Abstract
Within the last two decades, genomics and bioinformatics have profoundly impacted our understanding of the molecular mechanisms of Parkinson's disease (PD). From the description of the first PD gene in 1997 until today, we have witnessed the emergence of new technologies that have revolutionized our concepts to identify genetic mechanisms implicated in human health and disease. Driven by the publication of the human genome sequence and followed by the description of detailed maps for common genetic variability, novel applications to rapidly scrutinize the entire genome in a systematic, cost-effective manner have become a reality. As a consequence, about 30 genetic loci have been unequivocally linked to the pathogenesis of PD highlighting essential molecular pathways underlying this common disorder. Herein we discuss how neurogenomics and bioinformatics are applied to dissect the nature of this complex disease with the overall aim of developing rational therapeutic interventions.
Applications to scrutinize the entire genome in a systematic, cost-effective manner have revealed about 30 genetic loci that are unequivocally linked to the pathogenesis of Parkinson's disease.
“Knowing is not enough; we must apply. Willing is not enough; we must do.”
The pathogenesis of Parkinson's disease (PD) as we understand it today includes a broad spectrum of metabolic pathways, from oxidative stress caused by mitochondrial dysfunction, inflammation, abnormal protein metabolism, and aging (reviewed in Dawson and Dawson 2003; Dauer and Przedborski 2003). Most of these pathophysiological links with PD are the result of studying the functional consequences of gene mutations implicated in familial PD. With the introduction of modern genomic technologies numerous genetic risk loci involved in the more common nonfamilial form of PD are also being uncovered identifying novel pathways, raising new research questions and hopes to better understand and treat this neurodegenerative condition more effectively.
In this article, we will introduce some of the main concepts and discussions in PD genetics, outline the current status of PD genomics, discuss the impact of new sequencing technologies, and chart the necessary steps for translating these findings into targeted therapeutics.
“MENDELIAN” VERSUS COMPLEX DISEASE: SIMILAR IDEAS, DIFFERENT CONCEPTS
For simplicity, geneticists have separated genetic diseases into two main categories. The first one refers to “Mendelian” diseases such as Huntington's disease, muscular dystrophy, and cystic fibrosis. “Mendelian” diseases are defined as the classical familial forms of disease in which the underlying genetic defect causes disease in a large proportion of mutation carriers and therefore typical inheritance patterns can be inferred. Previously, this disease category was the mainstay of genetic research; this is primarily because of the fact that until recently the standard genetic approach for disease gene discovery was a linkage study, a technique that relies on ascertaining large families with multiple affected individuals. For the most part, this approach was very successful with nearly 3000 Mendelian disorders deciphered to date (Lander 2011); however, a linkage study design is only applicable to a small proportion of human ailments that are typically rather rare in the general population. Studying the genetics of complex diseases, which constitute the second disease category, proved to be more challenging. Indeed, PD is a good example of a complex neurodegenerative disease; only a small subset of PD patients report a positive family history of PD, and it is not surprising that the first mutations identified as a cause for PD were identified in this small subset of patients. In the vast majority of patients, however, a family history of PD is absent and the etiology is less clear. It is in these patients that most of the research in the last few years has been focused.
The differences in studying complex disease as opposed to familial disease lie within the techniques used, the study designs, the amount at which a particular genetic variant confers risk to disease, and the mechanistic ideas of disease pathogenesis. To illustrate these concepts we have to introduce some technical terms.
The distinction between association and causation. When a genetic variant is investigated for a potential contribution to disease, a number of questions need to be addressed. Is the variant commonly present in the population? If so, is the frequency of this variant significantly different in cases versus controls? If this is the observation, significant association with a particular phenotype has been determined. It is important to emphasize that establishing significant association should not be misconstrued as drawing inferences about causation. For example, the APOE ε4 allele on chromosome 19 has been consistently associated with increased risk for developing Alzheimer's disease, but carrying this risk allele is neither necessary nor sufficient to cause disease. Although an association signal sometimes implies genes or genetic regions that play a causative role in the pathogenesis of disease, it is not appropriate to assume that this applies to all instances in which associations are observed.
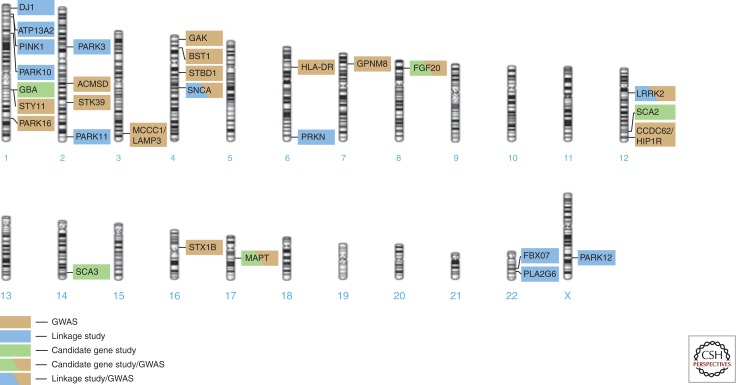
“Common disease–common variant hypothesis” versus “common disease–rare variants hypothesis.” In contrast to monogenic diseases in which a single mutation is sufficient to cause disease, complex diseases are thought to be caused by a combination of multiple genetic, environmental, and stochastic factors. Two distinctive concepts have been postulated for the detection of genetic variants underlying common, complex diseases. The “common disease–common variant hypothesis” posits that multiple, common small-risk variants of small effect size interact to cause common disease (Reich and Lander 2001). This hypothesis is the core basis for genome-wide association studies (GWAS), a study design that relies on testing several hundred thousand common genetic variants throughout the human genome in large case-control cohorts. Over the past few years, hundreds of new gene loci and pathways, including sixteen PD loci (Fig. 1) (Simon-Sanchez et al. 2009; Consortium IPDG 2011; Consortium IPDG unpubl.), have been implicated with various human disease traits using a GWAS design (an updated catalog of identified genetic risk loci can be found at www.genome.gov/gwastudies/).
Figure 1.
An overview of the genetic loci implicated in the pathogenesis of PD. The position of each locus relative to the ideogram of each chromosome is depicted. The background color of each box indicates the method that was used to identify this locus. Abbreviation: GWAS, genome-wide association study.
Despite the interest of the immediate past, heritability estimates have shown that large proportions of genetic risk underlying complex disease have not yet been explained (Manolio et al. 2009). In PD for example only ∼60% of heritability is understood, depending on the population studied (Consortium IPDG 2011). This “missing heritability” is at the center of much of the current debate, and possible explanations that have been brought forward include lack of power to detect common low-risk variants, rare variants, gene–gene interactions, gene–environment interactions, structural variants such as deletions or duplications, and inversions (Manolio et al. 2009). In particular the “common disease–rare variants hypothesis” has gained much traction, mainly because of the introduction of advanced sequencing technologies that allows cost-effective sequencing of entire genomes. An early lesson learned from these next-generation sequencing technologies is that there are numerous rare variants in the human genome, which have not yet been systematically explored. It is hoped that over the next couple of years more insights will be gained into the pathogenic relevance of rare genetic variability.
DISSECTING THE GENETICS OF COMPLEX DISEASE: THE REVOLUTION OF GENOMICS AND BIOINFORMATICS
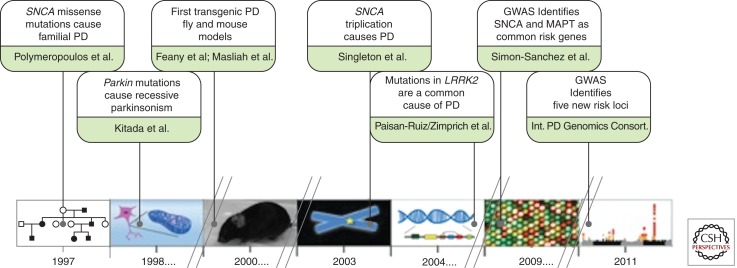
Genomic research in the past few years has been defined by the rapid integration of technological advances as the immediate ramifications of the human genome project. To reconstruct the developments, successes and challenges in genomic PD research, we will point out some of the past milestones of the genomics revolution, and discuss the events and developments achieved so far (Fig. 2 illustrates the selected landmark discoveries in genomic PD research).
Figure 2.
Highlights of key genomic discoveries in PD over the past decade and a half.
The year 1997 marks the starting point for PD genomics. Using a linkage study approach, Polymeropoulos et al. (1997) reported the discovery of missense mutations in the SNCA gene, coding for α-synuclein, to underlie a rare familial form of PD. This finding was crucial in that it provided clear evidence that there are genetic forms of disease, a view of which was evolving from the concept that PD was only a nongenetic disease. In consequence of this seminal work, the availability of a disease-causing gene allowed the generation of cell- and animal-based model systems for studying the functional mechanisms in disease pathogenesis (Feany and Bender 2000; Masliah et al. 2000). Moreover, subsequent screening studies showed that variability at the SNCA locus not only plays a role in this familial form of PD, but is also associated with risk for disease in sporadic cases (Farrer et al. 2001; Maraganore et al. 2006). Soon after the discovery of SNCA other “Mendelian” PD genes (Parkin, PINK1, DJ1, and LRRK2) were revealed using a linkage mapping design (KItada et al. 1998; Valente et al. 2001; Bonifati et al. 2003; Paisan-Ruiz et al. 2004; Zimprich et al. 2004).
In contrast to the advances in dissecting the genetics of rare familial forms of PD, the genetic factors influencing common sporadic cases remained enigmatic. The publication of the human genome sequence in 2001 marked an exciting turning point (Lander et al. 2001; Venter et al. 2001). For the first time researchers had the opportunity to examine the sequence of an entire human genome, and four years later a detailed catalog of common genetic variants (the HapMap) became available (http://hapmap.ncbi.nlm.nih.gov/). This catalog was the starting point for studying the genomics of complex diseases. Soon microarray platforms for genotyping hundreds of thousands of these common variants throughout the human genome were developed and genome-wide association testing became a reality. The revolutionary aspect of this new technology was that it provided a cost effective tool to rapidly scan hundreds of thousands of variants in the genome in thousands of individuals. In PD, genome-wide association strategies have been remarkably successful; because of this new technology the number of risk loci implicated in PD pathogenesis has doubled over the last 3 years (Fig. 2).
As new sequencing technologies for sequencing entire genomes emerge, the era of GWAS is diminishing in importance. In the same way as GWAS was the standard technology used to search for common risk variants, next-generation sequencing technologies are going to determine the exploration of rare genetic variability and of structural genomic rearrangements. With costs of sequencing dropping and the speed of genomic data generation reaching unprecedented scales, data handling and analysis have become big challenges in modern PD research. In fact, the generation of genomic data has accelerated so rapidly that the amount of data produced exceeds the exponential growth of computer processing speed known as Moore's law. In other words, the bottleneck for genomic discovery is no longer generating extensive datasets, but rather storing and analyzing the information; this problem has even generated interest in the lay press (Pollack 2011). To put the advances of genomics into perspective, it took several thousand researchers 13 years to sequence the first human genome at a cost of $3 billion (http://www.genome.gov/); today, one technician can sequence an entire genome in less than a week for about $5000. With even faster sequencing technologies about to be released, the $1000 genome, a commonly used benchmark at which genomic sequencing is considered economical for routine diagnostic testing, is imminent (Mardis 2006).
At the same time as genomics hastily moves on, the innovation of computational medicine and its diverse applications, commonly referred to as bioinformatics, is confronted with challenging demands to develop user-friendly methods to parse, analyze, and share genomic data. Automation of sequence data filtering, alignment with reference genomes, variant calling and statistical analyses across various platforms still pose a daunting task. Moreover, in depth analyses of epistatic effects, or gene–gene interactions, as well as investigations of gene–environment interactions, which are at the center of research ambitions aimed at resolving the complete genomic architecture of complex diseases, depend on sophisticated bioinformatics algorithms.
INFORMATICS OF GENOMICS, TRANSCRIPTOMICS, PROTEOMICS, AND METABOLOMICS
Developing comprehensive informatics tools requires, concurrent with the development of genome-centric algorithms, a deeper understanding of the functional consequences of genetic variability on disease vulnerability. In other words, how do we translate the relatively invariant nature of our genomes into what is surely a dynamic, evolving risk of disease? This understanding involves connecting genetic information onto the full molecular space, which includes genetics, epigenetics (i.e., noncoding changes such DNA methylation, chromatin conformation, noncoding RNAs—all affecting the read-out of the genome), gene expression, protein expression, and the resultant metabolic output of all of these processes. So far, technological advances in gene expression analyses (RNA expression or transcriptomics) have paralleled those in genome analyses, as similar chemistries allow for the rapid, high-throughput ascertainment of both DNA and RNA. For example, studies have begun to link data from PD GWAS to specific gene expression and epigenetic changes in brain tissue, as was recently reported by the International Parkinson’s Disease Genomics Consortium (2011). However, moving forward will involve a more complex understanding of the relationships between the static genome, the epigenome, and RNA expression, including regulatory RNAs such as small interfering RNA (siRNA) and microRNA. Additionally, the technologies for detecting and quantifying nucleic acids are progressing, as are methods for detecting and quantifying proteins (proteomics) and metabolites (metabolomics). Thus, the next step forward is coupling the information in the nucleic acid space to the proteomic (both at the transcriptional and posttranscriptional levels) and the metabolomic spaces. Collectively, we refer to the science and technology of measuring these biological molecules as “-omics”—genomics, epigenomics, transcriptomics, proteomics, and metabolomics. The ongoing revolution in “-omics” technologies, coupled with advances in bioinformatics, has greatly expanded our ability to link genetic architecture to its functional output, namely the expression of genes, proteins, and metabolites. Thus, a key challenge to the field is to detect and quantitatively measure the full complement of biological molecules and to integrate these into a meaningful understanding of both normal function and dysfunction (e.g., disease).
Although “-omics”-driven research holds great promise in understanding disease biology, enthusiasm should be tempered. The current state-of-the-art technologically to meaningfully interpret large and diverse datasets is limiting. We greatly expand the complexity of analyses as we integrate data across multiple domains, which includes not just those from the molecular space (e.g., RNA, protein, and metabolite expression), but also the clinical space, which includes neuroimaging among other sources. For example, how does genetic risk translate from the subcellular organelle (e.g., mitochondrion), cellular (e.g., midbrain dopaminergic neuron), circuit (e.g., nigrostriatal system), organ (central nervous system) levels to an individual's risk for developing PD? This is further complicated by the complex interactions we as individuals have with other organisms, be they the microbiome of our gastrointestinal systems to the human interactions that create our social and environmental communities. Indeed, the relevance of this holistic view is exemplified by the pioneering work of neuropathologist, Heiko Braak. On examining human postmortem tissues, Braak proposed a new framework for PD wherein the disease process begins outside the midbrain, the traditional locus of vulnerable nigrostriatal dopaminergic neuronal cell bodies that are the uniformly affected in PD (Braak et al. 2002, 2003a). Braak's most recent concept is that PD could be initiated in the enteric nervous system with ascending pathology to involve the dorsal motor nucleus of the vagus (Braak et al. 2003b, reviewed in Hawkes et al. 2010). As we test new hypotheses of pathogenesis such as these and others additional data will be needed, which will further increase informatic complexity. One possibility is that the informatic challenges will require the integration of -omics data with other data to derive empirically testable biological networks—those that explain an individual's risk for PD. Once risk is stratified, we may then be positioned to consider earlier interventions that could alter the natural history of PD.
IDENTIFICATION OF ABERRANT NETWORK ACTIVITIES
As discussed above, increasing evidence indicates that complex diseases such as PD are associated with multiple genetic polymorphisms that are postulated to affect biological networks (Schadt 2009; Tan et al. 2009; Meyerson et al. 2010). Biological networks represent series of actions among molecules that lead to a certain specific change in a cell (Croft et al. 2011). These networks may subserve many categories of cellular activities (Chang et al. 2009; Wang et al. 2009), including cell signaling networks, protein–protein interactions, and metabolic networks (Kim et al. 2010). Identifying aberrancies involving small numbers of genes in specific biological networks may lead to more precise diagnosis and treatment of a disease. However, because tens or hundreds of genes are often involved, conventional experimental systems developed for identifying aberrant gene(s) are inadequate for deciphering aberrancies in network activity. High throughput technologies enable the simultaneous detection of a large number of alterations in molecular components, or nodes in network parlance. As noted above, these high throughput technologies were developed to fill this gap and the first successful use of technology was in DNA sequencing (Sanger et al. 1977), which later developed into a gambit of genomics. As other -omics technologies developed and enabled the simultaneous detection of a large number of alterations in protein expression and metabolites, the correlations and dependencies between molecular components have become complex. However, these studies, when understood in a network context, offer a systems level perspective on processes underling disease initiation and progression.
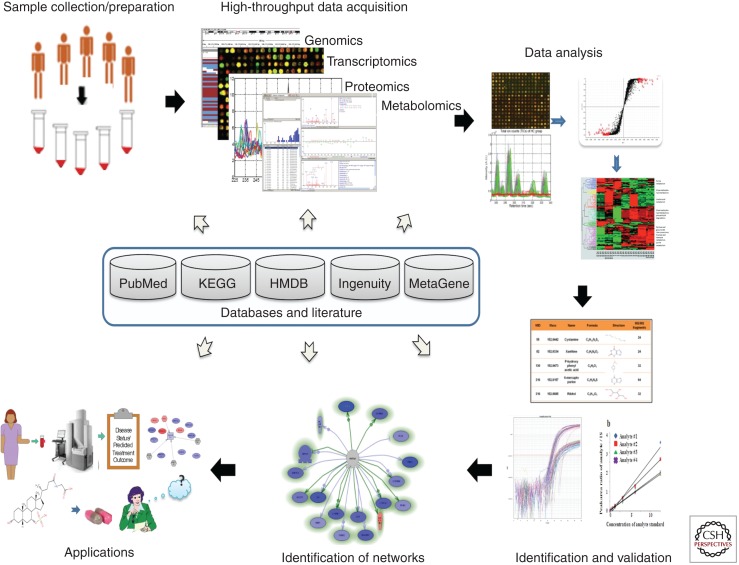
Analyses of high dimensional data using robust new informatics tools allows for the integration of these different data sources to yield new information about network functions and dysfunctions. Figure 3 depicts the workflow of a typical study involving high dimensional -omics data to identify aberrant network activities. Such data analyses often reveal that our current understanding of molecular and chemical biology underlying cellular functions remains incomplete (Ochs et al. 2011). However, an increasingly greater number of open-access databases complement the analysis of aberrant network activities from these high dimensional data (Peri et al. 2003; Kanehisa et al. 2004; Schaefer et al. 2009). In addition, numerous tools have been developed for visually exploring and analyzing biological networks, including Cytoscape (Smoot et al. 2010), VisANT (Hu et al. 2009), GeneGO (http://www.genego.com/), Ingenuity (http://www.ingenuity.com/), and Pathway Studio (Nikitin et al. 2003).
Figure 3.
A hypothetical systems based approach to identify aberrant networks of disease. Data (including biological, clinical, imaging) and samples are collected from a population. High-dimensional -omics data are acquired, integrated with clinical data, analyzed, and validated to identify networks involved in disease. Genomics will predict aberrant networks, whereas transcriptomics, proteomics, and metabolomics will report the outcomes of these networks. In turn, these networks and the aberrant nodes that are perturbed in disease can then be used to develop biomarkers, prognostic markers (e.g., markers that report disease progress or therapeutic efficacy), and rational therapeutics. The process is not inherently unidirectional nor is it intended to be single pass. Instead, as technologies improve, the process can be employed in an iterative fashion to refine nodes within aberrant disease networks and to generate better biomarkers, targets, and therapies. The approach is predicated on robust bioinformatics, and analytics that are critical to our abilities translate high-dimensional data to our understanding of disease and its treatment. (Image is from Wang et al. 2012; reprinted, with permission, from the author.)
In recent years, questions have been raised as to how genetic differences between individuals lead to differences in disease networks and, thus, to differences in phenotypes (Taylor et al. 2009; Vaske et al. 2010). Data driven computational models, such as PARADIGM (Vaske et al. 2010) and ResponseNet (Yeger-Lotem et al. 2009) among others (Amit et al. 2009; Tan et al. 2009; Kreeger and Lauffenburger 2010; Chien et al. 2011), have been especially useful in this area. For example, progress has been made in applying information about aberrant networks to the identification of functional modules; that is, groups of biological entities (e.g., gene, protein) that perform biological tasks (e.g., protein degradation) that are dependent on each constituent part (Qiu et al. 2009; Wu et al. 2010). However, sequenced mutations, copy number alterations, gene fusion events, or epigenetic changes are not well represented in these models. Nevertheless, information derived from putative aberrant network activities will facilitate the detection of biomarkers (Singh et al. 2009), the identification of novel drug targets (Andre et al. 2009), the classification of disease types (Gatza et al. 2010), and the prediction of clinical outcomes (Taylor et al. 2009; Cerami et al. 2010; Chen et al. 2010; Gatza et al. 2010). Increasing use of -omics data driven methodologies for identifying biomarkers and therapeutic targets will also include machine learning methods (Andre et al. 2009; Singh et al. 2009), graph theory (Taylor et al. 2009), and statistical methods (Singh et al. 2009).
Obviously, understanding of the aberrancies in biological networks responsible for complex diseases is far from complete and the use of high dimensional data together with large-scale biological databases (e.g., protein–protein interaction and pathway databases) will be crucial for uncovering aberrant biological processes. However, the challenge is to accommodate the large volume of -omics data, which is growing exponentially. Additionally, much work needs to be done to identify the subnetworks of metabolic reactions associated with diseases such as PD. Moreover, a reliable computational approach to identify subnetwork-associated disease processes is currently limited by the incompleteness of the available interactome maps and limitations of the existing tools.
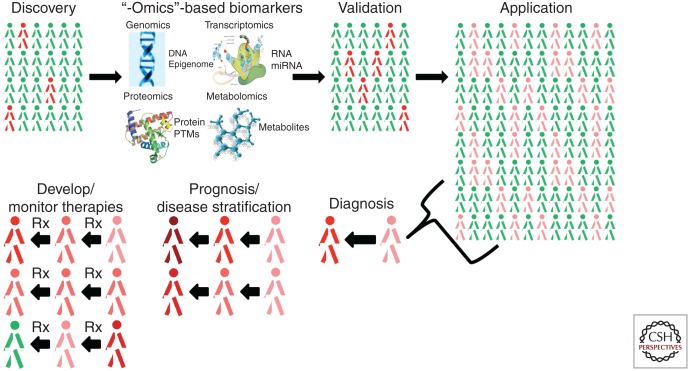
As widely acknowledged, gaining an integrated understanding of the interactions among the genome, proteome, metabolome, and environment as mediated by the underlying cellular network, may offer a basis for future advances. However, some of the most difficult problems in this area include discovering dynamic (rather than static) processes in cells, connecting molecular level network activities to functional behavior at the cellular level, developing data-driven computational models that reflect the causal relationships between molecules (including those designated as drug targets), biomarkers (as potential read-outs of network modulation as would be the case with a disease modifying therapy), measuring the consequent changes that occur in cellular dynamic processes, and predicting the impact of an intervention on the system to effect a change consistent with a beneficial outcome. An especially challenging focus of importance is to find ways to model changes in biological entities which could affect the dynamics of the biological process using large scale and diverse -omics data. To accommodate these challenges, network-based research is shifting toward integrated multiple networks or networks composed of heterogeneous large-scale data elements. To process large-scale -omics data, we need to develop next-generation algorithms and tools to study the relationships between aberrant human genes, proteins, and interactome networks. It is in this context that we delineate new disease-associated biological molecules in relation to disease-specific networks, that we understand how network perturbations can lead to disease, and that we use this knowledge to develop better diagnostics and therapeutics for disease. It is in the integration of these various datasets both temporally and spatially that we begin to see the emergent properties of disease-specific networks and that we then use this knowledge to modify disease natural history (Fig. 4).
Figure 4.
Application of “-omics” based biomarker strategy to discover, validate, and apply molecular profiles to disease diagnosis, prognosis, and therapeutic development. Biomarker discovery efforts follow a predictable model. A discovery cohort of case (red) and control (green) subjects is amassed. Biological samples along with clinical, demographic, and other data are collected. High-dimensional genomic, transcriptomic, proteomic, and metabolomic data are generated and integrated with clinical data to elucidate the dynamic networks and their critical nodes that contribute to risk and evolution of disease. These pathways are then validated in a separate cohort of cases and controls. Robust, sensitive, and specific profiles can then be applied on a population scale to provide readouts for individuals' risk (pink) for disease. These profiles can be informative to disease diagnosis (pink to red), to prognosis and disease stratification (affected individuals with different temporal progression and severity), and in developing and monitoring therapies that slow, halt, or reverse disease progression. Additional historical (environment, lifestyle), clinical, and imaging data (e.g., PET, SPECT) will be integrated with molecular pathway data that will also be informative in disease diagnosis, stratification, and therapies. (PTMs, posttranslational modifications.) Images of myoglobin structure (http://en.wikipedia.org/wiki/File:Myoglobin.png) and ribosome/mRNA translation (http://en.wikipedia.org/wiki/File:Ribosome_mRNA_translation_en.svg) have been released to the public domain.
INFORMATICS IN BIOMARKER DISCOVERY
In the era of -omics, moving forward in the clinical management of PD will require the development of specific and sensitive biomarkers that could detect disease early, define a prognostic trajectory of disease, and/or provide an indicator of response to therapy. Ideally, these biomarkers would be present early enough in the disease course that interventions could halt progression or even reverse damage. Risk alleles in and of themselves are biomarkers, measurable entities that denote risk for disease; however, they do so in a nonspecific and nonsensitive manner. The technologies outlined above provide a way forward as we link genetic information to the other molecular and organismal levels, translating disease risk to disease-specific networks. In fact, a number of studies have used various -omics approaches to identify potential PD biomarkers (reviewed in Caudle et al. 2010). As we increase our understanding of these disease-specific networks, we can begin to identify other tissues which may be affected and which could be used as surrogates for on-going monitoring of disease processes within the CNS. For example, induced pluripotent stem (iPS) cells derived from patients may become an important resource not only for understanding disease biology (Park et al. 2008) and fueling biomarker elucidation, but also as a strategic part of therapeutics development (Wernig et al. 2008; Cooper et al. 2010). A more immediate application of recent advances in genomics and informatics to biomarker development is as an initial screen for risk. As more risk loci are identified and as the costs of screening decline, more widespread screening of populations of individuals will become feasible. Although this would not be a particularly sensitive or specific screen, it would provide for an economical triaging of high-risk subjects for more in-depth screening. It is also important to note that modalities other than molecular markers will form an important component of our biomarker armamentarium and that these modalities will need to be integrated within the network-centric approach. These modalities range from the relatively inexpensive and insensitive (history of constipation; hyposmia) to the expensive and sensitive (PET or SPECT neuroimaging).
As noted above, GWAS have been performed to reveal the genetic association of disease with SNPs. Despite the success of GWAS, the heritability of common disorders such as PD cannot be fully explained by the genes that have been discovered. A similar pattern is seen when we expand our search for biomarkers of disease risk to the other -omics. A number of reasons can be ascribed to the limitations of these studies for common diseases with complex traits. One explanation is that the current biostatistical analyses are agnostic or unbiased and thus ignore what is known about disease pathology. In addition, the linear modeling in GWAS analyses usually considers only one SNP at a time, whereas ignoring the genomic and epigenomic factors of each SNP. However, recently, there has been a shift away from this approach toward a more holistic one that recognizes the complexity of the genotype–phenotype relationship that is likely characterized by significant genetic heterogeneity and gene–gene and gene–environment interactions. Furthermore, strategies have been used to iteratively mine GWAS data, including the identification of potential PD targets and pathways using meta-analyses of previous GWAS and neuronal transcriptomic profiling studies (Zheng et al. 2010; Edwards et al. 2011).
The limitations of a linear model and other parametric statistical approaches have motivated the development of data mining and machine learning methods (Hastie et al. 2009). The advantage of these computational approaches is that they make fewer assumptions about the functional form of the model and the effects being modeled. In effect, data mining and machine learning methods are much more consistent with the notion of having the data direct the model, rather than forcing the data to fit a predetermined model. Several recent reviews highlight the need for these newer methods, including machine learning approaches such as random forests (RFs) and multifactor dimensionality reduction (MDR), which have been developed to address some of these issues (Tarca et al. 2007; Ressom et al. 2008; Moore 2010; Sun 2010). It is also clear that evolving informatics approaches will play an important role in addressing the complexity of the underlying molecular basis of many common human diseases. These methodologies have the potential to identify other molecular species (proteins, metabolites), which could serve as disease biomarkers in addition to the genome and interactome. In identifying nodal proteins, proteins that are present at the nodes of a network, proteomics approach provides a great platform, which typically assesses proteins in an unbiased fashion and provides the means to study the proteomic profile of a complex biological system on a large scale. Several technologies continue to evolve that are composed of integrated technical components, including separation technology, mass spectrometry (MS), and bioinformatics data processing. With advances in analytical technology and statistical analyses, several studies have set out to develop proteomic “molecular profiles” of PD tissues, including blood, CSF, and postmortem brain (reviewed in Caudle et al. 2010). Similar methodologies and analytical approaches are being used in the search for “metabolic profiles” of PD, which include compounds such as lipids, amino acids, fatty acids, amines, alcohols, sugars, organic phosphates, hydroxyl acids, aromatics, purines, and other high abundance or clinically important molecules; however, these databases are incomplete for secondary metabolites, drugs, and environmental compounds.
As technology and analytic methods improve, we will generate more complete annotations of the genomic, transcriptomic, proteomic, and metabolic spaces, which would greatly enhance the analysis of specific pathways and molecules involved in PD and would yield additional insight into the pathogenesis of the disease. In addition, it would provide a platform for future meta-analytic studies, which could assuage much of the between-study variability currently encountered when analyzing multiple studies. However, despite the advance in -omics, there are still several issues that need to be addressed and resolved. Most importantly, issues around data integration, analysis, and interpretation pose a great challenge, especially in context of ever expanding data being generated.
TRANSLATING GENOMICS INTO RATIONAL THERAPEUTICS
Drug discovery is the process by which new drugs are identified. The traditional method relies on trial-and-error in testing chemical substances against purified molecules and cultured cells, and subsequently examining their effects on a model of disease before taking such a candidate into clinical studies. However, in the last few decades, a new approach, termed rational drug discovery (RDD) has been adopted, which relies on characterized molecular mechanisms of disease. RDD posits that modulation of a specific target, putatively causal in disease pathogenesis, will have therapeutic value. This raises two fundamental questions: What is a specific target? and How is a modulator of this target found? The first question is at the core of drug discovery.
Current drug discovery assumes that diseases can be characterized by a faulty protein structure or aberrant expression of a protein encoded by a variant gene, and that identification of candidate drugs that modulate the activity of the proteins will have an effect on phenotype and/or disease outcome. This view is exemplified by expectations heralded with human genome sequencing technology, in which it was estimated that about 8000 genes would be available as drug targets (Imming et al. 2006). Currently, the number of drug targets correlated with genetic variation or polymorphisms is ∼220 (Russ and Lampel 2005). As we move from monogenic diseases to complex diseases, there is no consensus as to how genetics will inform drug discovery. However, it is expected that discovery of aberrant genetic networks, populated by aberrant proteomic and metabolic nodes, will provide target candidates for therapeutics development. It is in this arena that disease specific networks and nodes can be used for the rational design of common and even personalized therapeutics.
From the above, it should be clear that defining a disease target is not formulaic. However, once a target is chosen, its prosecution unfolds in one of two ways: target-based or ligand-based drug discovery. Target based-drug discovery starts with the three-dimensional structure of a target. If a three-dimensional structure of the target is not available, it may be empirically determined by crystallography or generated informatically via homology modeling using proteins with similar domains as a template.
Alternatively, ligand-based drug discovery starts with structural information of a known or predicted ligand. In either case, a pharmacophore is designed as bait. A pharmacophore is an abstract description of the molecular features that are required for interaction between a ligand and a target. More specifically, the IUPAC defines a pharmacophore to be “an ensemble of steric and electronic features that are necessary to ensure the optimal supramolecular interactions with a specific biological target and to trigger (or block) its biological response” (Wermuth et al. 1998). Because target/ligand interactions are “polar positive,” “polar negative,” or “hydrophobic,” typical features considered in designing a pharmacophore are hydrophobic, aromatic, a hydrogen bond acceptor, a hydrogen bond donor, cation, or anion moieties. Because ligand-based drug discovery relies on knowledge of other molecules known to bind a biological target, the minimum necessary structural characteristics derived from these molecules are used in designing the pharmacophore, which then can be used to identify similar compounds via screening of chemical libraries or through de novo synthesis. The basic principle is that similar molecules behave similarly. In other words, similar chemical groups and entities will have similar biological effects. This gives rise to the concept of the structure-activity relationship, or SAR. As three-dimensional structures of biological targets increase, and detailed information about molecular interactions between ligand and target become available, the application of SAR has become more complex. Drug discovery has evolved with the use of high performance computing to enable computer aided drug design (CADD) and sophisticated statistical algorithms and molecular dynamic tools to provide quantitative methodologies for SAR (QSAR) to rank order the potential potency of a number of biologically similar compounds. Once a series of compounds is identified using the above approach, they are labeled as “hits,” which are ready to be tested in biological assay systems. As illustrated by the analytical bottlenecks in PD genomics and biomarker discovery, the evolution of bioinformatics will be critical to the successful development of improved PD therapeutics.
The role of bioinformatics in connecting aberrant networks and nodes fundamental to PD pathogenesis with validated targets developed through computational chemistry and target modeling is anticipated to accelerate the process of drug discovery.
THE ROAD AHEAD …
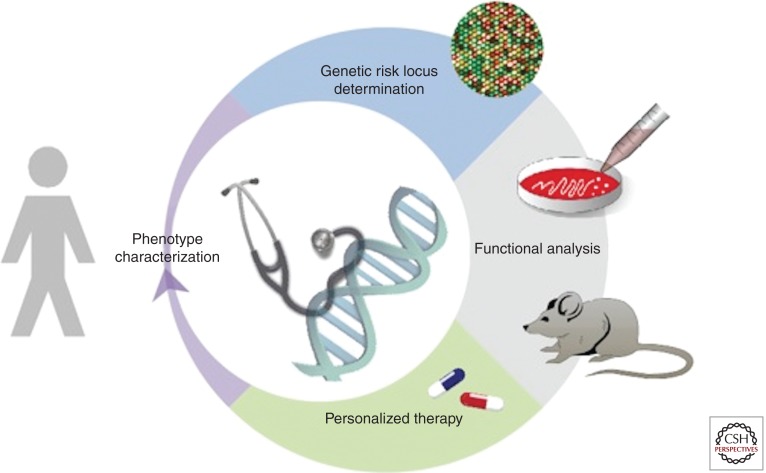
The main aim of genomic research is to identify pathways that are suitable for targeted therapeutic interventions to prevent, slow, halt, or reverse neurodegenerative disease processes. To that end, the success of translational research rests on the resolution of the complex genomic architecture of human disease, translating this to understanding aberrant networks and nodes associated with disease, and implementing this knowledge in the rational design of therapeutics, which could be tailored to the individual (Personalized Therapy, Fig. 5). However, this success is not only dependent on advancement of technologies and their applications. Success will also depend on regional, national, and even international collaborative efforts. In much the same way that the neurogenetics field has evolved, moving forward will require the collective efforts of scientists, clinicians, healthcare providers, policy makers, and importantly, patients. The impact of these efforts will also go beyond translational research and therapeutics development. Given the potential to revolutionize medicine, a host of societal issues will need to be addressed, including socioeconomic, ethical, clinical acceptance, medical education, cost effectiveness, and regulatory considerations.
Figure 5.
Genomics will play an integral role in the development of personalized therapeutics. The availability of detailed phenotype data from large patient/control cohorts is an important prerequisite for high-throughput genetic screening studies, including GWAS and genomic sequencing. After genetic risk loci have been dissected, in silico, in vitro, and in vivo analyses establish the underlying functional pathways and help to posit targets for rational, personalized therapies.
ACKNOWLEDGMENTS
This work is supported in part by DAMD17-03-1-0009, W81XWH-09-1-0103, and W81XWH-09-1-0107 (H.J.F.).
Footnotes
Editor: Serge Przedborski
Additional Perspectives on Parkinson's Disease available at www.perspectivesinmedicine.org
REFERENCES
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. 2009. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res 15: 441–451 [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, et al. 2003. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299: 256–259 [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Bratzke H, Hamm-Clement J, Sandmann-Keil D, Rub U 2002. Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson's disease (preclinical and clinical stages). J Neurol 249: III/1–5 [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E 2003a. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging 24: 197–211 [DOI] [PubMed] [Google Scholar]
- Braak H, Rub U, Gai WP, Del Tredici K 2003b. Idiopathic Parkinson's disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J Neural Transm 110: 517–536 [DOI] [PubMed] [Google Scholar]
- Caudle WM, Bammler TK, Lin Y, Pan S, Zhang J 2010. Using “omics” to define pathogenesis and biomarkers of Parkinson’s disease. Expert Rev Neurother 10: 925–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Demir E, Schultz N, Taylor BS, Sander C 2010. Automated network analysis identifies core pathways in glioblastoma. PLoS ONE 5: e8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Carvalho C, Mori S, Bild AH, Gatza ML, Wang Q, Lucas JE, Potti A, Febbo PG, West M, et al. 2009. A genomic strategy to elucidate modules of oncogenic pathway signaling networks. Mol Cell 34: 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sam L, Huang Y, Lee Y, Li J, Liu Y, Xing HR, Lussier YA 2010. Protein interaction network underpins concordant prognosis among heterogeneous breast cancer signatures. J Biomed Inform 43: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CH, Sun YM, Chang WC, Chiang-Hsieh PY, Lee TY, Tsai WC, Horng JT, Tsou AP, Huang HD 2011. Identifying transcriptional start sites of human microRNAs based on high-throughput sequencing data. Nucleic Acids Res 392: 9345–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O, Hargus G, Deleidi M, Blak A, Osborn T, Marlow E, Lee K, Levy A, Perez-Torres E, Yow A, et al. 2010. Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol Cell Neurosci 45: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. 2011. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res 39: D691–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Przedborski S 2003. Parkinson’s disease: Mechanisms and models. Neuron 39: 889–909 [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL 2003. Molecular pathways of neurodegeneration in Parkinson's disease. Science 302: 819–822 [DOI] [PubMed] [Google Scholar]
- Edwards YJ, Beecham GW, Scott WK, Khuri S, Bademci G, Tekin D, Martin ER, Jiang Z, Mash DC, ffrench-Mullen J, et al. 2011. Identifying consensus disease pathways in Parkinson's disease using an integrative systems biology approach. PLoS ONE 6: e16917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M, Maraganore DM, Lockhart P, Singleton A, Lesnick TG, de Andrade M, West A, de Silva R, Hardy J, Hernandez D 2001. α-Synuclein gene haplotypes are associated with Parkinson’s disease. Hum Mol Genet 10: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Feany MB, Bender WW 2000. A Drosophila model of Parkinson’s disease. Nature 404: 394–398 [DOI] [PubMed] [Google Scholar]
- Gatza ML, Lucas JE, Barry WT, Kim JW, Wang Q, Crawford MD, Datto MB, Kelley M, Mathey-Prevot B, Potti A, et al. 2010. A pathway-based classification of human breast cancer. Proc Natl Acad Sci 107: 6994–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J 2009. The elements of statistical learning: Data mining, inference, and prediction, 2nd ed Springer, New York [Google Scholar]
- Hawkes CH, Del Tredici K, Braak H 2010. A timeline for Parkinson’s disease. Parkinsonism Relat Disord 16: 79–84 [DOI] [PubMed] [Google Scholar]
- Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C 2009. VisANT 3.5: Multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res 37: W115–W121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imming P, Sinning C, Meyer A 2006. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 5: 821–834 [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium 2005. Haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Parkinson Disease Genomics Consortium, Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simón-Sánchez J, Schulte C, Lesage S, et al. 2011. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: A meta-analysis of genome-wide association studies. Lancet 377: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Parkinson's Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) 2011. A two-stage meta-analysis identifies several new loci for Parkinson's disease. PLoS Genet 7: e1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TY, Kim HU, Lee SY 2010. Data integration and analysis of biological networks. Curr Opin Biotechnol 21: 78–84 [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392: 605–608 [DOI] [PubMed] [Google Scholar]
- Kreeger PK, Lauffenburger DA 2010. Cancer systems biology: A network modeling perspective. Carcinogenesis 31: 2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES 2011. Initial impact of the sequencing of the human genome. Nature 470: 187–197 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, et al. 2006. Collaborative analysis of α-synuclein gene promoter variability and Parkinson disease. JAMA 296: 661–670 [DOI] [PubMed] [Google Scholar]
- Mardis ER 2006. Anticipating the 1,000 dollar genome. Genome Biol 7: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L 2000. Dopaminergic loss and inclusion body formation in α-synuclein mice: Implications for neurodegenerative disorders. Science 287: 1265–1269 [DOI] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696 [DOI] [PubMed] [Google Scholar]
- Moore JH 2010. Detecting, characterizing, and interpreting nonlinear gene-gene interactions using multifactor dimensionality reduction. Adv Genet 72: 101–116 [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I 2003. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics 19: 2155–2157 [DOI] [PubMed] [Google Scholar]
- Ochs MF, Karchin R, Ressom H, Gentleman R 2011. Identification of aberrant pathway and network activity from high-throughput data—workshop introduction. Pac Symp Biocomput 2011: 364–368 [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. 2004. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44: 595–600 [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ 2008. Disease-specific induced pluripotent stem cells. Cell 134: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. 2003. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13: 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack A 2011. DNA sequencing caught in deluge of data. The New York Times, November 30 [Google Scholar]
- Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. 1997. Mutation in the α-synuclein gene identified in families with Parkinson’s disease. Science 276: 2045–2047 [DOI] [PubMed] [Google Scholar]
- Qiu YQ, Zhang S, Zhang XS, Chen L 2009. Identifying differentially expressed pathways via a mixed integer linear programming model. IET Syst Biol 3: 475–486 [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES 2001. On the allelic spectrum of human disease. Trends Genet 17: 502–510 [DOI] [PubMed] [Google Scholar]
- Ressom HW, Varghese RS, Zhang Z, Xuan J, Clarke R 2008. Classification algorithms for phenotype prediction in genomics and proteomics. Front Biosci 13: 691–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Lampel S 2005. The druggable genome: An update. Drug Discov Today 10: 1607–1610 [DOI] [PubMed] [Google Scholar]
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M 1977. Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265: 687–695 [DOI] [PubMed] [Google Scholar]
- Schadt EE 2009. Molecular networks as sensors and drivers of common human diseases. Nature 461: 218–223 [DOI] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH 2009. PID: The Pathway Interaction Database. Nucleic Acids Res 37: D674–D679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. 2009. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 15: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T 2010. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV 2010. Multigenic modeling of complex disease by random forests. Adv Genet 72: 73–99 [DOI] [PubMed] [Google Scholar]
- Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R 2009. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal 2: ra39. [DOI] [PubMed] [Google Scholar]
- Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S 2007. Machine learning and its applications to biology. PLoS Comput Biol 3: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL 2009. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 27: 199–204 [DOI] [PubMed] [Google Scholar]
- Valente EM, Bentivoglio AR, Dixon PH, Ferraris A, Ialongo T, Frontali M, Albanese A, Wood NW 2001. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35–p36. Am J Hum Genet 68: 895–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM 2010. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26: 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wang CC, Cirit M, Haugh JM 2009. PI3K-dependent cross-talk interactions converge with Ras as quantifiable inputs integrated by Erk. Mol Syst Biol 5: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Marian C, Ressom HW 2012. Identification of aberrant pathways and network activities from high-throughput data. Brief Bioinform 10.1093/bib/bbs001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermuth CG, Ganellin CR, Lindberg P, Mitscher LA 1998. Glossary of terms used in medicinal chemistry (IUPAC Recommendations 1998). Pure Appl Chem 70: 1129–1143 [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R 2008. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci 105: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Feng X, Stein L 2010. A human functional protein interaction network and its application to cancer data analysis. Genome Biol 11: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, et al. 2009. Bridging high-throughput genetic and transcriptional data reveals cellular responses to α-synuclein toxicity. Nat Genet 41: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. 2010. PGC-1α, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med 2: 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. 2004. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 44: 601–607 [DOI] [PubMed] [Google Scholar]