Abstract
Tau represents the subunit protein of one of the major hallmarks of Alzheimer disease (AD), the neurofibrillary tangles, and is therefore of major interest as an indicator of disease mechanisms. Many of the unusual properties of Tau can be explained by its nature as a natively unfolded protein. Examples are the large number of structural conformations and biochemical modifications (phosphorylation, proteolysis, glycosylation, and others), the multitude of interaction partners (mainly microtubules, but also other cytoskeletal proteins, kinases, and phosphatases, motor proteins, chaperones, and membrane proteins). The pathological aggregation of Tau is counterintuitive, given its high solubility, but can be rationalized by short hydrophobic motifs forming β structures. The aggregation of Tau is toxic in cell and animal models, but can be reversed by suppressing expression or by aggregation inhibitors. This review summarizes some of the structural, biochemical, and cell biological properties of Tau and Tau fibers. Further aspects of Tau as a diagnostic marker and therapeutic target, its involvement in other Tau-based diseases, and its histopathology are covered by other chapters in this volume.
Tau, a neuronal protein that aggregates in AD, is natively unfolded, can adopt many conformations, is subject to multiple posttranslational modifications, and interacts with many cellular components.
Curiously, “Tau” the protein and “Tau” the lepton were discovered in the same year, 1975 (Weingarten et al. 1975; Perl et al. 1975). They have fascinated cell biologists and elementary particle physicists since then, yielded one Nobel prize, and produced more than 5000 hits in Pubmed, including more than 100 for the decay of the Higgs boson into Tau pairs. For cell biologists, Tau was one of the first microtubule-associated proteins (MAPs) to be characterized, named by Marc Kirschner when his team was searching for factors that promote the self-assembly of tubulin into microtubules (hence Tau = Tubulin binding protein). This started a line of research centered around the cell biological role of Tau as a stabilizer of microtubules in neurons and other cells, with important roles in cell differentiation and polarization. Early milestones in this research were the biochemical characterization of Tau (Cleveland et al. 1977a,b), its up-regulation, along with tubulin, during neuronal differentiation (Drubin and Kirschner 1986), the demonstration that it was mainly an axonal protein in mature neurons (in contrast to the dendritic MAP2; Binder et al. 1985), and the cloning and isoform characterization of Tau from mouse, cow, and human (Goedert et al. 1989; Himmler 1989; Lee et al. 1988).
A second line of Tau research was triggered by basic neurological research to identify the components of the abnormal protein deposits found in the brains of Alzheimer disease patients. One of them, Aβ, was identified as the core protein of extracellular amyloid plaques by molecular cloning (Kang et al. 1987), and the other, Tau, as the core of intracellular neurofibrillary tangles by antibody reactivity (Brion et al. 1985; Grundke-Iqbal et al. 1986; Kosik et al. 1986; Wood et al. 1986). These discoveries led to concerted efforts by cell biologists and neuroscientists to elucidate the physiological and pathological properties of Tau. Human Tau and its splicing isoforms were identified (Goedert et al. 1988), Tau-specific antibodies against normal and diseased states were developed (Kosik et al. 1988; Wolozin et al. 1986), abnormal posttranslational modifications of Tau were identified (e.g., phosphorylation sites, kinases, phosphatases; Biernat et al. 1992; Hanger et al. 1992; Goedert et al. 1994), and the principles of abnormal aggregation emerged (Wischik et al. 1988; Wille et al. 1992).
Over the years, interest in Tau waxed and waned, depending on new discoveries. After the initial excitement, it took a slump when it became clear that familial Alzheimer disease (AD) was caused by mutations affecting amyloid precursor protein (APP) processing, and therefore Tau pathology appeared secondary to Aβ pathology. Tau research was boosted by the discovery that a number of neurodegenerative diseases displayed Tau deposits independently of Aβ amyloid (e.g., PiD, PSP), including frontotemporal dementias caused by Tau mutations (FTDP17; see Goedert et al. 2011). These observations established a role for Tau in its own right as a disease-causing agent. Why Tau and Aβ enter a special relationship in AD is still a matter of debate, but recent results from transgenic mice suggest that Tau pathology is not simply a downstream consequence of Aβ pathology, but necessary for the appearance of Aβ-induced toxicity (for a review, see Morris et al. 2011).
Regarding therapy, the Tau-based research has led to several approaches (reviewed by Schneider and Mandelkow 2008; Lee et al. 2011). They are directed against hyperphosphorylation (e.g., the search for kinase inhibitors or phosphatase enhancers), aggregation (e.g., aggregation inhibitors), compounds promoting microtubule stability (to compensate for Tau dysfunction), and Tau-based immunotherapy. So far, no treatment has arrived in the clinic.
A number of excellent reviews on the biology and pathology of Tau have appeared over the past few years (Cassimeris and Spittle 2001; Garcia and Cleveland 2001; Terwel et al. 2002; Dehmelt and Halpain 2005; Andreadis 2006; Ballatore et al. 2007; Gotz et al. 2007, 2010; Iqbal and Grundke-Iqbal 2008; Schneider and Mandelkow 2008; Sergeant et al. 2008; Aguzzi and Rajendran 2009; Spires-Jones et al. 2009; Iqbal et al. 2009; Wolfe 2009; Goedert et al. 2010; Morris et al. 2011; Salminen et al. 2011). This brief review will cover a few salient aspects, with an emphasis on Tau structure and interactions.
TAU DOMAINS
Human Tau is encoded on chromosome 17q21 (Neve et al. 1986). The protein occurs mainly in the axons of the CNS and consists largely of six isoforms generated by alternative splicing (Goedert et al. 1989). They differ by the presence or absence of two near-amino-terminal inserts of 29 residues each, encoded by exons 2 and 3, and by one of the repeats (R2, 31 residues) in the carboxy-terminal half. Different names are in use, derived from inserts/repeats, number of residues, or clone names, as summarized in Table 1.
Table 1.
Isoforms of Tau protein
| Clone | Inserts/repeats | Number of amino acids (AA) | MW (kDa) |
|---|---|---|---|
| htau40 | 2N4R | 441 | 45.9 |
| htau39 | 2N3R | 410 | 42.6 |
| htau34 | 1N4R | 412 | 43.0 |
| htau37 | 1N3R | 381 | 39.7 |
| htau24 | 0N4R | 383 | 40.0 |
| htau23 | 0N3R | 352 | 36.7 |
| big Tau | 2N4R + exon 4a | 695 | 72.7 |
There are additional minor isoforms in the CNS (see review by Andreadis 2006), and a “big Tau” isoform occuring predominantly in the peripheral nervous system (PNS), equivalent to 2N4R plus 242 residues from exon 4a (Couchie et al. 1992; Goedert et al. 1992). The organization of other mammalian Taus (e.g., mouse, rat, cow) is similar, with only a few alterations, mostly in the amino-terminal region, for example, mouse Tau is 11 residues shorter (ranging from 341 to 430 residues) and has a carboxy-terminal half identical to human Tau (Lee et al. 1988).1 Tau, together with MAP2 and MAP4, forms a family of proteins with similar domain structure, with up to five repeats of ∼31 residues in the carboxy-terminal half and an amino-terminal half of variable size (MAP2, 1858 residues [4R], mostly in neuronal dendrites; MAP4, 1152 residues [4R], ubiquitous in many cell types; Cassimeris and Spittle 2001; Dehmelt and Halpain 2005; Doll et al. 1993).
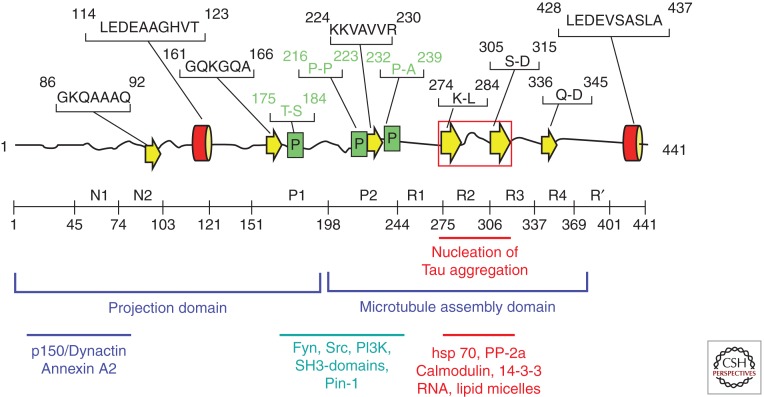
Domains of Tau (Fig. 1) can be defined on the basis of their microtubule interactions and/or their amino acid character. Chymotryptic cleavage in vitro behind Y197 generates two fragments whose carboxy-terminal part binds to microtubules and promotes their assembly, and hence is termed the “assembly domain” (Steiner et al. 1990). The amino-terminal fragment does not bind to microtubules but projects away from the microtubule surface and hence is termed “projection domain” (Hirokawa et al. 1988). A more detailed analysis, based on nuclear magnetic resonance (NMR) spectroscopy, confirms and refines these features: Pronounced interactions with microtubules (MT) occur beween Tau residues 200 and 400 (Mukrasch et al. 2005; Sillen et al. 2007) and thus cover the repeat domain plus the adjacent flanking domains of ∼40 residues each, consistent with binding studies (see below). However, weaker interactions are also distributed elsewhere in the carboxy-terminal half of Tau (Mukrasch et al. 2009).
Figure 1.
Domains and structural elements in Tau. Top: Representation of Tau deduced from NMR (Mukrasch et al. 2009). Most of the chain is unfolded (black lines), with a few short and transient elements of secondary structure (α-helix red, β-strand yellow, poly-proline helix green). The red box indicates the region of the two hexapeptide motifs responsible for Tau aggregation. Middle: Domain subdivision (following Gustke et al. 1994). The carboxy-terminal half promotes microtubule assembly, and the amino-terminal half projects out from the microtubule surface. N1, N2 and R2 may be absent owing to alternative splicing. R1–R4 represent the repeat domain; together with the flanking domains, this represents the microtubule interaction domain. Bottom: Approximate location of interaction sites with other proteins.
The overall amino acid composition is unusually hydrophilic, consistent with the unfolded character of the protein. In full-length Tau (2N4R) there are 80 S or T residues, 56 negative (D + E), 58 positive (K + R), and eight aromatic (5 Y, 3 F, and no W). Thus the protein has an overall basic character, but the amino-terminal ∼120 residues (including the two inserts) are predominantly acidic, and the carboxy-terminal ∼40 residues are roughly neutral. This asymmetry of charges is important for interactions with microtubules and other partners, internal folding, and Tau aggregation. The middle region (AA 150–240) contains numerous prolines, many of them as SP or TP motifs (targets of proline-directed kinases), up to seven PXXP motifs (binding sites of proteins with SH3 domains); hence this region is considered as a “proline-rich domain.” Another region (∼400) containing several prolines is downstream of the repeat domain. These proline-rich domains have acquired a special importance in the field because they harbor many epitopes of antibodies that become hyperphosphorylated in AD, often at SP or TP motifs, and are therefore diagnostic for the disease state, both in human patients and in animal models.
TAU STRUCTURE
Because of its hydrophilic character, Tau does not adopt the compact folded structure typical of most cytosolic proteins. In fact, evidence from various biophysical methods (circular dichroism, NMR and small angle X-ray scattering [SAXS]) show that the entire Tau molecule is “natively unfolded” or “intrinsically disordered” (Schweers et al. 1994; Mukrasch et al. 2009). This means that the polypeptide chain is highly flexible and mobile; there is only a low content of secondary structures (α-helix, β-strand, poly-proline II helix), which are, moreover, transient (Fig. 1, top). This corresponds to the observation that Tau can fulfill its physiological function of stabilizing MTs even after harsh treatment (heat, acid), which forms the basis of the biochemical preparation of assembly-competent Tau (Fellous et al. 1977). The loose disordered character of Tau is revealed by its unusually large extent in solution, sweeping out a volume ∼27 times that of an equivalent compact molecule (Mylonas et al. 2008). This illustrates that Tau could form transient interactions with a multiplicity of other proteins in the crowded environment of a cell. In spite of the disorder on the local level, Tau shows a preference for global interactions between domains that can be likened to a “paperclip,” where the amino-terminal, carboxy-terminal, and repeat domains approach each other (Jeganathan et al. 2006).
TAU AND MICROTUBULES
Microtubules are protein polymers of the cytoskeleton with diverse cellular tasks, best known for their role in stabilizing cell shape, mitosis, and as tracks for intracellular transport by motor proteins. Microtubules can be prepared from brain extracts by repetitive cycles of assembly and disassembly which can be monitored conveniently by light scattering (Gaskin et al. 1974; Weisenberg 1972). Thereby one copurifies many proteins termed microtubule-associated proteins, including Tau. Tau and other MAPs stabilize microtubules by binding to the MT surface and promote their self-assembly from tubulin subunits, but they are not essential for microtubule structure. Reported dissociation constants range from ∼0.02 to ∼1 µm, depending on isotype, mutations, phosphorylation and other modifications, and method of determination (Cleveland et al. 1977a; Butner and Kirschner 1991; Goode and Feinstein 1994; Gustke et al. 1994; Hong et al. 1998; Makrides et al. 2004; Sillen et al. 2007). Generally, Tau mutations (i.e., those occurring in FTDP17) tend to weaken the binding to microtubules somewhat (Hong et al. 1998; Barghorn et al. 2000); phosphorylation can have small or major inhibitory effects, depending on the type and number of sites (e.g., phosphorylation at sites inside the repeat domain has a larger effect than at sites outside; Biernat et al. 1993). Isoforms with more repeats (e.g., 2N4R) tend to bind more strongly than shorter ones (e.g., 0N3R).
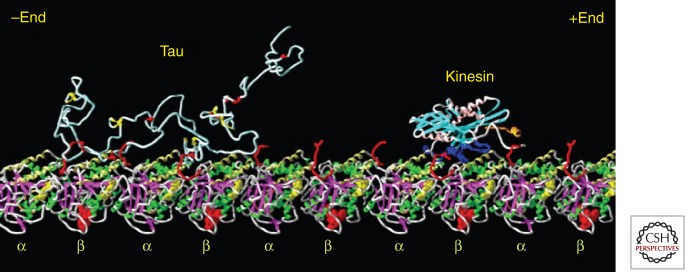
The exact binding site of Tau on MT is not known, despite various attempts by different imaging techniques, and despite the fact that the structure of microtubules is known at high resolution (Fig. 2; Nogales et al. 1999). The uncertainty is largely due to Tau’s loose and natively unfolded structure, which obliterates imaging contrast, both for negative staining or cryo-EM and atomic force microscopy (AFM) imaging (Al-Bassam et al. 2002; Santarella et al. 2004; Schaap et al. 2007). In contrast, other well-folded MT-interacting proteins have been located with high precision, for example, kinesin and doublecortin (Kikkawa et al. 2000; Wendt et al. 2002; Moores et al. 2004; Santarella et al. 2004). The presence of up to four repeats in Tau has led to the idea that Tau repeats and tubulin subunits bind to each other in a commensurate fashion, but this is not supported by other data. For example, Fauquant et al. (2011) recently achieved very tight microtubule binding (Kd ∼ low nanomolar range) for the 117 residue Tau fragment 208–324 (corresponding roughly to domains P2–R3 which stretch across two adjacent αβ-heterodimers of tubulin). This argues that the central part of Tau is aligned along protofilaments, consistent with cryo-electron microscopy data.
Figure 2.
Visualization of Tau and kinesin bound to microtubules. The diagram shows a Tau molecule and a kinesin motor domain bound to a microtubule protofilament (row of αβ-tubulin heterodimers). All molecules are in the same size range (∼350–450 residues), but tubulin and kinesin are compactly folded. Tau is not and therefore occupies a much larger volume, loosely filled with polypeptide chain and highly mobile. Structures modeled after Nogales et al. (1999) (tubulin), Sack et al. (1997) (kinesin), and Hoenger et al. (1998) (docking of kinesin on microtubule). Tau is shown as a random coil; its microtubule-bound conformation is not known. (Figure composed by A. Marx.)
In neurons, Tau is strongly substoichiometric compared with tubulin (∼20–40 µm tubulin compared with ∼1 µm Tau or less [Cleveland et al. 1977b; Hiller and Weber 1978]); in vitro binding studies show saturation at Tau:tubulin ratios up to ∼0.5, that is, about one Tau molecule for two tubulin αβ-heterodimers (Gustke et al. 1994; Makrides et al. 2004), and this ratio holds for Tau molecules with different numbers of repeats, consistent with the model of Knossow and colleagues (Fauquant et al. 2011). The highly acidic carboxy-terminal ∼30 residues of tubulin are important for tight binding (Littauer et al. 1986), but they are also natively unfolded and thus structurally invisible. Quick-freeze deep-etching reveals stubs of Tau’s projection domain ∼18 nm in length, with a low stoichiometry (0.2; Hirokawa et al. 1988). This projection is similar to the length of the repeat domain seen by the glycerol spray technique (Wille et al. 1992), but given that the radius of gyration in solution is only ∼6.5 nm, these values probably represent highly extended extremes.
In considering functions of Tau with regard to MTs, it is useful to distinguish direct and indirect interactions. Direct interactions include the binding, stabilization, and promotion of MT assembly that can be modulated by Tau and its phosphorylation (Brandt et al. 2005; Dolan and Johnson 2010). This function requires the MT-binding domain (repeats + flanking domains), but not necessarily the projection domain. A more subtle effect is the protection of microtubule ends against length fluctuations (dynamic instability), that is their ability to shrink and grow in a stochastic manner. Control of the rate of “catastrophe” and “rescue” is important for cellular organization (Hoogenraad and Bradke 2009), and overstabilization by Tau can impair cell viability (Panda et al. 2003; Thies and Mandelkow 2007; this effect is exploited in cancer chemotherapy by using drugs such as taxol; Wilson and Jordan 2004).
Indirect interactions affect other proteins that may or may not interact with MT by themselves, and may require the projection domain of Tau. An example is the spacer function of MAPs, which helps to establish a “clear zone” around microtubules in cells. This spacer function is pronounced for large MAPs (e.g., MAP2), whereas Tau and its cousin MAP2c allow a much closer apposition, suggesting that the long and acidic projection domains of MAP2/MAP4 are responsible for mutual repulsion (Chen et al. 1992; Umeyama et al. 1993). Another indirect function is the inhibition of MT-dependent transport by motor proteins, which is based on the competition between motors (kinesin, dynein) and MAPs for binding sites on the MT surface (Seitz et al. 2002; Stamer et al. 2002; Dixit et al. 2008). Thirdly, because MTs are distributed throughout cellular space, they provide natural anchoring structures for cell components in general. A classical example is the association of cAMP-dependent kinase (PKA) with the projection domain of MAP2 via the RII-docking subunit (Obar et al. 1990), which is important for phosphorylation of cAMP response element binding protein and neurite outgrowth (Harada et al. 2002). Various other kinases can be found enriched in MAP-microtubule fractions, for example, GSK3β, cdk5, MAP kinase, and others (Mandelkow et al. 1992; Morishima-Kawashima and Kosik 1996). In particular this includes major brain tyrosine kinases (e.g., Fyn, Src, Lck, Abl) that can bind to the PXXP motifs in the proline-rich domain of Tau through their SH3 domain (Fig. 1, bottom; Bhaskar et al. 2005; Williamson et al. 2008; Ittner et al. 2010).
STRUCTURE OF TAU FIBERS (PHFs)
Given that Tau is hydrophilic, unstructured, and dynamic, its aggregation into seemingly well-ordered and periodic fibers in AD is counterintuitive. In fact, the solubility of Tau in vitro is ∼ millimolar, much higher than its concentration in cells (∼ micromolar), which explains in part why it has taken a long time to discover assembly conditions in vitro (Fig. 3; Wille et al. 1992; Crowther et al. 1994; Wilson and Binder 1995). However, two factors contribute to pushing Tau toward aggregation. One is the charge compensation of the basic middle part of Tau by polyanions. In vitro this can be achieved by sulfated glycosaminoglycans (e.g., heparin, heparan sulfate, etc.; Goedert et al. 1996; Perez et al. 1996), which helps to overcome the nucleation barrier. A similar effect can be induced by nucleic acids (Kampers et al. 1996), acidic lipid micelles (e.g., made from arachidonic acid; Wilson and Binder 1997), acidic peptides (Friedhoff et al. 1998) or even carboxylated microbeads (Chirita et al. 2005). In neurons, the inducers of Tau fibrillization are not known, but acidic cofactors are likely (intriguingly, Tau can occur in nuclei and near ribosomes, rich in nucleic acids; Papasozomenos and Binder 1987). Of special interest is the case of microtubules that represent an array of acidic proteins, owing to their highly charged carboxy-terminal peptides. Tau binds strongly to the MT surface, but surprisingly does not self-assemble; rather, the MT surface can be overloaded with a coat of Tau molecules in a nonfilamentous form (Ackmann et al. 2000). Since the Tau–Tau interaction site largely overlaps with the Tau–MT interaction site (Mukrasch et al. 2005), it appears that MTs override Tau’s capacity for fibrillization, perhaps by stabilizing a nonaggregant conformation. Thus, MTs act effectively as chaperones for Tau to prevent their abnormal aggregation.
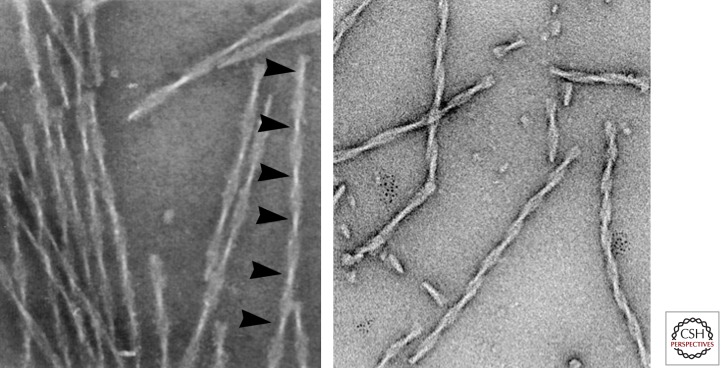
Figure 3.
Tau fibers. Left: Twisted fibers appearing as “paired helical filaments” isolated from Alzheimer brain tissue, with ∼80 nm periodicity (arrowheads). Right: Fibers assembled in vitro from the proaggregant Tau repeat domain (K18ΔK280). Note the similarity of the twisted structures, even though the repeat domain contains only ∼27% of the full-length protein. (Micrographs by E.M. Mandelkow and S. Barghorn.)
The second factor essential for Tau assembly is its propensity for β-structure, encoded in short hexapeptide motifs at the beginning of R2 and R3 (VQIINK and VQIVYK; von Bergen et al. 2000). Disruption of these motifs (e.g., by proline mutations) abrogates Tau’s tendency to aggregate, not only in vitro, but also in cell and animal models (Khlistunova et al. 2007; Mocanu et al. 2008; von Bergen et al. 2001). Conversely, strengthening the β-propensity by mutations (e.g., ΔK280 or P301L) accelerates aggregation in vitro and in animal models. The small size of the β-motifs, embedded in a disordered protein, made it difficult to detect their role, so that the nature of Tau as an “amyloid” protein remained controversial for a long time. However, the β-structure has now been verified by circular dichroism and FTIR (Barghorn et al. 2004), NMR (Mukrasch et al. 2009), electron diffraction (Berriman et al. 2003), X-ray fiber diffraction (Giannetti et al. 2000), and even X-ray crystallography (Sawaya et al. 2007). While the presence of a cross-β structure in the core of PHFs is unambiguous, its arrangement in detail is still unknown. Electron paramagnetic resonance spectroscopy (EPR) studies suggest that the initial ∼20 residues of R2 and R3 are stacked axially in the extended β-structure and in register (Margittai and Langen 2004, 2006). This still leaves various options for the course of the polypeptide chain that are currently being explored by solid-state NMR (A Lange, MPI Göttingen, pers. comm.). These insights will help to define the 3D structure of the core of PHFs, which in turn will allow to design specific inhibitors of aggregation targetting the repeat domain (Pickhardt et al. 2005; Bulic et al. 2010; Lee et al. 2011). One recent example is the design of a capping peptide blocking the elongation of PHFs (Sievers et al. 2011).
Even if the 3D structure of the repeat domain in the PHF core were known, this would reveal only a minor part (<25%) of the structure of Tau fibers.2 The remainder, which includes the amino-terminal half and the carboxy-terminal tail, has been remarkably difficult to image by different types of microscopy and has been dubbed the “fuzzy coat” (Crowther and Wischik 1985; Wischik et al. 1988). This coat is still highly mobile even in the aggregated state (Sillen et al. 2005), similar to Tau itself and the projection domain of MT-bound Tau. It can be viewed as a “soft polymer brush” extending from the PHF core, and able to enter multiple interactions with other cellular components (Wegmann et al. 2010).
TAU AND AD
Tau is a major MAP in the brain, but in this regard it is not more or less interesting than other MAPs that have been discovered and classified over the years (MAP1, MAP2, MAP4, etc.; Cassimeris and Spittle 2001; Dehmelt and Halpain 2005). The major interest in Tau stems from its aggregation in AD and other tauopathies. There has been a debate on whether Tau is causal to the disease or just a byproduct of some disease process. For the case of AD the case is still open, and changes in Tau are mostly viewed as a consequence of Aβ pathology (Haass and Selkoe 2007).3 However, the discovery of mutations in the Tau gene causing frontotemporal dementias has confirmed a causative role of Tau in neurodegeneration (Hutton et al. 1998; Poorkaj et al. 1998; Spillantini et al. 1998), as well as the identification of Tau as one of the risk factors in PSP, PD and others (Hardy and Singleton 2008). Even in the context of AD, the active contribution of Tau was highlighted by animal models, suggesting that Tau is required for the induction of Aβ-induced toxicity (Roberson et al. 2007). In fact, an increased Tau level alone suffices as a risk factor, as demonstrated for the H1c haplotype (Myers et al. 2007). This provides a rationale for the quest for Tau-lowering drugs.
The intense search for disease-related properties of Tau has revealed numerous changes, compared with Tau in nondiseased neurons (Table 2). The problem is to determine which of these changes are causative for disease, whether they are specific, or whether they are simply consequences of an altered state of the cell. We will consider only a few of these mechanisms here. As mentioned above, no mutations in Tau are known that cause bona fide AD. Some changes are common to several tauopathies (e.g., aggregation), others occur not only in disease but also in physiological states. A case in point is phosphorylation: In AD the level is high (eight or more phosphates per Tau molecule, compared with only ∼two in normal adult brain; Kopke et al. 1993), but in normal fetal brain it is also high (∼four), and likewise it is high in hibernating animals (Hartig et al. 2007). Thus hyperphosphorylation is not an absolute indicator of a disease state but rather appears to reflect a state of the cell where the balance of kinases versus phosphatases is tipped in favor of phosphorylation; or conversely a low level of phosphorylation may simply reflect the activity of phosphatases (mainly PP2a) during preparation (Matsuo et al. 1994). The ready response of Tau to such a change is not surprising, considering its disordered structure and numerous potential phosphorylation sites. Other cellular targets should sense the change in biochemical potential in a similar fashion, and in fact this has been demonstrated, for example, for neurofilaments (Ishihara et al. 2001). An overall shift of phosphorylation potential can occur, for example, by a temperature drop during anesthesia or experimental diabetes, which reduces the activity of PP2a and thus mimics an Alzheimer-like phosphorylation state on Tau, reminiscent of the effects of aging (Planel et al. 2007a; Planel et al. 2007b; Veeranna et al. 2009). Conversely, heat stress and oxidative stress activate PP2a and thus generate a low state of Tau phoshorylation, which protects against DNA damage (Davis et al. 1997; Sultan et al. 2011). Therefore, efforts are underway to develop treatments for AD by reducing Tau phosphorylation, either by inhibiting key kinases (e.g., Ahn et al. 2005; Seabrook et al. 2007) or by activating phosphatases such as PP2a (Tanimukai et al. 2005).
Table 2.
Changes of Tau protein in tauopathies
| Mis-sorting (somatodendritic compartment) |
| (Hyper-)phosphorylation |
| Dissociation from microtubules |
| Aggregation (PHF) |
| Proteolytic processing |
| Ubiquitination |
| Glycation |
| Glycosylation |
| Oxidation |
| Nitration |
| Acetylation |
| Amino acid modifications |
| Altered isoform distribution |
| Mutations in Tau gene (FTDP17) |
The majority of potential P-sites on Tau are found to be phosphorylatable (>45; Hanger et al. 2007), and multiple kinases are able to phosphorylate Tau. A subset of sites have gained prominence because they are recognized by antibodies raised against AD Tau in a phosphorylation-dependent manner and are thus useful as diagnostic reagents. They include PHF1 (pS396 + pS404; Greenberg and Davies 1990), several of the AT series of antibodies (Biernat et al. 1992; Goedert et al. 1994), for example, AT8 (pS202 + pT205), AT180 (pT231 + pS235), AT270 (pT181) and AT100 (pT212 + pS214), and Sternberger monoclonal antibodies of the SMI series (e.g., SMI31, 33, 34; Lichtenberg-Kraag et al. 1992; for a complete list see www.alzforum.org). Most of these sites include SP or TP motifs and are therefore targets of proline-directed kinases (e.g., GSK3β, cdc2, cdk5, MAPK, JNK). Other nonproline directed sites include the KXGS motifs in the repeats whose phosphorylation by MARK or related kinases of the AMPK family (Drewes et al. 1997) strongly decreases the Tau–MT affinity, as recognized by antibody 12E8 (Seubert et al. 1995).
A primary preoccupation of “Tauists” over the years has been the search for functional consequences of the bewildering complexities of phosphorylation. This search has taken two main directions, for consequences regarding microtubule binding and for those affecting Tau aggregation. Owing to differences in experimental procedures and detection methods, it is difficult to summarize the body of data, but there appears to be a consensus that phosphorylation tends to weaken the Tau–microtubule affinity. Some sites are efficient in detaching Tau from MT, notably S262 in R1, phosphorylated by MARK and related kinases (Mandelkow et al. 2004), or S214 in the proline-rich domain, phosphorylated by PKA (Brandt et al. 1994; Illenberger et al. 1998). By contrast, proline-directed sites (SP or TP motifs) are prominently phosphorylated by the proline-directed kinases mentioned above, and although single sites have only a small effect, a combination of them can also considerably weaken the interactions with microtubules (Hernandez and Avila 2007; Sergeant et al. 2008; Stoothoff and Johnson 2005). For the disease process, the important implication is that Tau is protected against aggregation while bound to MT, but not when it is detached by phosphorylation.
For Tau aggregation the consequences of phosphorylation are even less clear. The fact that Tau phosphorylation precedes aggregation in AD has led to the assumption that phosphorylation drives Tau into aggregation, and certain highly phosphorylated states of Tau may be prone to aggregation (Iqbal et al. 2008). However, in vitro this cannot be confirmed in a general way; on the contrary, phosphorylation at certain sites (e.g., KXGS motifs) protects against aggregation (Schneider et al. 1999). By comparison, polyanionic cofactors are far superior to phosphorylation in stimulating Tau aggregation. In our view it is therefore questionable whether strategies to reduce phosphorylation (e.g., kinase inhibitors) will be effective in preventing Tau aggregation (even though they may be beneficial for other reasons).
Another major modification of Tau in AD is proteolytic cleavage. Tau contains many potential cleavage sites accessible to multiple proteases, yielding breakdown products that could be toxic in various ways. For example, cleavage of the tails by caspases (behind D421 or behind D13; Gamblin et al. 2003; Horowitz et al. 2004; Rissman et al. 2004) perturbs the paperclip folding of Tau (Jeganathan et al. 2006) and makes it more vulnerable to aggregation. Tau can be cleaved by calpain at a number of sites (Canu and Calissano 2003; Park and Ferreira 2005). This generates a metastable fragment A125–R230 that spans several PXXP motifs and therefore has the potential of scavenging SH3-containing proteins including tyrosine kinases (Garg et al. 2011). This Tau fragment of MW = 10.7 kDa or Mr = ∼17 kDa was thought to mediate Aβ toxicity (Park and Ferreira 2005) in neurons, but this remains a matter of debate. A third type of cleavage occurs by PSA (puromycin-sensitive aminopeptidase), discovered by a genomic screen for modifiers of tauopathy in flies (Karsten et al. 2006). This enzyme cleaves substrates into small peptides and may therefore contribute to the degradation of Tau, independently of the proteasome and/or autophagy (see below). Finally, an inducible cell model of Tau aggregation revealed a complex cleavage pattern whereby a cytosolic protease with thrombin-like characteristics cooperates with a lysosomal protease (cathepsin L) to generate a fragment S258–I360 with a high propensity for aggregation, sufficient to nucleate and coaggregate with endogenous Tau (Wang et al. 2009).
We will mention other biochemical modifications only in passing: Oxidation (which is prominent in aging neurons) affects in the first instance the two cysteines C291 (in R2) and C322 (in R3). They can be cross-linked to form dimers, which greatly enhances the rate of aggregation or as an intra-dimer disulfide bridge, which locks the molecule in a folded conformation (Schweers et al. 1995). Ubiquitination occurs at several lysines (K254, 257, 311, 317, perhaps also 280, 294, 343, 353) that are all in the repeat domain (Morishima-Kawashima et al. 1993). It participates in the protein triage process whereby the chaperone system decides whether to keep the protein for refolding or to send it to the proteasome for degradation (Petrucelli et al. 2004; Shimura et al. 2004b).
O-GlcNac glycosylation is found on many residues, especially in S/T-P motifs, as an alternative for phosphorylation. The reaction protects proteins against aberrant phosphorylation but becomes defective in AD, thus allowing an increase in phosphorylation (Li et al. 2006). Nitration of tyrosine residues occurs via peroxynitrate during oxidative damage (Reynolds et al. 2007). It changes Tau’s conformation and reduces MT binding, but also reduces aggregation. Four of the five Tyr residues can be nitrated (Y18, 29, 197, 394); remarkably, the only tyrosine in the repeat domain (Y310) is not affected, possibly because of a protective conformation within R3. Acetylation of lysines affects predominantly K280, impairs MT binding of Tau, promotes aggregation and potentially interferes with ubiquitination and thus degradation of Tau (Cohen et al. 2011; Min et al. 2010). Finally, because Tau and its aggregates are long-lived, they are subject to nonenzymatic modifications. One is glycation by AGEs (advanced glycation end products); this decreases MT binding, promotes aggregation, activates RAGE receptors and thus causes oxidative damage (Ledesma et al. 1994). Other time-dependent changes in Tau include deamidation, isomerization, cross-linking and nonenzymatic cleavage around Asp residues; this is the cause of the “high MW smear” that is typical of Alzheimer Tau preparations run on SDS gels (Watanabe et al. 2004).
The most obvious change of Tau in AD is its aggregation, noticed already by Alois Alzheimer more than 100 years ago. The principles of aggregation are now reasonably well understood from in vitro studies (see above), but it has been difficult to reproduce this in cell and animal models. This is not surprising given the slow growth of aggregates in aging human brains compared with the short lifetimes of cell cultures or transgenic mice. A breakthrough came with the introduction of Tau mutants discovered in FTDP17 (Lewis et al. 2000), because their aggregation was sufficiently fast to be observed in mice. Mouse models of AD and tauopathies have been reviewed extensively in the literature (Gotz and Ittner 2008; Morrissette et al. 2009). Here we focus on some lessons learned from inducible cell and mouse models: An inducible N2a cell model, expressing the Tau repeat domain in a “proaggregant” form (construct K18–ΔK280) that aggregates rapidly because of its high propensity for β structure, develops Tau fibers within 3 days after switching the Tau gene expression on. This is possible, despite the short time window, because endogenous proteases cleave out an even more proaggregant fragment that nucleates Tau fibers and coassembles intact Tau. When switching expression off, aggregates disappear within 4 days. showing that the cell retains the ability to recover. Comparison with an antiaggregant form of the Tau repeat domain (which does not form β structure because of proline mutations in the hexapeptide motifs) reveals that toxicity leading to cell death occurs only with β-structure and aggregation. Toxicity and aggregation can both be prevented by aggregation inhibitors that break β structure (von Bergen et al. 2001; Khlistunova et al. 2007).
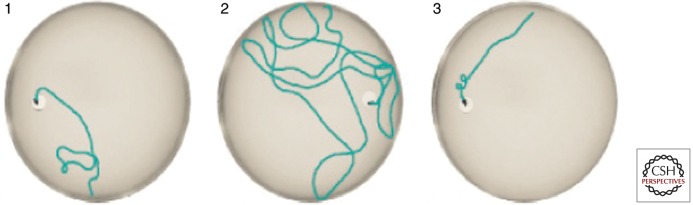
These results can be transferred, cum grano salis, to inducible transgenic mice expressing the same construct of Tau (Mocanu et al. 2008). Only proaggregant TauRD causes neurofibrillary tangles; these mice have cognitive deficits, correlating with loss of synapses and neurons and impaired LTP. Importantly, cognitive deficits and loss of synapses can be rescued by switching off the expression of the toxic protein (Fig. 5). Nevertheless, aggregates persist in the brain, but change their composition from mixed exogenous + endogenous Tau to endogenous mouse Tau only. The explanation is that proaggregant TauRD nucleates aggregation, but once the nucleation barrier has been overcome, endogenous Tau coaggregates, and these aggregates persist beyond the switch-off of exogenous Tau because endogenous Tau is still present (Sydow et al. 2011). This observation highlights the dynamic nature of Tau fibers and suggests the possibility of intervention by aggregation inhibitor compounds. Qualitatively similar results were obtained with another inducible mouse expressing Tau with the proaggregant mutation P301L (Santacruz et al. 2005). Here, too, toxicity and memory deficits were related to aggregation, and recovery of memory occurred after switch-off, yet aggregates tended to persist (in this case, the antiaggregant version of the protein was not tested). These results mean that the toxic agent is the continued expression of aggregation-prone Tau. Hyperphosphorylation is a consequence of this, but not the cause. A possible explanation is that exposure of β-strands in cells, ready to interact, represents a toxic burden to the cell and overexerts the cell’s chaperone-based defense system, which is designed to “detoxify” such exposed β-strands.
Figure 5.
Loss and recovery of memory in regulatable Tau-transgenic mice. The mice express the proaggregant Tau repeat domain (K18ΔK280). Expression is switched OFF in the presence of doxycyclin, and ON without doxycyclin in the drinking water (tet-off system; Gossen and Bujard 2002), and tested in the Morris Water Maze. Left: Control mouse learns position of hidden platform quickly, short length of swimming path. Center: Mouse after switching the Tau repeat domain ON for 12 months has severe learning deficits, and requires a long time and long path length to find the platform. Right: Same mouse as before, after switching Tau expression OFF again for 4 weeks. The memory has returned to control levels (details in Sydow et al. 2011).
DISTRIBUTION OF TAU
The bulk of Tau is located in the brain, specifically in neurons. It is ubiquitous in immature neurons but becomes axonal during maturation, accompanied by a shift toward higher-molecular-weight isoforms and reduced phosphorylation (Drubin and Kirschner 1986; Kosik et al. 1989). Even after maturation, low levels of Tau are present in other neuronal compartments, for example, the nucleus (Loomis et al. 1990; Sultan et al. 2011) and dendrites (Papasozomenos and Binder 1987), and in other brain cells, notably oligodendrocytes (Goldbaum et al. 2003; LoPresti et al. 1995). Moreover, Tau mRNA can be detected in many cell types (Gu et al. 1996), even in muscle fibers where Tau forms aggregates in inclusion body myositis (Askanas and Engel 2008).
The pathway of neuronal sorting is still incompletely understood; several mechanisms appear to contribute, for example, selective protein transport into axons and selective degradation in dendrites (Hirokawa et al. 1996; Nakata and Hirokawa 2003), selective axonal transport of Tau mRNA (Aronov et al. 2002), and selective up-regulation of translation in axons (Morita and Sobue 2009). Transport of Tau into axons occurs as part of the slow axonal transport component SCa (Mercken et al. 1995), probably as part of a (transient) complex with tubulin oligomers driven by kinesin, which provides directional bias and yet allows a high rate of diffusion (Baas and Buster 2004; Konzack et al. 2007).
The issue of axonal sorting is important because missorting of Tau into the somatodendritic compartment is recognized as one of the earliest signs of neurodegeneration in AD and in mouse models (Braak et al. 1994; Coleman and Yao 2003). Entry of Tau into dendrites (e.g., triggered by Aβ, glutamate, oxidative stress, etc.) causes a decay of dendritic spines mediated by Ca2+ influx through NMDA receptors and thus a decay of neuronal communication (Fig. 4; Mattson 2004; Shankar et al. 2007; Zempel et al. 2010).
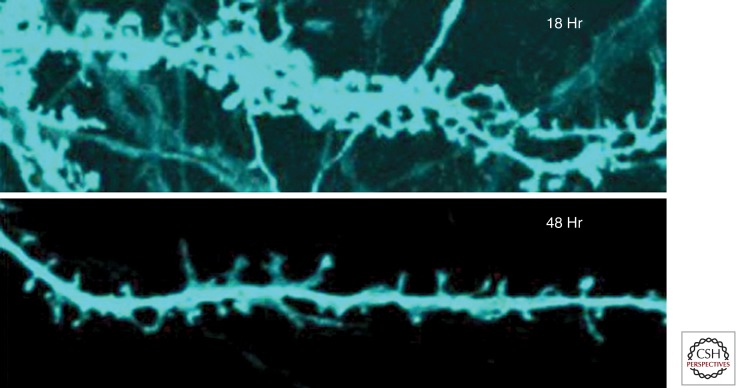
Figure 4.
Dendritic missorting of Tau and synaptic decay. Top: Mature primary rat hippocampal neuron at 21 DIV with numerous dendritic spines, 18 h after transfection with full-length Tau (2N4R, tagged with CFP, blue). Note that Tau has invaded dendritic shafts and spines. Bottom: At 48 h after transfection most spines have shrunk or disappeared. (Adapted from Thies and Mandelkow 2007; reprinted, with permission, from the author.)
Tau is not merely transported as a cargo but can actively interfere with microtubule-dependent traffic because it can compete with motors for binding to microtubules and thereby slow down both anterograde and retrograde transport by kinesin or dynein motors (Stamer et al. 2002; Dixit et al. 2008). Alternatively, Tau could inactivate selected motor complexes, by direct binding to kinesin (Utton et al. 2005), by sequestering the kinesin-associated protein JIP1 (in case of the K369I mutation; Ittner et al. 2009), or by activating phosphatase PP1 and kinase GSK3β through Tau’s amino-terminal 18-residues, leading to the release of cargo vesicles from kinesin light chains (Kanaan et al. 2011). Similarly, the amino-terminal domain of Tau binds to the p150 subunit of dynactin and thereby supports transport by dynein, which can be disrupted by the R5L mutation in Tau (Magnani et al. 2007). These modes of Tau-transport interference could contribute to the movement deficits observed in the first generation of Tau-transgenic mice with pan-neuronal expression of Tau (Lee et al. 2005; Terwel et al. 2002).
Tau is normally a cytosolic protein, but its occurrence outside cells has become important for studying and diagnosing the AD disease process. One reason is that Tau is elevated in the cerebrospinal fluid (CSF) at an early stage of AD and therefore serves as a biomarker (Hampel et al. 2010; Blennow et al. 2011). The origin of CSF-Tau is not clear, but dying and disintegrating neurons could contribute to it. In fact, recent microdialysis experiments suggest that Tau is even higher in the interstitial fluid (Yamada et al. 2011). Another recent development is derived from the earlier observation that Tau pathology spreads in the brain in a well-defined manner, so that its distribution can be correlated with the clinical stages of the disease (Braak stages; Braak and Braak 1991). One hypothesis is that affected neurons release pathological Tau, which is taken up by neighboring cells and thus spreads the pathology in a prion-like fashion (Clavaguera et al. 2009; Frost et al. 2009). Several mechanisms can be envisaged for this release and re-uptake (e.g., via exosomes; Aguzzi and Rajendran 2009). Alternatively, affected neurons could release factors (e.g., cytokines) that then challenge other neurons, either directly or via intermediate cells; an example is the cytosolic accumulation of Tau in neurons encountering activated microglia or exposed to tumor necrosis factor α (Gorlovoy et al. 2009).
TAU BINDING PARTNERS
Since Tau is highly flexible and carries many charges it has the capacity to interact with many partners in the crowded cytosol. This is one of the characteristic features of intrinsically disordered proteins (Dunker et al. 2008). Most of these interactions will be short-lived and difficult to detect. Table 3 presents selected Tau interacting proteins reported in the literature. Various methods to detect these have been employed so that the results are not strictly comparable (e.g., antibody pulldown, FRET, colocalization by microscopy, coassembly, yeast 2-hybrid, etc.).
Table 3.
Binding partners of Tau protein
| Binding partner | Binding region of Tau | Remarks | References |
|---|---|---|---|
| Microtubules | Repeat domain + flanking regions | Butner and Kirschner (1991); Gustke et al. (1994) | |
| Actin filaments | MAPs cross-link MT and actin fil.; Tau on actin fil. in growth cone; Tau in stress-induced actin–cofilin rods | Fulga et al. (2007); Griffith and Pollard (1978); Roger et al. (2004); Whiteman et al. (2009) | |
| Neurofilaments | MAPS cross-linking MT-neurofilaments | Aamodt and Williams (1984) | |
| Ribosomes | Colocalization with polysomes | Papasozomenos and Binder (1987) | |
| GSK3b | Amino-terminal; Exon 1–6 | In complex with presenilin | Sun et al. (2002); Takashima et al. (1998) |
| Fyn | Proline-rich domain | Tau binds to SH3 domain, truncated Tau causes accumulation of fyn in cell body | Belkadi and LoPresti (2008); Ittner et al. (2010); Klein et al. (2002); Lee et al. (1998); Reynolds et al. (2008) |
| Src | Proline-rich domain | Lee et al. (1998); Reynolds et al. (2008) | |
| Lck | Proline-rich domain | Lee et al. (1998); Reynolds et al. (2008) | |
| Pi3K | Proline-rich domain | Reynolds et al. (2008) | |
| JIP1 | Competes with kinesin in axonal transport | Ittner et al. (2009) | |
| PP-2a | Repeat domain + AA 221–242 | Facilitated binding with Pin1 | Lu et al. (1999); Sontag et al. (1999) |
| Hsp70/Hsc70 | Repeat domain | Sarkar et al. (2008); Wang et al. (2009) | |
| Hsp90 | VQIVYK motif involved | Dickey et al. (2007); Tortosa et al. (2009) | |
| Hsp27 | PHF Tau | Shimura et al. (2004a) | |
| CHIP | Dickey et al. (2007); Shimura et al. (2004b) | ||
| BAG-1 | Tau-BAG-1 association is Hsc70-dependent | Elliott et al. (2007) | |
| Ubiquitin | Repeat domain | Ubiquitination of PHF Tau | Morishima-Kawashima et al. (1993) |
| Dynactin | Amino-terminal, Exon1 + 4 | Disrupted by R5L mutation | Magnani et al. (2007) |
| Kinesin LC | Proline-rich domain | Phosphorylation-dependent | Utton et al. (2005) |
| Pin1 | Proline-rich domain, especially pT231–P232 site, also pT212–P213 site | Lu et al. (1999); Smet et al. (2004) | |
| FKBP52 | Through heat shock proteins (HSP) | Chambraud et al. (2010) | |
| FKBP51 | Hsp90 cochaperone | Jinwal et al. (2010) | |
| Cdc37 | Hsp90 cochaperone (interacts with Tau) | Jinwal et al. (2011) | |
| Calmodulin | Repeat domain, especially R3, AA 318–335 | Ca2+ dependent | Padilla et al. (1990); Lee and Wolff (1984); Baudier and Cole (1988) |
| S100b | Ca2+, Zn2+-dependent | Baudier and Cole (1988); Baudier et al. (1987) | |
| α-Synuclein | Repeat domain | Induces fibrillization of Tau | Giasson et al. (2003); Jensen et al. (1999) |
| 14-3-3 | Repeat domain, Proline-rich domain, S214, S324, S356 | Forms ternary complex with SGK1 and Tau | Chun et al. (2004); Hashiguchi et al. (2000); Sluchanko et al. (2009) |
| Presenilin | MTBR | Through GSK3 | Takashima et al. (1998) |
| ApoE | MTBR | Strittmatter et al. (1994) | |
| Annexin A2 | Involving R406 | Linking Tau to plasma membrane | Gauthier-Kemper et al. (2011) |
| Lipid bilayers | Repeat domain | Anionic lipid vesicles induce aggregation | Chirita et al. (2003); Elbaum-Garfinkle et al. (2010); Wilson and Binder (1997) |
| Membrane-associated proteins | Amino-terminal projection domain | Interaction with annexin A2, disrupted by R406W mutation. Interaction with Fyn at lipid rafts |
Brandt et al. (1995); Gauthier-Kemper et al. (2011); Williamson et al. (2008) |
| DNA | Tau protects DNA against heat and oxidative stress | Sultan et al. (2011) | |
| mRNA, tRNA, rRNA | Repeat domain | RNAs induce aggregation of Tau Tau on ribosomes, nucleolus |
Kampers et al. (1996) Loomis et al. (1990) |
By far the most predominant interaction of Tau is that with microtubules, so that the microtubule network can effectively be imaged by fluorescently labeled Tau, and Tau becomes randomized when MTs break down (curiously, databases of protein–protein interactions tend to ignore this major interaction partner of Tau). However, even with MTs the interaction is short-lived (dwell time ∼4s; Konzack et al. 2007; Samsonov et al. 2004), which explains why Tau can diffuse quite rapidly (diffusion constant D = ∼3 µm2/s). This interaction is based on the repeats + flanking domains, but can be interrupted by phosphorylation at key sites (e.g., KXGS motifs in repeats), which leads to the diffuse distribution of Tau in the cytosol and probably contributes to the somatodendritic missorting in degenerating neurons (Zempel et al. 2010). Interactions of Tau with other cytoskeletal elements have been reported as well, notably intermediate filaments (Aamodt and Williams 1984) and microfilaments (F-actin; Griffith and Pollard 1978; Roger et al. 2004). For example, Tau detached from MT by phosphorylation at the KXGS motifs tends to colocalize with actin filaments in growth cones (Biernat et al. 2002; Fulga et al. 2007) or in actin–cofilin rods (Whiteman et al. 2009). These non-MT interactions need not be direct, but could be mediated by other cytoskeleton-associated proteins, consistent with genetic evidence that links modifiers of Tau pathology to the actin network (SUT-1; Kraemer and Schellenberg 2007).
Binding sites of Tau have been reported for several kinases and phosphatases (Reynolds et al. 2008; Sontag et al. 1999). The binding sites lie in or near the MT-binding domain (Fig. 1). Here it is important to distinguish between short-lived interactions that lead to phosphorylation and the longer-lived interactions that characterize the complexes. Unlike MAP2, which has a recognition site for the RII subunit of PKA, Tau does not have a well-characterized binding site for kinase-regulatory proteins. The best-defined kinase interaction sites are the PXXP motifs in the proline-rich domain that can bind to SH3-containing proteins including tyrosine kinases Fyn, Src, or Lck (Reynolds et al. 2008; Williamson et al. 2008). Thus, overexpression of Tau fragments lacking the repeat domain, which stay in the cell body because they cannot be sorted into the axon, also leads to a local accumulation of Fyn (Ittner et al. 2010), and presumably other SH3-domain proteins. A role for Tau–Fyn interactions has been postulated for oligodendrocytes where the complex is transported into cell processes in a MT-dependent manner and is required for myelination (Belkadi and LoPresti 2008; Klein et al. 2002). In neurons, the Tau–Fyn complex becomes enriched in lipid rafts upon exposure to Aβ (Williamson et al. 2008). An analogous cotransport of Tau with Fyn has recently been postulated for neuronal dendrites and spines, enabling Fyn to phosphorylate NMDA receptors and to mediate Aβ-induced excitotoxicity. This way Tau might play a role in the regulation of neuronal network activity (Ittner et al. 2010; Morris et al. 2011).
A number of studies have dealt with the interactions of Tau with the chaperone system. Proteins such as hsp70 and their binding partners (e.g., CHIP) are thought to control the level of soluble Tau and thus help to prevent aggregation in a phosphorylation-dependent manner (Shimura et al. 2004a,b; Dickey et al. 2007; Petrucelli et al. 2004). The binding site has been mapped to the motif VQIVYK, the same as the site responsible for β-structure and aggregation (Sarkar et al. 2008). Since chaperones bind to short peptides with hydrophobic character, this finding suggests that the cell protects itself against aggregation by two types of “chaperones”—microtubules for the bound state, and hsp70 for the unbound state. Independently of this, there is a debate on whether Tau is degraded via the proteasome or the autophagy pathway, which may depend on cell type or phosphorylation state. However, it is noteworthy that the repeat domain possesses two motifs of the consensus type KVERQ, which interact with hsc70 for chaperone-mediated autophagy (Wang et al. 2009). Other interactors of Tau are linked to chaperone components, for example the prolyl isomerase Pin-1 (Liou et al. 2003), FKBP51, FKBP52 (Chambraud et al. 2010; Jinwal et al. 2010). Pin-1 isomerizes the motifs pT212-P and pS231-P in the proline-rich domain from cis to trans and thus allows the dephosphorylation by PP-2a and recovery of microtubule binding (Smet et al. 2004).
Among the nonprotein interaction partners of Tau, the interactions with the plasma membrane and with nuclear components have been intriguing over the years. The emerging picture is that a small fraction of cellular Tau binds to the membrane through membrane-associated proteins, for example, annexins or proteins of lipid rafts (Williamson et al. 2008; Gauthier-Kemper et al. 2011). Independently of that, Tau can also interact with anionic lipid bilayers and micelles, at least in vitro (Chirita et al. 2003; Elbaum-Garfinkle et al. 2010). The significance in a cellular context is not clear at present, but micelles of arachidonic acid have been suggested as nucleators of Tau aggregation (Wilson and Binder 1997). Likewise, the interaction with nucleic acids was suggested from the observation of nuclear Tau (Loomis et al. 1990); RNAs became interesting as potential nucleators of Tau aggregation (Kampers et al. 1996), but insights into functions came recently with the discovery that Tau can protect DNA against heat and oxidative damage (Sultan et al. 2011). Given Tau’s versatile structure, one can expect that the list of interactors will surely grow; for further details we refer to Table 3. The interaction pathways will hopefully reveal novel insights into approaches that suppress the toxic insults of Tau in the human brain.
ACKNOWLEDGMENTS
We are grateful to Alexander Marx for help with the compilation of material and stimulating discussions.
A note on “repeat” nomenclature. Lee et al. (1988) noticed the repeating character of the Tau sequence and defined as repeats the 18-AA stretches of the motifs VXSK to PGGG because of their high sequence homology. The intervening 13 or 14 residues were less conserved and were termed “interrepeats” (Goode and Feinstein 1994). Later, the comparison of 3R and 4R-isoforms revealed that the residues encoded by exon 10 correspond exactly to one repeat of 31 residues (VQIINK … PGGGS), suggesting this as a natural unit for the definition of a repeat (Himmler et al. 1989). In this paper we adhere to this definition, so that the four repeats of human Tau correspond to the sequences 244–274, 275–305 (exon 10), 306–336, and 337–368. Thus, one repeat in our definition corresponds roughly to one 13-AA interrepeat plus the following 18-AA repeat in the earlier definition.
A note on “PHF” terminology. The term “paired helical filament” (PHF) was introduced by Kidd (1963) to describe the filaments in AD neurons, long before their composition was known. The term describes the twisting structure with a crossover repeat of ∼80 nm and a stain-filled groove running along the center, which appears to divide the fiber into two subfibers ∼10 nm wide. Not all fibers have this appearance, some are “straight fibers” (SF), others have variable crossover repeats, but these differences appear to arise from variations in the packing of Tau molecules (Crowther 1991), depending on isoform composition and bound cofactors. There has been an ongoing debate on whether the two subfibers are genuine or merely staining artifacts. An alternative description is that of twisted ribbons, ∼22 nm wide and ∼10 nm thick (Moreno-Herrero et al. 2004; Pollanen et al. 1997; Wegmann et al. 2010). There is little evidence for the existence of physically separate subfibers; on the contrary, AFM imaging and force application suggest that subunits are stacked axially, not side by side (Wegmann et al. 2010). Thus, the term “Tau fibers” is more appropriate than “PHFs,” although the latter is probably too established to fade away.
A note on Tau misfolding. In pathological conditions, Tau is sometimes described as “misfolded,” implying that Tau could also be properly folded in normal conditions. However, because Tau is intrinsically disordered, this concept has little explanatory value. In fact, the best folded state of Tau is that in Tau fibers where at least the repeat domain has a well-defined structure, dominated by β-sheet interactions. In this sense, tauopathies are protein aggregation diseases, but not protein misfolding diseases.
Editors: Dennis J. Selkoe, Eckhard Mandelkow, and David M. Holtzman
Additional Perspectives on The Biology of Alzheimer Disease available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aamodt EJ, Williams RC Jr 1984. Microtubule-associated proteins connect microtubules and neurofilaments in vitro. Biochemistry 23: 6023–6031 [DOI] [PubMed] [Google Scholar]
- Ackmann M, Wiech H, Mandelkow E 2000. Nonsaturable binding indicates clustering of tau on the microtubule surface in a paired helical filament-like conformation. J Biol Chem 275: 30335–30343 [DOI] [PubMed] [Google Scholar]
- Aguzzi A, Rajendran L 2009. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64: 783–790 [DOI] [PubMed] [Google Scholar]
- Ahn JS, Radhakrishnan ML, Mapelli M, Choi S, Tidor B, Cuny GD, Musacchio A, Yeh LA, Kosik KS 2005. Defining Cdk5 ligand chemical space with small molecule inhibitors of tau phosphorylation. Chem Biol 12: 811–823 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA 2002. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A 2006. Misregulation of tau alternative splicing in neurodegeneration and dementia. Prog Mol Subcell Biol 44: 89–107 [DOI] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I 2002. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci 115: 3817–3827 [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK 2008. Inclusion-body myositis: Muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer’s and Parkinson’s disease brains. Acta Neuropathol 116: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Buster DW 2004. Slow axonal transport and the genesis of neuronal morphology. J Neurobiol 58: 3–17 [DOI] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, Trojanowski JQ 2007. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci 8: 663–672 [DOI] [PubMed] [Google Scholar]
- Barghorn S, Zheng-Fischhofer Q, Ackmann M, Biernat J, von Bergen M, Mandelkow EM, Mandelkow E 2000. Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39: 11714–11721 [DOI] [PubMed] [Google Scholar]
- Barghorn S, Davies P, Mandelkow E 2004. Tau paired helical filaments from Alzheimer’s disease brain and assembled in vitro are based on β-structure in the core domain. Biochemistry 43: 1694–1703 [DOI] [PubMed] [Google Scholar]
- Baudier J, Cole RD 1988. Interactions between the microtubule-associated tau proteins and S100b regulate tau phosphorylation by the Ca2+/calmodulin-dependent protein kinase II. J Biol Chem 263: 5876–5883 [PubMed] [Google Scholar]
- Baudier J, Mochly-Rosen D, Newton A, Lee SH, Koshland DE Jr, Cole RD 1987. Comparison of S100b protein with calmodulin: Interactions with melittin and microtubule-associated tau proteins and inhibition of phosphorylation of tau proteins by protein kinase C. Biochemistry 26: 2886–2893 [DOI] [PubMed] [Google Scholar]
- Belkadi A, LoPresti P 2008. Truncated Tau with the Fyn-binding domain and without the microtubule-binding domain hinders the myelinating capacity of an oligodendrocyte cell line. J Neurochem 107: 351–360 [DOI] [PubMed] [Google Scholar]
- Berriman J, Serpell LC, Oberg KA, Fink AL, Goedert M, Crowther RA 2003. Tau filaments from human brain and from in vitro assembly of recombinant protein show cross-β structure. Proc Natl Acad Sci 100: 9034–9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar K, Yen SH, Lee G 2005. Disease-related modifications in tau affect the interaction between Fyn and Tau. J Biol Chem 280: 35119–35125 [DOI] [PubMed] [Google Scholar]
- Biernat J, Mandelkow EM, Schroter C, Lichtenberg-Kraag B, Steiner B, Berling B, Meyer H, Mercken M, Vandermeeren A, Goedert M, et al. 1992. The switch of tau protein to an Alzheimer-like state includes the phosphorylation of two serine-proline motifs upstream of the microtubule binding region. EMBO J 11: 1593–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernat J, Gustke N, Drewes G, Mandelkow EM, Mandelkow E 1993. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: Distinction between PHF-like immunoreactivity and microtubule binding. Neuron 11: 153–163 [DOI] [PubMed] [Google Scholar]
- Biernat J, Wu YZ, Timm T, Zheng-Fischhofer Q, Mandelkow E, Meijer L, Mandelkow EM 2002. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol Biol Cell 13: 4013–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder LI, Frankfurter A, Rebhun LI 1985. The distribution of tau in the mammalian central nervous system. J Cell Biol 101: 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Blennow K, Zetterberg H, Fagan AM 2011. Fluid biomarkers in Alzheimer disease. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E 1991. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82: 239–259 [DOI] [PubMed] [Google Scholar]
- Braak E, Braak H, Mandelkow EM 1994. A sequence of cytoskeleton changes related to the formation of neurofibrillary tangles and neuropil threads. Acta Neuropathol 87: 554–567 [DOI] [PubMed] [Google Scholar]
- Brandt R, Lee G, Teplow DB, Shalloway D, Abdel-Ghany M 1994. Differential effect of phosphorylation and substrate modulation on tau’s ability to promote microtubule growth and nucleation. J Biol Chem 269: 11776–11782 [PubMed] [Google Scholar]
- Brandt R, Leger J, Lee G 1995. Interaction of tau with the neural plasma membrane mediated by tau’s amino-terminal projection domain. J Cell Biol 131: 1327–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Hundelt M, Shahani N 2005. Tau alteration and neuronal degeneration in tauopathies: Mechanisms and models. Biochim Biophys Acta 1739: 331–354 [DOI] [PubMed] [Google Scholar]
- Brion JP, Couck AM, Passareiro E, Flament-Durand J 1985. Neurofibrillary tangles of Alzheimer’s disease: An immunohistochemical study. J Submicrosc Cytol 17: 89–96 [PubMed] [Google Scholar]
- Bulic B, Pickhardt M, Mandelkow EM, Mandelkow E 2010. Tau protein and tau aggregation inhibitors. Neuropharmacology 59: 276–289 [DOI] [PubMed] [Google Scholar]
- Butner KA, Kirschner MW 1991. Tau protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol 115: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canu N, Calissano P 2003. In vitro cultured neurons for molecular studies correlating apoptosis with events related to Alzheimer disease. Cerebellum 2: 270–278 [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Spittle C 2001. Regulation of microtubule-associated proteins. Int Rev Cytol 210: 163–226 [DOI] [PubMed] [Google Scholar]
- Chambraud B, Sardin E, Giustiniani J, Dounane O, Schumacher M, Goedert M, Baulieu EE 2010. A role for FKBP52 in Tau protein function. Proc Natl Acad Sci 107: 2658–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kanai Y, Cowan NJ, Hirokawa N 1992. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature 360: 674–677 [DOI] [PubMed] [Google Scholar]
- Chirita CN, Necula M, Kuret J 2003. Anionic micelles and vesicles induce tau fibrillization in vitro. J Biol Chem 278: 25644–25650 [DOI] [PubMed] [Google Scholar]
- Chirita CN, Congdon EE, Yin H, Kuret J 2005. Triggers of full-length tau aggregation: A role for partially folded intermediates. Biochemistry 44: 5862–5872 [DOI] [PubMed] [Google Scholar]
- Chun J, Kwon T, Lee EJ, Kim CH, Han YS, Hong SK, Hyun S, Kang SS 2004. 14–3–3 Protein mediates phosphorylation of microtubule-associated protein tau by serum- and glucocorticoid-induced protein kinase 1. Mol Cells 18: 360–368 [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, et al. 2009. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol 11: 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Hwo SY, Kirschner MW 1977a. Physical and chemical properties of purified tau factor and the role of tau in microtubule assembly. J Mol Biol 116: 227–247 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Hwo SY, Kirschner MW 1977b. Purification of tau, a microtubule-associated protein that induces assembly of microtubules from purified tubulin. J Mol Biol 116: 207–225 [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM 2011. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun 2: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman PD, Yao PJ 2003. Synaptic slaughter in Alzheimer’s disease. Neurobiol Aging 24: 1023–1027 [DOI] [PubMed] [Google Scholar]
- Couchie D, Mavilia C, Georgieff IS, Liem RK, Shelanski ML, Nunez J 1992. Primary structure of high molecular weight tau present in the peripheral nervous system. Proc Natl Acad Sci 89: 4378–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA 1991. Straight and paired helical filaments in Alzheimer disease have a common structural unit. Proc Natl Acad Sci 88: 2288–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, Wischik CM 1985. Image reconstruction of the Alzheimer paired helical filament. EMBO J 4: 3661–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther RA, Olesen OF, Smith MJ, Jakes R, Goedert M 1994. Assembly of Alzheimer-like filaments from full-length tau protein. FEBS Lett 337: 135–138 [DOI] [PubMed] [Google Scholar]
- Davis DR, Anderton BH, Brion JP, Reynolds CH, Hanger DP 1997. Oxidative stress induces dephosphorylation of tau in rat brain primary neuronal cultures. J Neurochem 68: 1590–1597 [DOI] [PubMed] [Google Scholar]
- Dehmelt L, Halpain S 2005. The MAP2/Tau family of microtubule-associated proteins. Genome Biol 6: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, et al. 2007. The high-affinity HSP90–CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest 117: 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur EL 2008. Differential regulation of dynein and kinesin motor proteins by tau. Science 319: 1086–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan PJ, Johnson GV 2010. The role of tau kinases in Alzheimer’s disease. Curr Opin Drug Discov Devel 13: 595–603 [PMC free article] [PubMed] [Google Scholar]
- Doll T, Meichsner M, Riederer BM, Honegger P, Matus A 1993. An isoform of microtubule-associated protein 2 (MAP2) containing four repeats of the tubulin-binding motif. J Cell Sci 106: 633–639 [DOI] [PubMed] [Google Scholar]
- Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E 1997. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89: 297–308 [DOI] [PubMed] [Google Scholar]
- Drubin DG, Kirschner MW 1986. Tau protein function in living cells. J Cell Biol 103: 2739–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL 2008. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 18: 756–764 [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Ramlall T, Rhoades E 2010. The role of the lipid bilayer in tau aggregation. Biophys J 98: 2722–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Tsvetkov P, Ginzburg I 2007. BAG-1 associates with Hsc70.Tau complex and regulates the proteasomal degradation of Tau protein. J Biol Chem 282: 37276–37284 [DOI] [PubMed] [Google Scholar]
- Fauquant C, Redeker V, Landrieu I, Wieruzseski JM, Verdegem D, Laprevote O, Lippens G, Gigant B, Knossow M 2011. Systematic identification of tubulin interacting fragments of the microtubule-associated protein TAU leads to a highly efficient promoter of microtubule assembly. J Biol Chem 286: 33358–33368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellous A, Francon J, Lennon AM, Nunez J 1977. Microtubule assembly in vitro. Purification of assembly-promoting factors. Eur J Biochem 78: 167–174 [DOI] [PubMed] [Google Scholar]
- Friedhoff P, Schneider A, Mandelkow EM, Mandelkow E 1998. Rapid assembly of Alzheimer-like paired helical filaments from microtubule-associated protein tau monitored by fluorescence in solution. Biochemistry 37: 10223–10230 [DOI] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI 2009. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 284: 12845–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, Feany MB 2007. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol 9: 139–148 [DOI] [PubMed] [Google Scholar]
- Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, et al. 2003. Caspase cleavage of tau: Linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc Natl Acad Sci 100: 10032–10037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia ML, Cleveland DW 2001. Going new places using an old MAP: Tau, microtubules and human neurodegenerative disease. Curr Opin Cell Biol 13: 41–48 [DOI] [PubMed] [Google Scholar]
- Garg S, Timm T, Mandelkow EM, Mandelkow E, Wang Y 2011. Cleavage of Tau by calpain in Alzheimer’s disease: The quest for the toxic 17 kD fragment. Neurobiol Aging 32: 1–14 [DOI] [PubMed] [Google Scholar]
- Gaskin F, Cantor CR, Shelanski ML 1974. Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J Mol Biol 89: 737–755 [DOI] [PubMed] [Google Scholar]
- Gauthier-Kemper A, Weissmann C, Golovyashkina N, Sebo-Lemke Z, Drewes G, Gerke V, Heinisch JJ, Brandt R 2011. The frontotemporal dementia mutation R406W blocks tau’s interaction with the membrane in an annexin A2-dependent manner. J Cell Biol 192: 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannetti AM, Lindwall G, Chau MF, Radeke MJ, Feinstein SC, Kohlstaedt LA 2000. Fibers of tau fragments, but not full length tau, exhibit a cross β-structure: Implications for the formation of paired helical filaments. Protein Sci 9: 2427–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, Trojanowski JQ, Lee VM 2003. Initiation and synergistic fibrillization of tau and α-synuclein. Science 300: 636–640 [DOI] [PubMed] [Google Scholar]
- Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A 1988. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: Identification as the microtubule-associated protein tau. Proc Natl Acad Sci 85: 4051–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA 1989. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J 8: 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Crowther RA 1992. Cloning of a big tau microtubule-associated protein characteristic of the peripheral nervous system. Proc Natl Acad Sci 89: 1983–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Crowther RA, Cohen P, Vanmechelen E, Vandermeeren M, Cras P 1994. Epitope mapping of monoclonal antibodies to the paired helical filaments of Alzheimer’s disease: Identification of phosphorylation sites in tau protein. Biochem J 301: 871–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Jakes R, Spillantini MG, Hasegawa M, Smith MJ, Crowther RA 1996. Assembly of microtubule-associated protein tau into Alzheimer-like filaments induced by sulphated glycosaminoglycans. Nature 383: 550–553 [DOI] [PubMed] [Google Scholar]
- Goedert M, Clavaguera F, Tolnay M 2010. The propagation of prion-like protein inclusions in neurodegenerative diseases. Trends Neurosci 33: 317–325 [DOI] [PubMed] [Google Scholar]
- *.Goedert M, Ghetti B, Grazia Spillantini M 2011. Frontotemporal dementia: Implications for understanding Alzheimer disease. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbaum O, Oppermann M, Handschuh M, Dabir D, Zhang B, Forman MS, Trojanowski JQ, Lee VM, Richter-Landsberg C 2003. Proteasome inhibition stabilizes tau inclusions in oligodendroglial cells that occur after treatment with okadaic acid. J Neurosci 23: 8872–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Feinstein SC 1994. Identification of a novel microtubule binding and assembly domain in the developmentally regulated inter-repeat region of tau. J Cell Biol 124: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlovoy P, Larionov S, Pham TT, Neumann H 2009. Accumulation of tau induced in neurites by microglial proinflammatory mediators. FASEB J 23: 2502–2513 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H 2002. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet 36: 153–173 [DOI] [PubMed] [Google Scholar]
- Gotz J, Ittner LM 2008. Animal models of Alzheimer’s disease and frontotemporal dementia. Nat Rev Neurosci 9: 532–544 [DOI] [PubMed] [Google Scholar]
- Gotz J, Deters N, Doldissen A, Bokhari L, Ke Y, Wiesner A, Schonrock N, Ittner LM 2007. A decade of tau transgenic animal models and beyond. Brain Pathol 17: 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz J, Lim YA, Ke YD, Eckert A, Ittner LM 2010. Dissecting toxicity of tau and β-amyloid. Neurodegener Dis 7: 10–12 [DOI] [PubMed] [Google Scholar]
- Greenberg SG, Davies P 1990. A preparation of Alzheimer paired helical filaments that displays distinct tau proteins by polyacrylamide gel electrophoresis. Proc Natl Acad Sci 87: 5827–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith LM, Pollard TD 1978. Evidence for actin filament–microtubule interaction mediated by microtubule-associated proteins. J Cell Biol 78: 958–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI 1986. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci 83: 4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Oyama F, Ihara Y 1996. Tau is widely expressed in rat tissues. J Neurochem 67: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E 1994. Domains of tau protein and interactions with microtubules. Biochemistry 33: 9511–9522 [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ 2007. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat Rev Mol Cell Biol 8: 101–112 [DOI] [PubMed] [Google Scholar]
- Hampel H, Frank R, Broich K, Teipel SJ, Katz RG, Hardy J, Herholz K, Bokde AL, Jessen F, Hoessler YC, et al. 2010. Biomarkers for Alzheimer’s disease: Academic, industry and regulatory perspectives. Nat Rev Drug Discov 9: 560–574 [DOI] [PubMed] [Google Scholar]
- Hanger DP, Hughes K, Woodgett JR, Brion JP, Anderton BH 1992. Glycogen synthase kinase-3 induces Alzheimer’s disease-like phosphorylation of tau: Generation of paired helical filament epitopes and neuronal localisation of the kinase. Neurosci Lett 147: 58–62 [DOI] [PubMed] [Google Scholar]
- Hanger DP, Byers HL, Wray S, Leung KY, Saxton MJ, Seereeram A, Reynolds CH, Ward MA, Anderton BH 2007. Novel phosphorylation sites in tau from Alzheimer brain support a role for casein kinase 1 in disease pathogenesis. J Biol Chem 282: 23645–23654 [DOI] [PubMed] [Google Scholar]
- Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N 2002. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol 158: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Singleton A 2008. The HapMap: Charting a course for genetic discovery in neurological diseases. Arch Neurol 65: 319–321 [DOI] [PubMed] [Google Scholar]
- Hartig W, Stieler J, Boerema AS, Wolf J, Schmidt U, Weissfuss J, Bullmann T, Strijkstra AM, Arendt T 2007. Hibernation model of tau phosphorylation in hamsters: Selective vulnerability of cholinergic basal forebrain neurons—Implications for Alzheimer’s disease. Eur J Neurosci 25: 69–80 [DOI] [PubMed] [Google Scholar]
- Hashiguchi M, Sobue K, Paudel HK 2000. 14–3–3ζ is an effector of tau protein phosphorylation. J Biol Chem 275: 25247–25254 [DOI] [PubMed] [Google Scholar]
- Hernandez F, Avila J 2007. Tauopathies. Cell Mol Life Sci 64: 2219–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller G, Weber K 1978. Radioimmunoassay for tubulin: A quantitative comparison of the tubulin content of different established tissue culture cells and tissues. Cell 14: 795–804 [DOI] [PubMed] [Google Scholar]
- Himmler A 1989. Structure of the bovine tau gene: Alternatively spliced transcripts generate a protein family. Mol Cell Biol 9: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmler A, Drechsel D, Kirschner MW, Martin DW Jr 1989. Tau consists of a set of proteins with repeated C-terminal microtubule-binding domains and variable N-terminal domains. Mol Cell Biol 9: 1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Shiomura Y, Okabe S 1988. Tau proteins: The molecular structure and mode of binding on microtubules. J Cell Biol 107: 1449–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Funakoshi T, Sato-Harada R, Kanai Y 1996. Selective stabilization of tau in axons and microtubule-associated protein 2C in cell bodies and dendrites contributes to polarized localization of cytoskeletal proteins in mature neurons. J Cell Biol 132: 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Sack S, Thormahlen M, Marx A, Muller J, Gross H, Mandelkow E 1998. Image reconstructions of microtubules decorated with monomeric and dimeric kinesins: Comparison with x-ray structure and implications for motility. J Cell Biol 141: 419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, Geschwind DH, Bird TD, McKeel D, Goate A, et al. 1998. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 282: 1914–1917 [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Bradke F 2009. Control of neuronal polarity and plasticity—A renaissance for microtubules?. Trends Cell Biol 19: 669–676 [DOI] [PubMed] [Google Scholar]
- Horowitz PM, Patterson KR, Guillozet-Bongaarts AL, Reynolds MR, Carroll CA, Weintraub ST, Bennett DA, Cryns VL, Berry RW, Binder LI 2004. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer’s disease. J Neurosci 24: 7895–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. 1998. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393: 702–705 [DOI] [PubMed] [Google Scholar]
- Illenberger S, Zheng-Fischhofer Q, Preuss U, Stamer K, Baumann K, Trinczek B, Biernat J, Godemann R, Mandelkow EM, Mandelkow E 1998. The endogenous and cell cycle-dependent phosphorylation of tau protein in living cells: Implications for Alzheimer’s disease. Mol Biol Cell 9: 1495–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Grundke-Iqbal I 2008. Alzheimer neurofibrillary degeneration: Significance, etiopathogenesis, therapeutics and prevention. J Cell Mol Med 12: 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Alonso Adel C, Grundke-Iqbal I 2008. Cytosolic abnormally hyperphosphorylated tau but not paired helical filaments sequester normal MAPs and inhibit microtubule assembly. J Alzheimers Dis 14: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I 2009. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol 118: 53–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Higuchi M, Zhang B, Yoshiyama Y, Hong M, Trojanowski JQ, Lee VM 2001. Attenuated neurodegenerative disease phenotype in tau transgenic mouse lacking neurofilaments. J Neurosci 21: 6026–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Gotz J 2009. Phosphorylated Tau interacts with c-Jun N-terminal kinase-interacting protein 1 (JIP1) in Alzheimer disease. J Biol Chem 284: 20909–20916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wolfing H, Chieng BC, Christie MJ, Napier IA, et al. 2010. Dendritic function of tau mediates amyloid-β toxicity in Alzheimer’s disease mouse models. Cell 142: 387–397 [DOI] [PubMed] [Google Scholar]
- Jeganathan S, von Bergen M, Brutlach H, Steinhoff HJ, Mandelkow E 2006. Global hairpin folding of tau in solution. Biochemistry 45: 2283–2293 [DOI] [PubMed] [Google Scholar]
- Jensen PH, Hager H, Nielsen MS, Hojrup P, Gliemann J, Jakes R 1999. α-Synuclein binds to Tau and stimulates the protein kinase A-catalyzed tau phosphorylation of serine residues 262 and 356. J Biol Chem 274: 25481–25489 [DOI] [PubMed] [Google Scholar]
- Jinwal UK, Koren J, Borysov SI, Schmid AB, Abisambra JF, Blair LJ, Johnson AG, Jones JR, Shults CL, O’Leary JC, et al. 2010. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J Neurosci 30: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinwal UK, Trotter JH, Abisambra JF, Koren J, Lawson LY, Vestal GD, O’Leary JC, Johnson AG, Jin Y, Jones JR, et al. 2011. The Hsp90 kinase co-chaperone Cdc37 regulates tau stability and phosphorylation dynamics. J Biol Chem 286: 16976–16983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampers T, Friedhoff P, Biernat J, Mandelkow EM, Mandelkow E 1996. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett 399: 344–349 [DOI] [PubMed] [Google Scholar]
- Kanaan NM, Morfini GA, Lapointe NE, Pigino GF, Patterson KR, Song Y, Andreadis A, Fu Y, Brady ST, Binder LI 2011. Pathogenic forms of Tau inhibit kinesin-dependent axonal transport through a mechanism involving activation of axonal phosphotransferases. J Neurosci 31: 9858–9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B 1987. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325: 733–736 [DOI] [PubMed] [Google Scholar]
- Karsten SL, Sang TK, Gehman LT, Chatterjee S, Liu J, Lawless GM, Sengupta S, Berry RW, Pomakian J, Oh HS, et al. 2006. A genomic screen for modifiers of tauopathy identifies puromycin-sensitive aminopeptidase as an inhibitor of tau-induced neurodegeneration. Neuron 51: 549–560 [DOI] [PubMed] [Google Scholar]
- Khlistunova I, Pickhardt M, Biernat J, Wang Y, Mandelkow EM, Mandelkow E 2007. Inhibition of tau aggregation in cell models of tauopathy. Curr Alzheimer Res 4: 544–546 [DOI] [PubMed] [Google Scholar]
- Kidd M 1963. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature 197: 192–193 [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Okada Y, Hirokawa N 2000. 15 A resolution model of the monomeric kinesin motor, KIF1A. Cell 100: 241–252 [DOI] [PubMed] [Google Scholar]
- Klein C, Kramer EM, Cardine AM, Schraven B, Brandt R, Trotter J 2002. Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J Neurosci 22: 698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack S, Thies E, Marx A, Mandelkow EM, Mandelkow E 2007. Swimming against the tide: Mobility of the microtubule-associated protein tau in neurons. J Neurosci 27: 9916–9927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopke E, Tung YC, Shaikh S, Alonso AC, Iqbal K, Grundke-Iqbal I 1993. Microtubule-associated protein tau. Abnormal phosphorylation of a non-paired helical filament pool in Alzheimer disease. J Biol Chem 268: 24374–24384 [PubMed] [Google Scholar]
- Kosik KS, Joachim CL, Selkoe DJ 1986. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci 83: 4044–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS, Orecchio LD, Binder L, Trojanowski JQ, Lee VM, Lee G 1988. Epitopes that span the tau molecule are shared with paired helical filaments. Neuron 1: 817–825 [DOI] [PubMed] [Google Scholar]
- Kosik KS, Orecchio LD, Bakalis S, Neve RL 1989. Developmentally regulated expression of specific tau sequences. Neuron 2: 1389–1397 [DOI] [PubMed] [Google Scholar]
- Kraemer BC, Schellenberg GD 2007. SUT-1 enables tau-induced neurotoxicity in C. elegans. Hum Mol Genet 16: 1959–1971 [DOI] [PubMed] [Google Scholar]
- Ledesma MD, Bonay P, Colaco C, Avila J 1994. Analysis of microtubule-associated protein tau glycation in paired helical filaments. J Biol Chem 269: 21614–21619 [PubMed] [Google Scholar]
- Lee YC, Wolff J 1984. Calmodulin binds to both microtubule-associated protein 2 and tau proteins. J Biol Chem 259: 1226–1230 [PubMed] [Google Scholar]
- Lee G, Cowan N, Kirschner M 1988. The primary structure and heterogeneity of tau protein from mouse brain. Science 239: 285–288 [DOI] [PubMed] [Google Scholar]
- Lee G, Newman ST, Gard DL, Band H, Panchamoorthy G 1998. Tau interacts with src-family non-receptor tyrosine kinases. J Cell Sci 111: 3167–3177 [DOI] [PubMed] [Google Scholar]
- Lee VM, Kenyon TK, Trojanowski JQ 2005. Transgenic animal models of tauopathies. Biochim Biophys Acta 1739: 251–259 [DOI] [PubMed] [Google Scholar]
- *.Lee V, Brunden KR, Hutton M, Trojanowski JQ 2011. Developing therapeutic approaches to Tau, selected kinases, and related neuronal protein targets. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J, McGowan E, Rockwood J, Melrose H, Nacharaju P, Van Slegtenhorst M, Gwinn-Hardy K, Paul Murphy M, Baker M, Yu X, et al. 2000. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat Genet 25: 402–405 [DOI] [PubMed] [Google Scholar]
- Li X, Lu F, Wang JZ, Gong CX 2006. Concurrent alterations of O-GlcNAcylation and phosphorylation of tau in mouse brains during fasting. Eur J Neurosci 23: 2078–2086 [DOI] [PubMed] [Google Scholar]
- Lichtenberg-Kraag B, Mandelkow EM, Biernat J, Steiner B, Schroter C, Gustke N, Meyer HE, Mandelkow E 1992. Phosphorylation-dependent epitopes of neurofilament antibodies on tau protein and relationship with Alzheimer tau. Proc Natl Acad Sci 89: 5384–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou YC, Sun A, Ryo A, Zhou XZ, Yu ZX, Huang HK, Uchida T, Bronson R, Bing G, Li X, et al. 2003. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature 424: 556–561 [DOI] [PubMed] [Google Scholar]
- Littauer UZ, Giveon D, Thierauf M, Ginzburg I, Ponstingl H 1986. Common and distinct tubulin binding sites for microtubule-associated proteins. Proc Natl Acad Sci 83: 7162–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis PA, Howard TH, Castleberry RP, Binder LI 1990. Identification of nuclear tau isoforms in human neuroblastoma cells. Proc Natl Acad Sci 87: 8422–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, Binder LI 1995. Functional implications for the microtubule-associated protein tau: Localization in oligodendrocytes. Proc Natl Acad Sci 92: 10369–10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP 1999. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399: 784–788 [DOI] [PubMed] [Google Scholar]
- Magnani E, Fan J, Gasparini L, Golding M, Williams M, Schiavo G, Goedert M, Amos LA, Spillantini MG 2007. Interaction of tau protein with the dynactin complex. EMBO J 26: 4546–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides V, Massie MR, Feinstein SC, Lew J 2004. Evidence for two distinct binding sites for tau on microtubules. Proc Natl Acad Sci 101: 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Drewes G, Biernat J, Gustke N, Van Lint J, Vandenheede JR, Mandelkow E 1992. Glycogen synthase kinase-3 and the Alzheimer-like state of microtubule-associated protein tau. FEBS Lett 314: 315–321 [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Thies E, Trinczek B, Biernat J, Mandelkow E 2004. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J Cell Biol 167: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R 2004. Template-assisted filament growth by parallel stacking of tau. Proc Natl Acad Sci 101: 10278–10283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margittai M, Langen R 2006. Side chain-dependent stacking modulates tau filament structure. J Biol Chem 281: 37820–37827 [DOI] [PubMed] [Google Scholar]
- Matsuo ES, Shin RW, Billingsley ML, Van deVoorde A, O’Connor M, Trojanowski JQ, Lee VM 1994. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron 13: 989–1002 [DOI] [PubMed] [Google Scholar]
- Mattson MP 2004. Pathways towards and away from Alzheimer’s disease. Nature 430: 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercken M, Fischer I, Kosik KS, Nixon RA 1995. Three distinct axonal transport rates for tau, tubulin, and other microtubule-associated proteins: Evidence for dynamic interactions of tau with microtubules in vivo. J Neurosci 15: 8259–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, et al. 2010. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67: 953–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocanu MM, Nissen A, Eckermann K, Khlistunova I, Biernat J, Drexler D, Petrova O, Schonig K, Bujard H, Mandelkow E, et al. 2008. The potential for β-structure in the repeat domain of tau protein determines aggregation, synaptic decay, neuronal loss, and coassembly with endogenous Tau in inducible mouse models of tauopathy. J Neurosci 28: 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Perderiset M, Francis F, Chelly J, Houdusse A, Milligan RA 2004. Mechanism of microtubule stabilization by doublecortin. Mol Cell 14: 833–839 [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F, Perez M, Baro AM, Avila J 2004. Characterization by atomic force microscopy of Alzheimer paired helical filaments under physiological conditions. Biophys J 86: 517–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Titani K, Ihara Y 1993. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 10: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Kosik KS 1996. The pool of map kinase associated with microtubules is small but constitutively active. Mol Biol Cell 7: 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Sobue K 2009. Specification of neuronal polarity regulated by local translation of CRMP2 and Tau via the mTOR-p70S6K pathway. J Biol Chem 284: 27734–27745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M, Maeda S, Vossel K, Mucke L 2011. The many faces of tau. Neuron 70: 410–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette DA, Parachikova A, Green KN, LaFerla FM 2009. Relevance of transgenic mouse models to human Alzheimer disease. J Biol Chem 284: 6033–6037 [DOI] [PubMed] [Google Scholar]
- Mukrasch MD, Biernat J, von Bergen M, Griesinger C, Mandelkow E, Zweckstetter M 2005. Sites of tau important for aggregation populate {β}-structure and bind to microtubules and polyanions. J Biol Chem 280: 24978–24986 [DOI] [PubMed] [Google Scholar]
- Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M 2009. Structural polymorphism of 441-residue tau at single residue resolution. PLoS Biol 7: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, Lees A, Leung D, McKeith IG, Perry RH, et al. 2007. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis 25: 561–570 [DOI] [PubMed] [Google Scholar]
- Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI 2008. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry 47: 10345–10353 [DOI] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N 2003. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol 162: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RL, Harris P, Kosik KS, Kurnit DM, Donlon TA 1986. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res 387: 271–280 [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH 1999. High-resolution model of the microtubule. Cell 96: 79–88 [DOI] [PubMed] [Google Scholar]
- Obar RA, Collins CA, Hammarback JA, Shpetner HS, Vallee RB 1990. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature 347: 256–261 [DOI] [PubMed] [Google Scholar]
- Oddo S, Billings L, Kesslak JP, Cribbs DH, LaFerla FM 2004. Aβ immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron 43: 321–332 [DOI] [PubMed] [Google Scholar]
- Padilla R, Maccioni RB, Avila J 1990. Calmodulin binds to a tubulin binding site of the microtubule-associated protein tau. Mol Cell Biochem 97: 35–41 [DOI] [PubMed] [Google Scholar]
- Panda D, Samuel JC, Massie M, Feinstein SC, Wilson L 2003. Differential regulation of microtubule dynamics by three- and four-repeat tau: Implications for the onset of neurodegenerative disease. Proc Natl Acad Sci 100: 9548–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasozomenos SC, Binder LI 1987. Phosphorylation determines two distinct species of Tau in the central nervous system. Cell Motil Cytoskeleton 8: 210–226 [DOI] [PubMed] [Google Scholar]
- Park SY, Ferreira A 2005. The generation of a 17 kDa neurotoxic fragment: An alternative mechanism by which tau mediates β-amyloid-induced neurodegeneration. J Neurosci 25: 5365–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Valpuesta JM, Medina M, Montejo de Garcini E, Avila J 1996. Polymerization of tau into filaments in the presence of heparin: The minimal sequence required for tau-tau interaction. J Neurochem 67: 1183–1190 [DOI] [PubMed] [Google Scholar]
- Perl ML, Abrams GS, Boyarski AM, Breidenbach M, Briggs DD, Bulos F, Chinowsky W, Dakin JT, Feldman GJ, Friedberg CE, et al. 1975. Evidence for anomalous lepton production in e+–e− annihilation. Phys Rev Lett 35: 1489 [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al. 2004. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet 13: 703–714 [DOI] [PubMed] [Google Scholar]
- Pickhardt M, Gazova Z, von Bergen M, Khlistunova I, Wang Y, Hascher A, Mandelkow EM, Biernat J, Mandelkow E 2005. Anthraquinones inhibit tau aggregation and dissolve Alzheimer’s paired helical filaments in vitro and in cells. J Biol Chem 280: 3628–3635 [DOI] [PubMed] [Google Scholar]
- Planel E, Richter KE, Nolan CE, Finley JE, Liu L, Wen Y, Krishnamurthy P, Herman M, Wang L, Schachter JB, et al. 2007a. Anesthesia leads to tau hyperphosphorylation through inhibition of phosphatase activity by hypothermia. J Neurosci 27: 3090–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planel E, Tatebayashi Y, Miyasaka T, Liu L, Wang L, Herman M, Yu WH, Luchsinger JA, Wadzinski B, Duff KE, et al. 2007b. Insulin dysfunction induces in vivo tau hyperphosphorylation through distinct mechanisms. J Neurosci 27: 13635–13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollanen MS, Markiewicz P, Goh MC 1997. Paired helical filaments are twisted ribbons composed of two parallel and aligned components: Image reconstruction and modeling of filament structure using atomic force microscopy. J Neuropathol Exp Neurol 56: 79–85 [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD 1998. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol 43: 815–825 [DOI] [PubMed] [Google Scholar]
- Reynolds MR, Berry RW, Binder LI 2007. Nitration in neurodegeneration: Deciphering the “Hows” “nYs”. Biochemistry 46: 7325–7336 [DOI] [PubMed] [Google Scholar]
- Reynolds CH, Garwood CJ, Wray S, Price C, Kellie S, Perera T, Zvelebil M, Yang A, Sheppard PW, Varndell IM, et al. 2008. Phosphorylation regulates tau interactions with Src homology 3 domains of phosphatidylinositol 3-kinase, phospholipase Cγ1, Grb2, and Src family kinases. J Biol Chem 283: 18177–18186 [DOI] [PubMed] [Google Scholar]
- Rissman RA, Poon WW, Blurton-Jones M, Oddo S, Torp R, Vitek MP, LaFerla FM, Rohn TT, Cotman CW 2004. Caspase-cleavage of tau is an early event in Alzheimer disease tangle pathology. J Clin Invest 114: 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L 2007. Reducing endogenous tau ameliorates amyloid β-induced deficits in an Alzheimer’s disease mouse model. Science 316: 750–754 [DOI] [PubMed] [Google Scholar]
- Roger B, Al-Bassam J, Dehmelt L, Milligan RA, Halpain S 2004. MAP2c, but not tau, binds and bundles F-actin via its microtubule binding domain. Curr Biol 14: 363–371 [DOI] [PubMed] [Google Scholar]
- Sack S, Muller J, Marx A, Thormahlen M, Mandelkow EM, Brady ST, Mandelkow E 1997. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry 36: 16155–16165 [DOI] [PubMed] [Google Scholar]
- Salminen A, Ojala J, Kaarniranta K, Hiltunen M, Soininen H 2011. Hsp90 regulates tau pathology through co-chaperone complexes in Alzheimer’s disease. Prog Neurobiol 93: 99–110 [DOI] [PubMed] [Google Scholar]
- Samsonov A, Yu JZ, Rasenick M, Popov SV 2004. Tau interaction with microtubules in vivo. J Cell Sci 117: 6129–6141 [DOI] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science 309: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarella RA, Skiniotis G, Goldie KN, Tittmann P, Gross H, Mandelkow EM, Mandelkow E, Hoenger A 2004. Surface-decoration of microtubules by human tau. J Mol Biol 339: 539–553 [DOI] [PubMed] [Google Scholar]
- Sarkar M, Kuret J, Lee G 2008. Two motifs within the tau microtubule-binding domain mediate its association with the hsc70 molecular chaperone. J Neurosci Res 86: 2763–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. 2007. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447: 453–457 [DOI] [PubMed] [Google Scholar]
- Schaap IA, Hoffmann B, Carrasco C, Merkel R, Schmidt CF 2007. Tau protein binding forms a 1 nm thick layer along protofilaments without affecting the radial elasticity of microtubules. J Struct Biol 158: 282–292 [DOI] [PubMed] [Google Scholar]
- Schneider A, Mandelkow E 2008. Tau-based treatment strategies in neurodegenerative diseases. Neurotherapeutics 5: 443–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Biernat J, von Bergen M, Mandelkow E, Mandelkow EM 1999. Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry 38: 3549–3558 [DOI] [PubMed] [Google Scholar]
- Schweers O, Schonbrunn-Hanebeck E, Marx A, Mandelkow E 1994. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for β-structure. J Biol Chem 269: 24290–24297 [PubMed] [Google Scholar]
- Schweers O, Mandelkow EM, Biernat J, Mandelkow E 1995. Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc Natl Acad Sci 92: 8463–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabrook GR, Ray WJ, Shearman M, Hutton M 2007. Beyond amyloid: The next generation of Alzheimer’s disease therapeutics. Mol Interv 7: 261–270 [DOI] [PubMed] [Google Scholar]
- Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E 2002. Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J 21: 4896–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant N, Bretteville A, Hamdane M, Caillet-Boudin ML, Grognet P, Bombois S, Blum D, Delacourte A, Pasquier F, Vanmechelen E, et al. 2008. Biochemistry of Tau in Alzheimer’s disease and related neurological disorders. Expert Rev Proteomics 5: 207–224 [DOI] [PubMed] [Google Scholar]
- Seubert P, Mawal-Dewan M, Barbour R, Jakes R, Goedert M, Johnson GV, Litersky JM, Schenk D, Lieberburg I, Trojanowski JQ, et al. 1995. Detection of phosphorylated Ser262 in fetal tau, adult tau, and paired helical filament tau. J Biol Chem 270: 18917–18922 [DOI] [PubMed] [Google Scholar]
- Shankar GM, Bloodgood BL, Townsend M, Walsh DM, Selkoe DJ, Sabatini BL 2007. Natural oligomers of the Alzheimer amyloid-β protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J Neurosci 27: 2866–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Miura-Shimura Y, Kosik KS 2004a. Binding of tau to heat shock protein 27 leads to decreased concentration of hyperphosphorylated tau and enhanced cell survival. J Biol Chem 279: 17957–17962 [DOI] [PubMed] [Google Scholar]
- Shimura H, Schwartz D, Gygi SP, Kosik KS 2004b. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem 279: 4869–4876 [DOI] [PubMed] [Google Scholar]
- Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, Stevens JT, Munch J, Baker D, Eisenberg D 2011. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 475: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillen A, Leroy A, Wieruszeski JM, Loyens A, Beauvillain JC, Buee L, Landrieu I, Lippens G 2005. Regions of tau implicated in the paired helical fragment core as defined by NMR. Chembiochem 6: 1849–1856 [DOI] [PubMed] [Google Scholar]
- Sillen A, Barbier P, Landrieu I, Lefebvre S, Wieruszeski JM, Leroy A, Peyrot V, Lippens G 2007. NMR investigation of the interaction between the neuronal protein tau and the microtubules. Biochemistry 46: 3055–3064 [DOI] [PubMed] [Google Scholar]
- Sluchanko NN, Seit-Nebi AS, Gusev NB 2009. Phosphorylation of more than one site is required for tight interaction of human tau protein with 14–3–3ζ. FEBS Lett 583: 2739–2742 [DOI] [PubMed] [Google Scholar]
- Smet C, Sambo AV, Wieruszeski JM, Leroy A, Landrieu I, Buee L, Lippens G 2004. The peptidyl prolyl cis/trans-isomerase Pin1 recognizes the phospho-Thr212–Pro213 site on Tau. Biochemistry 43: 2032–2040 [DOI] [PubMed] [Google Scholar]
- Sontag E, Nunbhakdi-Craig V, Lee G, Brandt R, Kamibayashi C, Kuret J, White CL 3rd, Mumby MC, Bloom GS 1999. Molecular interactions among protein phosphatase 2A, tau, and microtubules. Implications for the regulation of tau phosphorylation and the development of tauopathies. J Biol Chem 274: 25490–25498 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B 1998. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci 95: 7737–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT 2009. Tau pathophysiology in neurodegeneration: A tangled issue. Trends Neurosci 32: 150–159 [DOI] [PubMed] [Google Scholar]
- Stamer K, Vogel R, Thies E, Mandelkow E, Mandelkow EM 2002. Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J Cell Biol 156: 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner B, Mandelkow EM, Biernat J, Gustke N, Meyer HE, Schmidt B, Mieskes G, Soling HD, Drechsel D, Kirschner MW, et al. 1990. Phosphorylation of microtubule-associated protein tau: Identification of the site for Ca2(+)-calmodulin dependent kinase and relationship with tau phosphorylation in Alzheimer tangles. EMBO J 9: 3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoothoff WH, Johnson GV 2005. Tau phosphorylation: Physiological and pathological consequences. Biochim Biophys Acta 1739: 280–297 [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Goedert M, Weisgraber KH, Dong LM, Jakes R, Huang DY, Pericak-Vance M, Schmechel D, Roses AD 1994. Isoform-specific interactions of apolipoprotein E with microtubule-associated protein tau: Implications for Alzheimer disease. Proc Natl Acad Sci 91: 11183–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan A, Nesslany F, Violet M, Begard S, Loyens A, Talahari S, Mansuroglu Z, Marzin D, Sergeant N, Humez S, et al. 2011. Nuclear tau, a key player in neuronal DNA protection. J Biol Chem 286: 4566–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Qureshi HY, Cafferty PW, Sobue K, Agarwal-Mawal A, Neufield KD, Paudel HK 2002. Glycogen synthase kinase-3β is complexed with tau protein in brain microtubules. J Biol Chem 277: 11933–11940 [DOI] [PubMed] [Google Scholar]
- Sydow A, Van der Jeugd A, Zheng F, Ahmed T, Balschun D, Petrova O, Drexler D, Zhou L, Rune G, Mandelkow E, et al. 2011. Tau-induced defects in synaptic plasticity, learning, and memory are reversible in transgenic mice after switching off the toxic Tau mutant. J Neurosci 31: 2511–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima A, Murayama M, Murayama O, Kohno T, Honda T, Yasutake K, Nihonmatsu N, Mercken M, Yamaguchi H, Sugihara S, et al. 1998. Presenilin 1 associates with glycogen synthase kinase-3β and its substrate tau. Proc Natl Acad Sci 95: 9637–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimukai H, Grundke-Iqbal I, Iqbal K 2005. Up-regulation of inhibitors of protein phosphatase-2A in Alzheimer’s disease. Am J Pathol 166: 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwel D, Dewachter I, Van Leuven F 2002. Axonal transport, tau protein, and neurodegeneration in Alzheimer’s disease. Neuromolecular Med 2: 151–165 [DOI] [PubMed] [Google Scholar]
- Thies E, Mandelkow EM 2007. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J Neurosci 27: 2896–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa E, Santa-Maria I, Moreno F, Lim F, Perez M, Avila J 2009. Binding of Hsp90 to tau promotes a conformational change and aggregation of tau protein. J Alzheimers Dis 17: 319–325 [DOI] [PubMed] [Google Scholar]
- Umeyama T, Okabe S, Kanai Y, Hirokawa N 1993. Dynamics of microtubules bundled by microtubule associated protein 2C (MAP2C). J Cell Biol 120: 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utton MA, Noble WJ, Hill JE, Anderton BH, Hanger DP 2005. Molecular motors implicated in the axonal transport of tau and α-synuclein. J Cell Sci 118: 4645–4654 [DOI] [PubMed] [Google Scholar]
- Veeranna, Yang DS, Lee JH, Vinod KY, Stavrides P, Amin ND, Pant HC, Nixon RA 2009. Declining phosphatases underlie aging-related hyperphosphorylation of neurofilaments. Neurobiol Aging 2010 10.1016/j.neurobiolaging.2009.1012.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E 2000. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming β structure. Proc Natl Acad Sci 97: 5129–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M, Barghorn S, Li L, Marx A, Biernat J, Mandelkow EM, Mandelkow E 2001. Mutations of tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local β-structure. J Biol Chem 276: 48165–48174 [DOI] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E 2009. Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing. Hum Mol Genet 18: 4153–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Hong WK, Dohmae N, Takio K, Morishima-Kawashima M, Ihara Y 2004. Molecular aging of tau: Disulfide-independent aggregation and non-enzymatic degradation in vitro and in vivo. J Neurochem 90: 1302–1311 [DOI] [PubMed] [Google Scholar]
- Wegmann S, Jung YJ, Chinnathambi S, Mandelkow EM, Mandelkow E, Muller DJ 2010. Human Tau isoforms assemble into ribbon-like fibrils that display polymorphic structure and stability. J Biol Chem 285: 27302–27313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW 1975. A protein factor essential for microtubule assembly. Proc Natl Acad Sci 72: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberg RC 1972. Microtubule formation in vitro in solutions containing low calcium concentrations. Science 177: 1104–1105 [DOI] [PubMed] [Google Scholar]
- Wendt TG, Volkmann N, Skiniotis G, Goldie KN, Muller J, Mandelkow E, Hoenger A 2002. Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J 21: 5969–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman IT, Gervasio OL, Cullen KM, Guillemin GJ, Jeong EV, Witting PK, Antao ST, Minamide LS, Bamburg JR, Goldsbury C 2009. Activated actin-depolymerizing factor/cofilin sequesters phosphorylated microtubule-associated protein during the assembly of alzheimer-like neuritic cytoskeletal striations. J Neurosci 29: 12994–13005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H, Drewes G, Biernat J, Mandelkow EM, Mandelkow E 1992. Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J Cell Biol 118: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R, Usardi A, Hanger DP, Anderton BH 2008. Membrane-bound β-amyloid oligomers are recruited into lipid rafts by a fyn-dependent mechanism. FASEB J 22: 1552–1559 [DOI] [PubMed] [Google Scholar]
- Wilson DM, Binder LI 1995. Polymerization of microtubule-associated protein tau under near-physiological conditions. J Biol Chem 270: 24306–24314 [DOI] [PubMed] [Google Scholar]
- Wilson DM, Binder LI 1997. Free fatty acids stimulate the polymerization of tau and amyloid β peptides. In vitro evidence for a common effector of pathogenesis in Alzheimer’s disease. Am J Pathol 150: 2181–2195 [PMC free article] [PubMed] [Google Scholar]
- Wilson L, Jordan MA 2004. New microtubule/tubulin-targeted anticancer drugs and novel chemotherapeutic strategies. J Chemother 16: 83–85 [DOI] [PubMed] [Google Scholar]
- Wischik CM, Novak M, Edwards PC, Klug A, Tichelaar W, Crowther RA 1988. Structural characterization of the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci 85: 4884–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS 2009. Tau mutations in neurodegenerative diseases. J Biol Chem 284: 6021–6025 [DOI] [PubMed] [Google Scholar]
- Wolozin BL, Pruchnicki A, Dickson DW, Davies P 1986. A neuronal antigen in the brains of Alzheimer patients. Science 232: 648–650 [DOI] [PubMed] [Google Scholar]
- Wood JG, Mirra SS, Pollock NJ, Binder LI 1986. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau). Proc Natl Acad Sci 83: 4040–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Cirrito JR, Stewart FR, Jiang H, Finn MB, Holmes B, Binder LI, Mandelkow EM, Diamond MI, Lee V.M.Y, Holtzman DM 2011. In vivo microdialysis reveals age-dependent decrease of brain interstitial tau levels in P301S human tau transgenic mice. J Neurosci 31: 13110–13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempel H, Thies E, Mandelkow E, Mandelkow EM 2010. Aβ oligomers cause localized Ca(2+) elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J Neurosci 30: 11938–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]