either inhibit translation or degrade the target mRNA, predictions from such correlations are likely to be fraught with errors. Such hurdles remain to be overcome, and the readers are referred to a recent review for a more in depth discussion (Lai, 2004).
RNA POLYMERASE II AS THE ENGINE FOR miRNA?
A question arises whether miRNAs are also transcribed via RNA polymerase III, in a mechanism similar to that utilized for siRNAs. Since mature (processed) miRNAs were similar in size (~21–25 nucleotides) to siRNAs, RNA polymerase III was inferred to be responsible for miRNA transcription.
The first characterized miRNAs were located within non-coding transcription units that were located between known protein-coding genes. Thus, these miRNAs were labeled as intergenic miRNAs. Subsequently, numerous miRNAs were discovered within introns of known protein-coding genes (Nelson et al., 2003; Kim and Nam, 2006; Lin et al., 2006). The latter were known as intronic miRNAs. These are commonly spliced off from the pre-mRNA transcript. In addition, certain sets of miRNAs were identified encoded within polycistronic transcription units, which enabled them to functionally coordinate their RNAi. Yet other miRNAs were expressed in a tissue-specific or development-specific manner.
The primary transcript of intergenic miRNA, known as pri-miRNA, can sometimes be 1 kb or longer. These pri-miRNAs are now known to contain cap structures and poly(A) tails, in a phenotype similar to mRNAs (Cai et al., 2004; Lee et al., 2004; Du and Zamore, 2005). Thus, evidence directs toward RNA Polymerase II–initiated transcription for intergenic miRNA.
However, even in the studies that characterized RNA polymerase II–driven transcription for intergenic miRNA, a considerable population of pri-miRNAs was identified that lacked the cap structure or the poly(A) tails (Lee et al., 2004). Thus, the possibility for involvement of alternate RNA polymerases in miRNA biogenesis still remains open.
Compounding the above problems is a serious lack of RNA polymerase II–type consensus sequences within the putative promoters identified thus far, for the known miRNA transcription units (Lee et al., 2004). Although several cis-regulatory elements have been putatively identified, lack of well-known transcription initiation elements (e.g., TATAA and CAAT), except for the presence of CG islands, points to the possibility that the miRNAs are perhaps transcribed from a type of TATA-independent promoter.
miRNA BIOGENESIS: THE CURRENT MODEL IN BRIEF
As endogenous gene products, all miRNAs [except for putative miRNAs of mitochondrial origin (Maniataki and Mourelatos, 2005)] originate in the nucleus. In mammals, a primary “precursor” miRNA (termed pri-miRNA) is first transcribed off noncoding transcription units, or spliced off from introns of pre-mRNA. Pri-miRNA is then processed by Drosha-type endonucleases. The processed miRNA (now termed pre-miRNA) is then exported from the nucleus via the receptor Exportin-5 (Fig. 1). Once in the cytoplasm, pre-miRNA is cleaved by Dicer-type nucleases to form mature, fully processed miRNA. It is then incorporated into the RNA-induced gene-silencing complex, which initiates RNAi in a manner similar to that for siRNA. Numerous reviews are available with detailed diagrammatic representations of the above scheme, and the readers are referred to them for further information (Du and Zamore, 2005; Ying and Lin, 2005; Lin et al., 2006; Tsuchiya et al., 2006).
SCREENING AND IDENTIFICATION OF NOVEL miRNA
Prior to the discovery of miRNA, RNA isolation techniques were optimized toward removal of small RNA (~200 bases or less) from a given RNA population, with the aim of purifying “intact” mRNA. The same protocols have now been modified to accommodate the retention and then purification of RNA in the size range of 10–200 bases. Currently, the first step in generating an “miRNA library” for screening purposes involves the isolation of miRNA, and then the isolated miRNA pool is size fractionated via polyacrylamide gel-electrophoresis (PAGE). Subsequently, small oligonucleotide adapters are ligated to miRNA ends, followed by RT-PCR amplification to generate sufficient template for cloning purposes (Berezikov et al., 2006a). The clone library is then sequenced to identify potential miRNA. Detailed protocols are available on numerous websites maintained by laboratories that work on miRNA, and by commercial sources (Ambion, Austin, TX, and Invitrogen, Rockville, MD, among others).
miRNAS FUNCTION AS BOTH ONCOGENES AND TUMOR SUPPRESSORS
Recent reports indicate that the biogenesis of specific miRNA moieties is tightly regulated. In turn, miRNAs regulate downstream gene (mRNA) expression, in parallel with cell differentiation and development (Pillai, 2005). Thus, there is high likelihood that disturbances in a cell's miRNA profile would lead to oncogenesis. In fact, this has been borne out recently in numerous studies. mir-155, one of the well-characterized miRNAs, is highly expressed in B-cell lymphomas. Transgenic mice that express mir-155 develop B-cell malignancy at an early stage mir-155 is now known to upregulate expression of Myc, a key gene that can lead to tumor promotion when aberrantly expressed [reviewed in Esquela-Kerscher and Slack (2006) and Calin and Croce (2006a)]. Another observation is that most annotated miRNAs can be mapped to fragile sites on the human genome, where a greater degree of genomic instability exists (Calin et al., 2004).
Thus, a stable miRNA profile may act as a global tumor suppressor repertoire within a given cell population. Support for this hypothesis came from a study involving B-cell leukemia (chronic lymphocytic leukemia), which often harbors deletions in two clustered miRNAs, mir-15a and mir-16-1 (Calin et al., 2002). These two miRNAs have been shown to downregulate Bcl-2 expression, a known anti-apoptotic gene, at a posttranscriptional level, and this repression is sufficient to induce apoptosis (Cimmino et al., 2005). Other key finding has been the discovery that human homologs of C. elegans let-7 family of miRNA regulate expression of Ras, of which mutations are common across many types of cancers (Esquela-Kerscher and Slack, 2006). Here as well, the respective miRNAs are located in fragile chromosomal sites of these malignancies (Calin et al., 2004).
The evidence outlined above points to two key facts: deletions in fragile chromosomal regions or translocations in chromosomal regions that harbor miRNAs (which can be considered as tumor suppressors) are frequently associated with cancers, while abnormal overexpression of miRNAs themselves (which can be considered as tumor promoters) can lead to oncogenesis.
miRNA EXPRESSION IN NORMAL BRAIN AND IN BRAIN TUMORS
miRNA profiles in normal brain
Several studies are available to date, where small-scale oligonucleotide arrays have been used analyze miRNA expression profiles in normal brain. These studies have profiled the miRNA expression patterns in normal brains of mammals, including those from murine, rat, primate, and human sources. For example, an analysis of murine brain miRNA has indicated miR-101, -127, -128, -131, and -132 to be brain-specific (Lagos-Quintana et al., 2002). A follow-up analysis (Sempere et al., 2004) of both murine and human brain expressed miRNAs have largely supported the above analysis, and also shown that additional miRNAs are also specifically expressed in the brain. Except for miR-183, which was found to be preferentially expressed in murine brain only, miR-9, -124-a, -124-b, -135, -153, and -219 were found to be distinctively expressed in human brain tissue. Other miRNAs found to be overexpressed in brain by at least twofold over the expression levels in other organs include miR-125-a, -125-b, -128, -132, -137, and -139.
Also found were exceptions to the 50-kb boundary for coordinated expression of miRNA that was described before (Baskerville and Bartel, 2005); for example, other brain-specific miRNA pairs (Baskerville and Bartel, 2005) miR-7/miR-9 and miR-128/miR-138, which are separated by > 750 kb and 8 Mb, respectively, were expressed in a coordinated manner in normal brain tissues.
Separate studies have indicated extensive regulation of miRNA expression patterns during brain development. Here, an oligonucleotide array with probes specific for 44 mature miRNAs that were expressed in both mouse and rat brain was used to profile miRNAs at various stages of murine brain development (Krichevsky et al., 2003; Kosik and Krichevsky, 2005). For example, miR-19b was expressed during prenatal stages, while miR-128 was expressed in the postnatal period. miR-9, -125b, -131, and -178 were expressed maximally at birth. miR-124a and -266 continued to increase throughout the embryonic stages and into postnatal stages.
Interestingly, the miR-9/miR-131 pair is encoded on multiple chromosomal loci identical in both mouse and human (Homo sapiens chromosomes 1, 5, and 15; Mus musculus chromosomes 3, 13, and 7). Therefore, the expression of these miRNAs can be independently regulated during brain development. Thus, it has been proposed that the pair may function as a double-stranded siRNA-like duplex, that is, as a double-stranded RNA. Alternatively, the two single-stranded miRNAs may harbor independent functions or have inter-dependent functions. As described previously, such similarities are not unique but, in fact, are common across the miRNA pool, with multiple miRNA encoded at distant loci having similar or closely similar sequences.
Microarray profiling of brain tumors
Despite the infancy of the miRNA field, several key studies on miRNA expression patterns in both low-grade and high-grade gliomas have been completed to date. Notable among these are methods to retrieve and analyze miRNA populations in archived brain tumor tissue specimens, where it has been demonstrated that miRNAs are left intact after standard formalin fixation and paraffin-embedding processes (Nelson et al., 2004, 2006). Thus, miRNAs can be effectively isolated and amplified from archival tissue banks. In one such study, miRNA populations were characterized on an “miRNA array” platform (RAKE: RNA-primed, array-based, Klenow Enzyme) for identification of expression patterns among and between different glioma subtypes. Here, the miRNAs were first hybridized to an array of immobilized oligomers followed by extension with Klenow fragment of DNA polymerase in the presence of either radiolabeled (Nelson et al., 2004, 2006) or biotinylated (Berezikov et al., 2006b) deoxynucleotides, under stringent conditions such that only perfectly matched miRNA–oligonucleotide pairings could form. The hybridized miRNAs were subsequently extended via the Klenow enzyme. Thus, an expression pattern for miRNAs can be generated in a strategy analogous to standard oligonucleotide microarrays.
miRNA expression in oligodendrogliomas
The above type of microarray analysis was first carried out on oligodendrogliomas, and contrasted with expression profiles of miRNA in both fetal and adult normal brain. The results indicated miR-124a and miR-125b to be highly expressed in both fetal and adult brains. miR-125b was also found to be highly expressed in oligodendrogliomas. Another miRNA, miR-9, was found to have increased expression in both fetal brain tissue and oligodendrogliomas but not in normal adult brain (Nelson et al., 2006).
miRNA expression in pituitary adenomas
As indicated previously, the vast majority of tumors that arise from the pituitary gland are usually benign. In pituitary adenomas, the loss of heterozygosity (LOH) in chromosome arm 13q is thought to be involved in tumorigenesis (Pei et al., 1995). Interestingly, mir-15a and miR-16-1, previously described to play a major role in B-cell chronic lymphocytic leukemia, are in fact located within this chromosomal arm. Thus, these two miRNAs were examined for their expression pattern in pituitary adenomas (Bottoni et al., 2005). In comparison to the normal pituitary gland, both miRNAs were found to be expressed at much lower levels in pituitary adenomas, where their expression inversely correlated with the size of the tumor. In addition, the miRNA expression levels were directly correlated with expression of the pro-inflammatory cytokine p43, which may inhibit tumor growth and sensitize tumors to TNF-α. These early results suggest that miR15a and miR16-1 may also play a pivotal role in regulating the growth of pituitary tumors.
miRNA expression in glioblastoma
Glioblastomas are the most malignant and lethal of all brain tumors. Therefore, a considerable effort has gone into identifying miRNA profiles of these poorly differentiated high-grade tumors. A comparison between primary GBMs, GBM cell-lines, fetal brain, and normal adult brain tissue via an miRNA array has indicated miR-21 to be a key candidate that is differentially expressed between the malignant and normal tissues (Chan et al., 2005). This study reported that miR-21 was highly expressed in GBMs, perhaps functioning as an oncogene/anti-apoptotic factor. Both miRNA arrays and follow-up northern blots indicated the high expression (5- to 100-fold) of miR-21 in primary GBMs and GBM cell lines. In contrast, miR-21 was expressed at a basal level in normal brain (both adult and fetal), and in oligodendrogliomas as well as in medulloblastomas. The function of miR-21 was tested via an “anti-sense” siRNA strategy involving complementary O-methyl-oligonucleotides to knockdown miR-21 levels. This resulted in a marked enhancement in apoptosis, indicating the contribution of miR-21 as an anti-apoptotic factor in GBMs. It remains to be established whether miR-21 has a direct effect on mRNAs that encode pro-apoptotic genes in glioblastomas.
Likewise, a second miRNA array profiling study of GBMs demonstrated a separate set of candidate miRNA genes to be expressed (Ciafre et al., 2005), with miR-221 being strongly upregulated in GBMs. Four other candidate miRNAs, miR-128, -181a, -181b, and -181c, were found to be downregulated. Conversely, this report failed to identify miR-21 as a key miRNA overexpressed in GBMs.
miRNA expression signatures of GBMs have also been analyzed by reverse-transcription via the use of stem-loop primers followed by real-time PCR (Ridzon et al., 2005). In this study, expression of 180 human miRNAs between normal brain and GBMs was analyzed to identify miRNA expression signatures. Most distinct changes (>10-fold) were observed in 14% of the analyzed miRNA, of which, 2% were upregulated in GBMs. The latter included miR-10a, -10b, and -96.
Most GBMs are refractory to chemotherapeutics, a primary reason for the poor prognosis of patients diagnosed with these tumors despite surgical intervention. To identify putative miRNA expression signatures that may impart drug resistance in recurrent GBMs, miRNA has been profiled from primary and recurrent GBMs, postexposure to two key alkylating chemotherapeutic agents used in glioma therapy, BCNU [1,3-bis(2-chloroethyl)-1-nitroso-urea] and Temozolomide (4-methyl-5-oxo-2,3,4,6,8-pentazabicyclo [4.3.0] nona-2,7,9-triene-9-carboxamide) (Winkler et al., 2005). A microarray of 226 precursor and mature miRNAs was used in this study, where cells from primary and recurrent GBM from the same patient were selected in vitro for drug resistance and analyzed. In preliminary studies reported for BCNU, the authors identified four miRNAs (let-7b, mir-125b-2, mir133a-1, and mir-183) that were differentially expressed in both primary vs. recurrent GBMs and BCNUsensitive vs. resistant cells. Of these, let-7b is mapped to chromosome 22q12, a region previously correlated by the same group to be correlated with GBM resistance to BCNU (Hank et al., 2006).
A better consensus of the candidate miRNAs that are differentially expressed in glioma will come with further array profiling studies. As a greater number of studies have been completed via siRNA profiling of glioma [reviewed in Mathupala et al. (2006)], correlations between known and experimentally verified siRNA templates, their target genes, and candidate miRNA may help experimentally identify candidate miRNA targets in glioma.
miRNAS AS A DIAGNOSTIC AND PROGNOSTIC TOOL
Currently, two main methods are in development for high-throughput miRNA profiling. First technique involves miRNA arrays based on traditional oligonucleotide array methods (Liu et al., 2004). Second technique, which may be more amenable in a clinical setting, is known as bead-based miRNA profiling (Lu et al., 2005). In this method, labeled miRNA-specific polystyrene beads, which are differentially color-coded with a duo of fluorescent labels, are used to trap and enumerate miRNA species within a given miRNA population via flow cytometry.
Since miRNAs are now known to play a crucial role as oncogenes or tumor suppressors, their expression profile may be developed as a highly sensitive diagnostic and prognostic tool. In fact, a comparison between mRNA profiling and miRNA profiling has indicated that the latter is more accurate in classifying highly malignant, poorly differentiated tumors (Lu et al., 2005). GBMs fit well into this type of solid tumors. Furthermore, it was recognized that the expression level of miRNA inversely correlated with the degree of differentiation within a given tissue of origin, which has been suggested to make the miRNA-based predictions more precise (Calin and Croce, 2006b).
As an ever growing number of miRNAs are identified to enhance the current databases of “tumor miRNA,” more statistically significant datasets will be available to improve the in silico predictive algorithms, and then link them to sensitive high-throughput flow cytometry–based analytical systems for clinically relevant rapid miRNA profiling. This may also provide a more accurate tool for both clinical diagnosis and prognosis of brain tumors, which can supplement the currently available histopathological grading and classification schemes.
miRNA AS A THERAPEUTIC TOOL AGAINST MALIGNANT GLIOMA—TECHNOLOGICAL HURDLES AND SOLUTIONS
miRNAs suffer from the same predicament that befalls any potential therapy utilizing siRNAs: that of efficient and targeted delivery to the brain in vivo [reviewed in Mathupala et al. (2006)]. The primary mode of delivering therapeutics to a tumor target is via the systemic route, that is, intravenous delivery. However, the CNS poses a special challenge, primarily due to the blood–brain barrier (BBB), the neuroprotective tissue layer that protects the CNS against both toxins and pathogens that enter the systemic circulation. Due to the molecular mass and the polar nature of miRNAs, the BBB will essentially be impenetrable in a systemic therapeutic setting. Thus, targeting of miRNAs to the brain will invariably involve penetrating the BBB, or placing the therapeutics behind the BBB, that is, intracranial placement of delivery mechanisms. Both these methods are discussed below in brief; the former method is known as intrathecal delivery, where the therapeutic agent is delivered directly to the cerebrospinal fluid via a shunted reservoir referred to as an Ommaya reservoir. The latter is placed below the scalp of the patient through which the therapeutic agent can be replenished, or used to monitor the cerebrospinal fluid status. Alternatively, the therapeutic agent can be staged postsurgery, within the resected tumor cavity, and applied in a programmed manner via continuous application under positive pressure. The latter is currently referred to as convection-enhanced delivery (CED), which can be applied via infusion pumps or via osmo-regulated delivery. A third strategy is the entrapment of therapeutic miRNA in biodegradable wafers, which can again be placed in the resected tumor cavity for localized diffusion into the residual tissue bed. The goal of these delivery methods is to achieve diffuse homogenous distribution of the therapeutic agent throughout the tumor and into the adjacent area of tumor-infiltrated brain parenchyma. Each of the above techniques is applicable in a future miRNA delivery strategy, with the therapeutic miRNAs been admixtured or conjugated with chemical moieties to enhance their cellular uptake (discussed below).
It should be noted that one technique that may be amenable for systemic application of miRNAs has been tested with siRNA, in which receptor-mediated transport across the BBB is utilized. Both immuno-liposomes, and nanoparticles that perhaps mimic low-density lipoproteins (LDL) have been utilized in these strategies (Zhang et al., 2003, 2004; Pardridge, 2004; Fountaine et al., 2005; Lesniak, 2005). Despite its impenetrable nature, the BBB is routinely compromised in patients with late-stage glioma. Thus, it may possible to deliver therapeutic dosages of miRNA via the systemic route, at least in these patients.
Also, a quick perusal of the potential “hit” sites per miRNA within the human genome (miRBase, V. 4, 2006; microRNA.sanger.ac.uk) indicates approximately 1000 potential targets per miRNA. This clearly outlines the necessity for further refinement of in silico methods for potential target prediction, and the paucity of our current knowledge in how the miRNAs are targeted to select mRNA transcripts. Thus, numerous hurdles, unrelated to the potential efficacy or specificity of miRNAs, remain to be overcome.
“ANTI-SENSE” miRNAS
One method that has shown efficacy in altering the cellular miRNA profile has been the introduction of “anti-sense” oligonucleotides from exogenous sources. These synthetic anti-miRNA oligonucleotides or AMOs (Esquela-Kerscher and Slack, 2006), chemically stabilized to enhance their in vivo half-life, are currently in preclinical studies. However, being synthetic, these also suffer from the same technical difficulties as siRNAs, for potential in vivo applications. A promising development that may help alleviate the in vivo “delivery problem” is the recent report that modification of these AMOs by conjugation with membrane lipids (which have been named antagomirs) enhances their in vivo uptake, in the absence of toxicity that is inherent to chemical-based in vivo delivery methods (Lorenz et al., 2004; Krutzfeldt et al., 2005).
SID-1–MEDIATED IN VIVO DELIVERY OF miRNA?
Systemic RNAi deficient-1 (SID-1) was initially identified as a transmembrane protein that mediates RNAi in C. elegans. It was later recognized as mediating intercellular transport of double-stranded RNA (Feinberg and Hunter, 2003). Its mammalian homolog, when expressed in human cells, was recently shown to enhance cellular uptake of siRNA resulting in enhanced siRNA-mediated gene silencing (Duxbury et al., 2005). The data suggest that SID-1 perhaps functions as a pore or channel that facilitates uptake of double-stranded RNA into cells. Thus, future strategies where SID-1 expression is first upregulated in the targeted tumor tissue prior to localized administration of miRNA may help alleviate the in vivo delivery issues that currently plague clinical applicability of miRNA.
COMPETITION BETWEEN ENDOGENOUS AND THERAPEUTIC miRNAS
As discussed earlier, pre-miRNA is an endogenous product transcribed off the host's genomic DNA. For prolonged therapy, it would be necessary to introduce exogenous genetic constructs where both types of transcription will occur in the nucleus. Thus, both types of pre-miRNAs will be destined to the cytoplasm for processing by Dicer to initiate RNAi. A recent study indicates that overexpression of exogenous miRNA in this manner has the potential to saturate trans-nuclear export machinery (i.e., in this case, Exportin-5–mediated miRNA translocation), leading to competition with the normal export of endogenous pre-miRNAs, resulting in adverse effects in the targeted tissue as well as on the organism (Grimm et al., 2006; Marsden, 2006).
DIRECT TARGETING/ACTIVATION OF miRNA GENES FOR THERAPY?
Currently identified miRNAs can be broadly categorized as those that promote tumor initiation or growth (i.e., oncogenes), or those that act as “tumor suppressors.” Thus, in a therapeutic modality similar to classical oncogenes or tumor suppressors, it should be possible to inhibit or activate the respective miRNA in a future therapeutic strategy (outlined in Fig. 2). Thus, a “gain of function” by replacing deleted or mutated miRNA tumor suppressors, and induction of “loss of function” by inhibiting miRNA transcription are two strategies that can be employed to modulate their cellular expression.
FIG. 2.
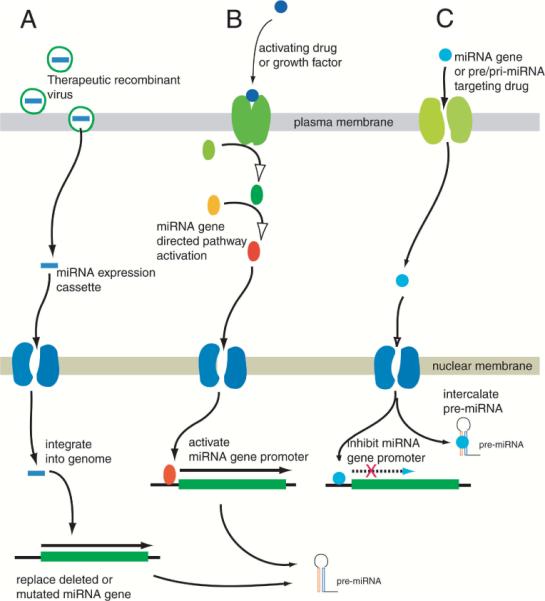
Possible pathways for therapeutic targeting of miRNA genes: Three schema can be visualized for therapeutic targeting: (A) Recombinant virions capable of integrating into the genome can be used as a therapeutic vehicle to replace “tumor-suppressor-type” miRNA genes that were lost due to genomic instability, or to introduce miRNA-generating constructs that can target specific genes; (B) Small-molecule drugs or growth factor components can be applied to activate specific miRNA gene promoters to activate or upregulate tumor-suppressor-type genes; (C) Small-molecule drugs can be designed to target “oncogenic-type” miRNA gene promoters to inhibit transcription, or they can be designed to target pre-miRNA themselves via intercalation or cross-linking. However, at present, the paucity of information on cis-elements of miRNA gene promoters, or the solution structure of pri-miRNA or pre-miRNA, remains to be overcome for schemes B and C to be of therapeutic utility.
CONCLUSIONS
Despite success at the laboratory bench, a key difficulty that has dogged RNAi studies with siRNA technology has been their in vivo targeting to distant organs within an organism. The latter is crucial for bringing RNAi technology to the bedside. Several preclinical studies, including those involving primates, have demonstrated the efficacy of RNAi in vivo (Zimmermann et al., 2006). However, as with any field in its infancy, adverse reactions to RNAi therapy have also been reported (Grimm et al., 2006). It is hoped that with the advent of miRNA, which can be considered an “ortholog” of siRNA, RNAi can be successfully maintained in vivo in a tumor-specific manner for prolonged clinical efficacy. A crucial feature that needs to be elucidated is how the expression of miRNA is regulated, as the promoters of “miRNA transcription units” still remain to be mapped in detail with regard to the key response elements and their trans-activators. Once these are identified, it may become a matter of exposing the diseased tissue or organs to cell-permeable small-molecule drugs that can activate or silence expression of key miRNA either via direct intercalation or by regulating the miRNA gene promoters. Success in such a strategy for gene therapy would provide clinicians an additional repertoire of promising therapeutic agents that can be used in parallel to currently available drugs that act upon various signal transduction cascades within malignant cells.
ACKNOWLEDGMENTS
Research support for the corresponding author was provided by a grant from the National Cancer Institute/National Institutes of Health (CA 116257), the Fund for Medical Research and Education (FMRE), Wayne State University, and by a gift from the Marvin E. Klein MD Charitable Trust.
REFERENCES
- AMBROS V, BARTEL B, BARTEL DP, BURGE CB, CARRINGTON JC, CHEN X, DREYFUSS G, EDDY SR, GRIFFITHS-JONES S, MARSHALL M, MATZKE M, RUVKUN G, TUSCHL T. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMERICAN BRAIN TUMOR ASSOCIATION . A Primer of Brain Tumors. Des Plaines, IL: 2006. pp. 13–17. www.abta.org/buildingknowledge5.htm. [Google Scholar]
- BASKERVILLE S, BARTEL DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEREZIKOV E, CUPPEN E, PLASTERK RH. Approaches to microRNA discovery. Nat. Genet. 2006a;38(Suppl):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- BEREZIKOV E, VAN TG, VERHEUL M, VAN DE BJ, VAN LL, VOS J, VERLOOP R, VAN DE WM, GURYEV V, TAKADA S, VAN ZONNEVELD AJ, MANO H, PLASTERK R, CUPPEN E. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006b;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTTONI A, PICCIN D, TAGLIATI F, LUCHIN A, ZATELLI MC, GLI UBERTI EC. miR-15a and miR-16-1 down-regulation in pituitary adenomas. J. Cell Physiol. 2005;204:280–285. doi: 10.1002/jcp.20282. [DOI] [PubMed] [Google Scholar]
- CAI X, HAGEDORN CH, CULLEN BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALIN GA, CROCE CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006a;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- CALIN GA, CROCE CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006b;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- CALIN GA, DUMITRU CD, SHIMIZU M, BICHI R, ZUPO S, NOCH E, ALDLER H, RATTAN S, KEATING M, RAI K, RASSENTI L, KIPPS T, NEGRINI M, BULLRICH F, CROCE CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALIN GA, SEVIGNANI C, DUMITRU CD, HYSLOP T, NOCH E, YENDAMURI S, SHIMIZU M, RATTAN S, BULLRICH F, NEGRINI M, CROCE CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CENTRAL BRAIN TUMOR REGISTRY OF THE UNITED STATES . Primary Brain Tumors in the United States, Statistical Report (1998–2002) Chicago, IL: 2005–2006. www.cbtrus.org/reports/reports.html. [Google Scholar]
- CHAN JA, KRICHEVSKY AM, KOSIK KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- CIAFRE SA, GALARDI S, MANGIOLA A, FERRACIN M, LIU CG, SABATINO G, NEGRINI M, MAIRA G, CROCE CM, FARACE MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- CIMMINO A, CALIN GA, FABBRI M, IORIO MV, FERRACIN M, SHIMIZU M, WOJCIK SE, AQEILAN RI, ZUPO S, DONO M, RASSENTI L, ALDER H, VOLINIA S, LIU CG, KIPPS TJ, NEGRINI M, CROCE CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DU T, ZAMORE PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- DUXBURY MS, ASHLEY SW, WHANG EE. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem. Biophys. Res. Commun. 2005;331:459–463. doi: 10.1016/j.bbrc.2005.03.199. [DOI] [PubMed] [Google Scholar]
- DYKXHOORN DM, LIEBERMAN J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Annu. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- ESQUELA-KERSCHER A, SLACK FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- FEINBERG EH, HUNTER CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- FILIPOWICZ W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- FILIPOWICZ W, JASKIEWICZ L, KOLB FA, PILLAI RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- FOUNTAINE TM, WOOD MJ, WADE-MARTINS R. Delivering RNA interference to the mammalian brain. Curr. Gene Ther. 2005;5:399–410. doi: 10.2174/1566523054546206. [DOI] [PubMed] [Google Scholar]
- GRIFFITHS-JONES S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- GRIFFITHS-JONES S, GROCOCK RJ, VAN DS, BATEMAN A, ENRIGHT AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRIMM D, STREETZ KL, JOPLING CL, STORM TA, PANDEY K, DAVIS CR, MARION P, SALAZAR F, KAY MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- HANK NC, SHAPIRO JR, SCHECK AC. Over-representation of specific regions of chromosome 22 in cells from human glioma correlate with resistance to 1,3-bis(2-chloroethyl)-1-nitrosourea. BMC Cancer. 2006;6:2. doi: 10.1186/1471-2407-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEMMATI HD, NAKANO I, LAZAREFF JA, MASTERMAN-SMITH M, GESCHWIND DH, BRONNER-FRASER M, KORNBLUM HI. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IGNATOVA TN, KUKEKOV VG, LAYWELL ED, SUSLOV ON, VRIONIS FD, STEINDLER DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- INTERNATIONAL AGENCY FOR RESEARCH ON CANCER . In: IARC Scientific Publication 155: Cancer Incidence in Five Continents. Parkin DM, Whelan SL, Ferlay J, Teppo L, Thomas DB, editors. Vol. VIII. Lyon Cedex, France: 2003. http://www.iarc.fr/IARCPress/pdfs/index.php. [Google Scholar]
- KIM VN, NAM JW. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- KLEIHUES P, CAVANEE WK. World Health Organization Classification of Tumors: Pathology and Genetics: Tumours of the Nervous System. IARC Press; Lyon: 2000. [Google Scholar]
- KOSIK KS, KRICHEVSKY AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- KRICHEVSKY AM, KING KS, DONAHUE CP, KHRAPKO K, KOSIK KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROL J, KRZYZOSIAK WJ. Structural aspects of microRNA biogenesis. IUBMB Life. 2004;56:95–100. doi: 10.1080/15216540410001670142. [DOI] [PubMed] [Google Scholar]
- KRUTZFELDT J, RAJEWSKY N, BRAICH R, RAJEEV KG, TUSCHL T, MANOHARAN M, STOFFEL M. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- LAGOS-QUINTANA M, RAUHUT R, YALCIN A, MEYER J, LENDECKEL W, TUSCHL T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- LAI EC. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE RC, FEINBAUM RL, AMBROS V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- LEE Y, KIM M, HAN J, YEOM KH, LEE S, BAEK SH, KIM VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEGENDRE M, LAMBERT A, GAUTHERET D. Profile-based detection of microRNA precursors in animal genomes. Bioinformatics. 2005;21:841–845. doi: 10.1093/bioinformatics/bti073. [DOI] [PubMed] [Google Scholar]
- LESNIAK MS. Novel advances in drug delivery to brain cancer. Technol. Cancer Res. Treat. 2005;4:417–428. doi: 10.1177/153303460500400409. [DOI] [PubMed] [Google Scholar]
- LIN SL, MILLER JD, YING SY. Intronic microRNA (miRNA) J. Biomed. Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU CG, CALIN GA, MELOON B, GAMLIEL N, SEVIGNANI C, FERRACIN M, DUMITRU CD, SHIMIZU M, ZUPO S, DONO M, ALDER H, BULLRICH F, NEGRINI M, CROCE CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LORENZ C, HADWIGER P, JOHN M, VORNLOCHER HP, UNVERZAGT C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- LU J, GETZ G, MISKA EA, VAREZ-SAAVEDRA E, LAMB J, PECK D, SWEET-CORDERO A, EBERT BL, MAK RH, FERRANDO AA, DOWNING JR, JACKS T, HORVITZ HR, GOLUB TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- MANIATAKI E, MOURELATOS Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARSDEN PA. RNA interference as potential therapy—not so fast. N. Engl. J. Med. 2006;355:953–954. doi: 10.1056/NEJMcibr063864. [DOI] [PubMed] [Google Scholar]
- MATHUPALA SP, GUTHIKONDA M, SLOAN AE. RNAi based approaches to the treatment of malignant glioma. Technol. Cancer Res. Treat. 2006;5:261–269. doi: 10.1177/153303460600500313. [DOI] [PubMed] [Google Scholar]
- MELTZER PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–746. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- MITTAL V. Improving the efficiency of RNA interference in mammals. Nat. Rev. Genet. 2004;5:355–365. doi: 10.1038/nrg1323. [DOI] [PubMed] [Google Scholar]
- NATIONAL BRAIN TUMOR FOUNDATION . The Essential Guide to Brain Tumors. San Francisco, CA: 2004. pp. 18–23. http://www.braintumor.org/patient_info/publications/brochures/documents/guide_wpics.pdf. [Google Scholar]
- NELSON P, KIRIAKIDOU M, SHARMA A, MANIATAKI E, MOURELATOS Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- NELSON PT, BALDWIN DA, KLOOSTERMAN WP, KAUPPINEN S, PLASTERK RH, MOURELATOS Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NELSON PT, BALDWIN DA, SCEARCE LM, OBERHOLTZER JC, TOBIAS JW, MOURELATOS Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- PARDRIDGE WM. Intravenous, non-viral RNAi gene therapy of brain cancer. Expert. Opin. Biol. Ther. 2004;4:1103–1113. doi: 10.1517/14712598.4.7.1103. [DOI] [PubMed] [Google Scholar]
- PEI L, MELMED S, SCHEITHAUER B, KOVACS K, BENEDICT WF, PRAGER D. Frequent loss of heterozygosity at the retinoblastoma susceptibility gene (RB) locus in aggressive pituitary tumors: evidence for a chromosome 13 tumor suppressor gene other than RB. Cancer Res. 1995;55:1613–1616. [PubMed] [Google Scholar]
- PILLAI RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJEWSKY N. microRNA target predictions in animals. Nat. Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- REINHART BJ, SLACK FJ, BASSON M, PASQUINELLI AE, BETTINGER JC, ROUGVIE AE, HORVITZ HR, RUVKUN G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- RIDZON D, TAN R, NGUYEN J, BROOMER A, CHEN C, AMBROS V, ISRAEL M. MicroRNA expression signatures in human glioblastoma multiforme brain tumors. European Society of Human Genetics (ESHG) Annual Meeting; Prague, Czech Republic. 2005. docs.appliedbiosystems.com/pebiodocs/00114298.pdf. [Google Scholar]
- SEMPERE LF, FREEMANTLE S, PITHA-ROWE I, MOSS E, DMITROVSKY E, AMBROS V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SINGH SK, CLARKE ID, TERASAKI M, BONN VE, HAW KINS C, SQUIRE J, DIRKS PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- TSUCHIYA S, OKUNO Y, TSUJIMOTO G. Micro-RNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J. Pharmacol. Sci. 2006;101:267–270. doi: 10.1254/jphs.cpj06013x. [DOI] [PubMed] [Google Scholar]
- WANG X, ZHANG J, LI F, GU J, HE T, ZHANG X, LI Y. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610–3614. doi: 10.1093/bioinformatics/bti562. [DOI] [PubMed] [Google Scholar]
- WILLIAMS RW, HERRUP K. The control of neuron number. Annu. Rev. Neurosci. 1988;11:423–453. doi: 10.1146/annurev.ne.11.030188.002231. [DOI] [PubMed] [Google Scholar]
- WINKLER JA, HENDRICKS WP, JOHNSON D, XIA J, SCHECK AC. Global microRNA expression analysis in human malignant glioma cells reveal novel miRNA-mediated therapy-resistance mechanisms. American Association for Cancer Research (AACR) 96th Annual Meeting; Anaheim, CA. 2005. www.genosensorcorp.com/pdf/96th%20AACR%20presentation.pdf. [Google Scholar]
- YING SY, LIN SL. Intronic microRNAs. Biochem. Biophys. Res. Commun. 2005;326:515–520. doi: 10.1016/j.bbrc.2004.10.215. [DOI] [PubMed] [Google Scholar]
- ZHANG Y, BOADO RJ, PARDRIDGE WM. In vivo knockdown of gene expression in brain cancer with intravenous RNAi in adult rats. J. Gene Med. 2003;5:1039–1045. doi: 10.1002/jgm.449. [DOI] [PubMed] [Google Scholar]
- ZHANG Y, ZHANG YF, BRYANT J, CHARLES A, BOADO RJ, PARDRIDGE WM. Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin. Cancer Res. 2004;10:3667–3677. doi: 10.1158/1078-0432.CCR-03-0740. [DOI] [PubMed] [Google Scholar]
- ZIMMERMANN TS, LEE AC, AKINC A, BRAMLAGE B, BUMCROT D, FEDORUK MN, HARBORTH J, HEYES JA, JEFFS LB, JOHN M, JUDGE AD, LAM K, MCCLINTOCK K, NECHEV LV, PALMER LR, RACIE T, ROHL I, SEIFFERT S, SHANMUGAM S, SOOD V, SOUTSCHEK J, TOUDJARSKA I, WHEAT AJ, YAWORSKI E, ZEDALIS W, KOTELIANSKY V, MANOHARAN M, VORNLOCHER HP, MACLACHLAN I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]


