Abstract
Retroviruses are distinguished from other viruses by two characteristic steps in the viral replication cycle. The first is reverse transcription, which results in the production of a double-stranded DNA copy of the viral RNA genome, and the second is integration, which results in covalent attachment of the DNA copy to host cell DNA. The initial catalytic steps of the integration reaction are performed by the virus-encoded integrase (IN) protein. The chemistry of the IN-mediated DNA breaking and joining steps is well worked out, and structures of IN-DNA complexes have now clarified how the overall complex assembles. Methods developed during these studies were adapted for identification of IN inhibitors, which received FDA approval for use in patients in 2007. At the chromosomal level, HIV integration is strongly favored in active transcription units, which may promote efficient viral gene expression after integration. HIV IN binds to the cellular factor LEDGF/p75, which promotes efficient infection and tethers IN to favored target sites. The HIV integration machinery must also interact with many additional host factors during infection, including nuclear trafficking and pore proteins during nuclear entry, histones during initial target capture, and DNA repair proteins during completion of the DNA joining steps. Models for some of the molecular mechanisms involved have been proposed, but important details remain to be clarified.
HIV integration occurs more frequently in chromosomal regions that are actively transcribed. The process is mediated by HIV-encoded integrase as well as several host-encoded factors (e.g., LEDGF/p75).
Integration of a DNA copy of the viral genome into a host cell chromosome is an essential step in the retroviral replication cycle (Varmus et al. 1989; Coffin et al. 1997). Once integrated, the proviral DNA is replicated along with cellular DNA during cycles of cell division, as with any cellular gene. The provirus serves as the template for transcription of viral RNAs. Some viral RNAs are translated to yield the viral proteins, whereas a portion of the full-length viral RNA is recruited to serve as genomic RNA in progeny virions.
Integration is mediated by the virus-encoded IN protein, which is introduced into cells during infection along with reverse transcriptase, the viral RNA, and other proteins as a part of the viral core. After the viral DNA is synthesized by reverse transcription in the cytoplasm, it stably associates with IN and other proteins as a high-molecular-weight nucleoprotein complex that is later transported to the nucleus for subsequent integration. The mechanism of integration has been extensively studied and the basic biochemistry is quite well understood. Recently, structural studies of the active nucleoprotein complexes of IN bound to viral DNA (intasomes) have also made great progress.
Integration occurs precisely at the termini of the viral DNA but integration can take place at many locations in the host genome. Most positions in chromosomal DNA can serve as integration acceptor sites, but there are distinct regional preferences that differ among groups of retroviruses. Some of these preferences appear to involve chromatin-associated factors that also interact with IN. Understanding targeting is especially important because of the application of retroviral insertion in gene therapy, where adverse events have been associated with integration of retroviral vectors near proto-oncogenes (Howe et al. 2008; Hacein-Bey-Abina et al. 2010). The integration step is also the target of FDA-approved inhibitors (discussed by Arts and Hazuda 2011).
This review takes advantage of data from both HIV and model retroviruses because important advances came from both. First we review the evolution of models for retroviral integration, then our present picture of the biochemical steps of the integration pathway, and lastly integration in the cellular context.
HISTORY
Howard Temin’s provirus hypothesis holds that the viral RNA introduced into cells during infection becomes converted to DNA by reverse transcription. Viral DNA is then integrated into the host genome. This explained how cells transformed by Rous sarcoma virus (RSV) could stably maintain the transformed state in the absence of viral replication (Temin 1976). Temin’s hypothesis was vindicated by the discovery of reverse transcriptase (Baltimore 1970; Temin and Mizutani 1970) and the physical characterization of integrated viral DNA in the genome of infected cells (Weiss et al. 1984; Coffin et al. 1997).
How does the viral DNA become integrated? The first sighting of the protein we now call IN was as a nuclease activity associated with cores of avian retroviral particles (Grandgenett et al. 1978). Genetic studies later defined the IN coding region and the ends of the viral DNA to be important for integration (Donehower and Varmus 1984; Panganiban and Temin 1984; Schwartzberg et al. 1984). In these studies, cloned copies of the viral DNA were modified in vitro, and the modified DNA was then introduced into cells by transfection, allowing production of viral particles. The phenotype of the mutant viral stocks could then be characterized by infecting fresh cells. Mutation of the 3′ region of the pol gene resulted in viruses that were able to enter cells and carry out reverse transcription normally, but failed to integrate the reverse-transcribed DNA. This region of pol encodes a protein, now called IN. Another group of mutants with an essentially identical phenotype mapped to the ends of the viral DNA at the sites that become joined to host DNA on integration (Panganiban and Temin 1983; Murphy and Goff 1992; Murphy et al. 1993; Du et al. 1997). Studies of the synthesis of the viral proteins showed that IN is cleaved from the gag-pol polyprotein precursor by the virus-encoded protease to yield an independent protein. These studies indicated that IN likely acts on the ends of the viral DNA but could not reveal the mechanisms involved. The presence of circular forms of viral DNA in infected cells initially suggested that retroviral integration might proceed via a circular DNA intermediate, as was known to be the case for bacteriophage λ, but this idea was later refuted by biochemical studies, as described below.
KEY ADVANCES
Biochemical Studies of the Integration Mechanism
Demonstration that PICs Isolated from Infected Cells Are Competent for Integration In Vitro
The first biochemical studies of the integration reaction used viral replication intermediates purified from infected cells as a source of the integration machinery. Studies were initially performed using Moloney murine leukemia virus as a model system (Brown et al. 1987), soon followed by similar experiments with HIV (Ellison et al. 1990; Farnet and Haseltine 1990). The viral DNA made by reverse transcription within the cytoplasm was found to be part of a large nucleoprotein complex, the preintegration complex (PIC), which is derived from the core of the infecting virion (Bowerman et al. 1989). On incubation in vitro with a target DNA in the presence of a Mg2+ ion, the viral DNA within PICs efficiently integrates into the target DNA.
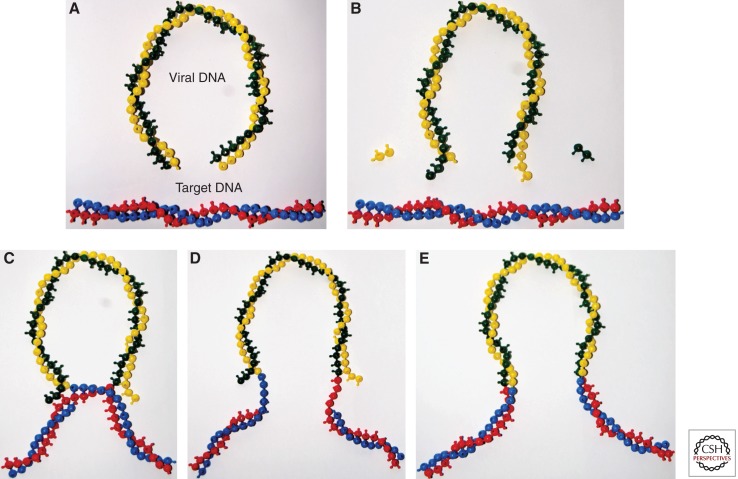
Analysis of the structure of the integration intermediates produced in these reactions (Fujiwara and Mizuuchi 1988; Brown et al. 1989) revealed the DNA cutting and joining steps of integration (Fig. 1). In the first step (3′ end processing), two nucleotides are in most cases removed from each 3′ end of the blunt-ended linear viral DNA. The resulting 3′ ends of the viral DNA in all cases terminate with the conserved CA-3′ sequence. In the second step (DNA-strand transfer), these 3′ ends attack a pair of phosphodiester bonds on opposite strands of the target DNA, across the major groove. In the resulting integration intermediate the 3′ ends of the viral DNA are covalently joined to the target DNA. The single-strand gaps and the two-nucleotide overhang at the 5′ ends of the viral DNA must be repaired by cellular enzymes to complete integration. The sites of joining on the two target DNA strands are separated by four nucleotides in the case of murine leukemia virus (MLV), resulting in a four-nucleotide duplication of target DNA flanking the integrated proviral DNA. For HIV, the sites are five base pairs apart, resulting in a five base-pair duplication.
Figure 1.
DNA breaking and joining reactions mediating DNA integration. DNA bases are shown by balls in the snap-together models, although the HIV DNA (10 kb) and the cellular chromosome (megabases) are not shown to scale. (A) The linear blunt-ended viral DNA (green and yellow) and target DNA (blue and red). (B) 3′ end processing. Two nucleotides are in most cases removed from each 3′ end of the viral DNA. (C) The 3′ ends generated by 3′ processing attack a pair of phosphodiester bonds in the target DNA. The sites of attack on the two target DNA strands are separated by five nucleotides in the case of HIV-1. The 3′ ends of the viral DNA are joined to the 5′ ends of the target DNA at the site of integration. The 5′ ends of the viral DNA are not joined to target DNA in the intermediate. (D) Completion of provirus formation requires removal of the two unpaired bases at the 5′ ends of the viral DNA, filling in the single-strand gaps between viral and target DNA and ligation of the 5′ ends of the viral DNA to target DNA. IN catalyzes the 3′ processing and DNA-strand transfer steps to form the integration intermediate. Subsequent steps are thought to be catalyzed by cellular enzymes. (E) The integrated provirus.
Development of In Vitro Integration Assays Using Purified IN
PICs contain many viral and cellular proteins, as shown, for example, by immunoprecipitation assays (Farnet and Haseltine 1991b; Bukrinsky et al. 1993; Li et al. 2001). However, possible roles of these proteins in integration are difficult to assess, in part because the low abundance of PICs in extracts of infected cells complicates analysis. Simpler in vitro systems were therefore required to identify the minimal set of proteins required for integration. The next advance was the discovery that cell extracts containing PICs or detergent-disrupted virus particles could promote in vitro integration of linear plasmid DNA with terminal sequences that mimic the viral DNA ends (Fujiwara and Craigie 1989), albeit with low efficiency. It was later shown using the same assay that HIV-1 IN protein alone was able to support in vitro DNA integration (Bushman et al. 1990). The discovery that IN alone is sufficient to promote not only 3′ end processing (Katzman et al. 1989), but also in vitro integration (Craigie et al. 1990; Katz et al. 1990) of oligonucleotides matching the viral DNA ends opened the door to detailed biochemical studies of the integration reaction. The high efficiency of these simplified assay systems allowed the products to be directly detected by physical methods such as gel electrophoresis and provided the foundation for the development of high-throughput screens that later led to the development of IN inhibitors (Arts and Hazuda 2011).
Although highly efficient, the oligonucleotide assay system initially lacked the full fidelity of integration in vivo. Many reaction products resulted from integration of only a single viral DNA end into one strand of target DNA, rather than concerted integration of pairs of viral DNA ends. Subsequent improvements have enabled in vitro concerted integration of both ends by HIV-1 IN, although still with somewhat low efficiency (Hindmarsh et al. 1999; Sinha et al. 2002; Li and Craigie 2005; Sinha and Grandgenett 2005). Under these conditions, IN forms a stable synaptic complex (SSC) with a pair of viral DNA ends that is an intermediate on the reaction pathway (Li et al. 2006), and in which 3′ end processing occurs on both viral DNA ends.
Integration Mechanism: Similarities to DNA Transposition
A mechanistic connection between DNA transposition and retroviral DNA integration was first suggested by the short duplication of target DNA sequences that flank integrated proviruses and integrated transposons (Ju and Skalka 1980; Shimotohno et al. 1980). In the case of DNA transposons, this duplication was known to arise by staggered cleavage of the target DNA and subsequent repair of the resulting single-strand gaps between transposon DNA and target DNA. In contrast to many DNA recombinases, these transposases splice the transposon DNA into the new target DNA by a one-step transesterification mechanism. In this reaction, the 3′ end of the viral or transposon DNA acts as a nucleophile in the attack on the target DNA backbone, breaking the target DNA and joining the viral DNA all in a single step. This is in contrast to two-step reactions involving a covalent intermediate between DNA and protein as is found in the tyrosine and serine recombinase families (reviewed in Mizuuchi 1992).
The oligonucleotide integration assay allowed the number of reaction steps to be counted in the HIV-1 DNA-strand transfer reaction using stereochemically marked phosphate atoms in target DNA substrates. The result was that integration proceeded in a single step, implicating a direct transesterification (Engelman et al. 1991), as had been previously shown for bacteriophage Mu transposase (Mizuuchi and Adzuma 1991). Reactions involving a covalent protein-DNA intermediate, in contrast, require two steps. Subsequent structural studies revealed that retroviral IN and transposases of the D,D,E family, which includes bacteriophage Mu and Tn5, share a common active site organization in which the conserved acidic amino acids bind two divalent metal atoms, confirming earlier suggestions based on mutagenesis data (Engelman and Craigie 1992; Kulkosky et al. 1992; van Gent et al. 1992; Leavitt et al. 1993).
Structural Studies of IN
In early studies, partial proteolysis of HIV IN protein revealed three structurally distinct domains (Engelman and Craigie 1992), and expression of these domains individually yielded proteins that were amenable to study. The central core domain, which from biochemical studies was expected to contain the active site (Engelman and Craigie 1992; Kulkosky et al. 1992; van Gent et al. 1992; Bushman et al. 1993; Leavitt et al. 1993), remained poorly soluble but a single amino change markedly improved solubility (Jenkins et al. 1995) and enabled crystallization. The structure (Dyda et al. 1994) confirmed the triad of acidic residues, the D,D-35-E motif identified from mutagenesis studies, to be key active site residues. The structure of the ASV IN core domain was determined soon after (Bujacz et al. 1995). The IN core structures belong to the superfamily of a functionally diverse group of polynucleotidyl transferases, which includes prokaryotic and HIV RNase H enzymes, Holiday junction resolvase RuvC, and the phage Mu transposase catalytic domain (Rice et al. 1996), collectively referred to as “RNase H superfamily.”
The structures of the isolated catalytic domains provided no clue as to how IN interacts with DNA to position a pair of active sites with the correct spacing for DNA-strand transfer. Both HIV and ASV IN fragments crystallized as dimers, with similar dimer interfaces. However, the pairs of active sites were on opposite faces of the roughly spherical complexes with a separation incompatible with the five-nucleotide spacing of the sites of catalysis in the target DNA.
Structures were also solved for the amino-terminal and carboxy-terminal domains. The amino-terminal domain, refolded in the presence of zinc, was initially solved by nuclear magnetic resonance (NMR) (Cai et al. 1997; Eijkelenboom et al. 1997). It consists of a bundle of α helices with coordination of a single Zn2+ ion stabilizing the structure. The carboxy-terminal domain structure was also solved by NMR, revealing an SH3-like β barrel (Eijkelenboom et al. 1995; Lodi et al. 1995). Subsequently, two-domain structures, amino-terminal plus catalytic domain or catalytic domain plus carboxy-terminal domain, were solved for several retroviral INs (reviewed in Chiu and Davies 2004; Jaskolski et al. 2009). On the basis of these partial structures, numerous models were proposed for the IN complex with viral DNA ends. However, the differences in the arrangement of domains between the various partial structures indicated that structures including viral DNA would be required to understand the organization of the active complex.
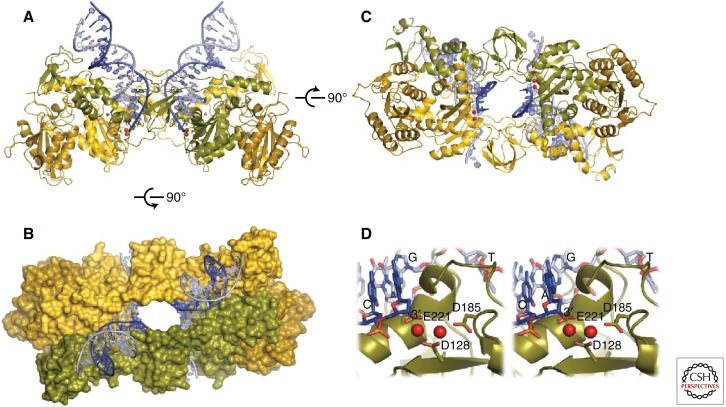
Determination of the structure of prototype foamy virus (PFV) IN in complex with viral DNA (Hare et al. 2010; Maertens et al. 2010) provided a major breakthrough in understanding IN function. The PFV intasome is comprised of a homotetramer of IN assembled on viral DNA ends (see Movie 1). Within it, the IN domains have essentially the same structures as observed in the isolated domains previously. The tetramer has a dimer-of-dimers architecture, where the individual dimers are formed via the canonical catalytic domain dimerization interface. The tetramerization interface includes contacts between the amino-terminal domain from one monomer and the catalytic core domain of the opposing dimer. Isomorphous domain-domain interfaces have been observed in crystal structures containing two-domain constructs of HIV-1 and visna virus INs (Wang et al. 2001; Hare et al. 2009). However, the conformation of the tetramerization interface and mutual orientation of the catalytic domain dimers could not have been predicted based on existing partial structures. The tetramer is held together not only by protein–protein contacts but also protein–DNA interactions that together bury more than 10,000 Å2 of molecular surface (Fig. 2). Two of the monomers extend across the center of symmetry in the complex, thus tightly linking the two halves. The intertwined nature of protein–protein and protein–DNA contacts is reminiscent of the structures of Tn5 and Mos1 transpososomes (Davies et al. 2000; Richardson et al. 2009). The PFV structure also confirmed that the IN active site binds a pair of divalent metal cations (Fig. 2D). A notable feature of the PFV intasome crystals is that amino- and carboxy-terminal domains of the IN subunits that do not participate in the tetramerization interface are disordered. The functions of these four domains remain to be determined.
Figure 2.
Structure of the complex of PFV IN and viral DNA (Hare et al. 2010). (A,B) Two views of the IN-DNA complex. (C) Top view of the complex, colored to emphasize that two monomers cross the center of symmetry and link up the two halves of the complex. (D) Stereo pair showing the structure of the active site, including two magnesium atoms bound to the three conserved acidic amino acids that comprise the active site.
Movie 1.
Complex of PFV integrase bound to DNA. Movie provided by Peter Cherepanov.
Crystallization of the PFV intasome also provided critical new information on the mechanism of action of IN inhibitors such as raltegravir. Structures of intasome/drug complexes revealed that not only does raltegravir binding block access of target DNA to the active site, but also displaces the 3′ end of the viral DNA from the active site, thereby disrupting catalysis. The active site region of PFV and HIV-1 IN are sufficiently similar that interactions of HIV-1 IN with inhibitors can be modeled based on the PFV structure (Krishnan et al. 2010). However, some resistance mutations map to regions that are dissimilar between the two proteins, so structures of the HIV-1 intasome will ultimately be required to fully understand the mechanism of drug resistance.
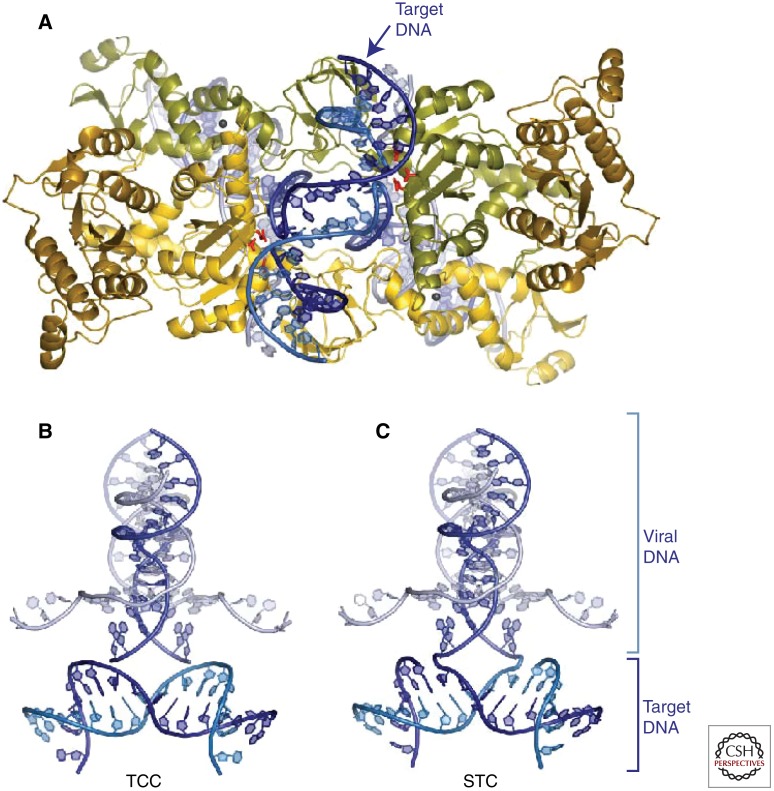
The PFV intasome presents a surface that can accommodate target DNA without any significant conformational changes, suggesting that the viral nucleoprotein complex does not undergo major structural rearrangements between 3′ processing and DNA-strand transfer. However, because the active sites of the intasome are separated by some 25 Å, a significant deformation of target DNA is required for the active sites to access the scissile phosphodiesters (Fig. 3; see Movie 2) (Maertens et al. 2010). The target DNA-containing PFV structures explained the early observations that retroviral INs are biased toward deformed target DNA (Pryciak and Varmus 1992; Pruss et al. 1994; Katz et al. 1998).
Figure 3.
Target DNA capture by the complex of PFV IN and viral DNA (Maertens et al. 2010). (A) Overview of the complex, showing target DNA in blue. (B,C) Two views of the DNA only, highlighting formation of the initial covalent link between the viral DNA 3′ ends and target DNA 5′ ends.
Movie 2.
Initial DNA breaking and joining reactions mediating integration, inferred from PFV integrase/DNA complex structures. Movie provided by Peter Cherepanov.
Integration in the Cellular Context
Nuclear Localization
HIV can infect nondividing cells, implying that PICs must cross the nuclear membrane to carry out integration. The mechanism of nuclear localization has not been fully clarified (for recent reviews see Fassati 2006; Suzuki and Craigie 2007). Briefly, several known components of PICs exhibit nuclear localization properties when fused to a polypeptide that does not normally localize to the nucleus. However, mutating these determinants individually in the context of the PIC does not abolish nuclear localization. Proposed viral determinants include HIV-1 MA, CA, and IN proteins, and in addition, the three-stranded DNA flap structure generated at the central polypurine tract.
Several genome-wide screens have been performed to identify genes that when reduced in dosage using small interfering RNA (siRNA), diminish HIV infection, and several of the identified host factors have been mapped to the integration part of the viral replication cycle (Brass et al. 2008; Konig et al. 2008; Zhou et al. 2008). Among these, several nuclear pore proteins appear to also be important not only for nuclear localization but also for efficient integration after nuclear entry, suggesting possible coupling of nuclear translocation and integration. Some cellular proteins implicated as important include transportin 3 (product of the TNPO3 gene) and Nup358 (product of the RANBP2 gene). Surprisingly, an amino acid substitution in the CA protein (N74D) was able to abrogate sensitivity to a knockdown of TNPO3, but created sensitivities to knockdown of several other nuclear pore proteins (Lee et al. 2010). These data suggest that PICs may interact with specific nuclear pore proteins during nuclear transit, and that there may be multiple redundant pathways accessible to PICs for nuclear entry.
Multiple Fates of the Viral DNA in the Nucleus
After the HIV PIC enters the nucleus, the viral DNA must become integrated into chromosomal DNA of the host for productive infection to proceed. However, the viral DNA can also undergo several circularization reactions that do not support subsequent replication and represent dead ends for the virus (Farnet and Haseltine 1991a). The two ends of the viral DNA can be ligated to each other, probably following dissociation of the PIC proteins, to yield 2-long long terminal repeat (LTR) circles. Inactivation of the host cell nonhomologous DNA end-joining (NHEJ) components Ku70/80, ligase IV, and XRCC4 blocks 2-LTR circle formation, implicating that these factors are involved in the circularization reaction (Li et al. 2001; Jeanson et al. 2002). Circles with one LTR copy can also be detected. These can be formed either by recombination between the LTRs within the nucleus, possibly involving action of the cellular MRN complex (Mre11, Rad50, and NBS1) (Kilzer et al. 2003), or as stalled products of reverse transcription that failed to complete the final steps of strand displacement synthesis (Hu and Hughes 2011). In addition, the viral DNA can use itself as an integration target, resulting in either circles with an inverted segment, or pairs of smaller circles, depending on whether each 3′ end joins initially to the same or a different DNA strand (Shoemaker et al. 1980; Farnet and Haseltine 1991a). The cellular DNA condensing protein BAF can block autointegration in vitro, suggesting that tight packaging of the viral DNA in a protein complex may protect against autointegration (Lee and Craigie 1998).
Integration Target Site Selection
Once in the nucleus, integration requires capture of host cell DNA sequences by viral PICs and completion of the chemical steps of integration. The nature of favored and disfavored target sites for retroviral integration has been the topic of close study (for reviews see Varmus et al. 1989; Coffin et al. 1997; Ciuffi and Bushman 2006). Early analysis of the DNA sequences at junctions between proviral DNA and host DNA showed that host cell sequences at the point of integration differed among proviral isolates, indicating that the integration reaction was not highly sequence specific, although close analysis subsequently showed weakly conserved sequences at target sites (Varmus et al. 1989; Stevens and Griffith 1994; Coffin et al. 1997; Holman and Coffin 2005; Wu et al. 2005; Berry et al. 2006). Early studies of gammaretroviruses suggested that integration might be favored near DNAse I hypersensitive sites in vertebrate genomes, indicating a possible association with open chromatin (Varmus et al. 1989; Coffin et al. 1997).
The development of methods for studying integration in vitro using PICs or purified IN allowed target site selection to be analyzed in reconstituted reactions. Following the idea that chromatin packing could obstruct integration, DNA templates wrapped in nucleosomes were tested as in vitro integration targets, which surprisingly showed that integration was actually favored in nucleosomal DNA (Pryciak and Varmus 1992; Pryciak et al. 1992; Pruss et al. 1994). Mapping of favored sites indicated that sharp DNA bends in the nucleosome structure were particularly favored targets (Pruss et al. 1994). Several studies have shown that DNA distortion can promote integration (Bushman and Craigie 1992; Bor et al. 1995; Katz et al. 1998), perhaps because completing the reaction cycle requires DNA distortion, so that kinking the DNA on the nucleosome may lower the activation energy. Consistent with these observations, the target DNA in the PFV target capture and strand transfer complexes is significantly deformed to position the scissile phosphates close to the two active sites (Maertens et al. 2010). The deformation is more drastic than the relatively smooth curvature of DNA on the nucleosome surface, suggesting that some remodeling of nucleosome structure may be required to facilitate integration. In addition, the presence of several sequence-specific DNA-binding proteins on integration targets was shown to obstruct integration in vitro (Pryciak and Varmus 1992; Bor et al. 1995), probably by steric occlusion.
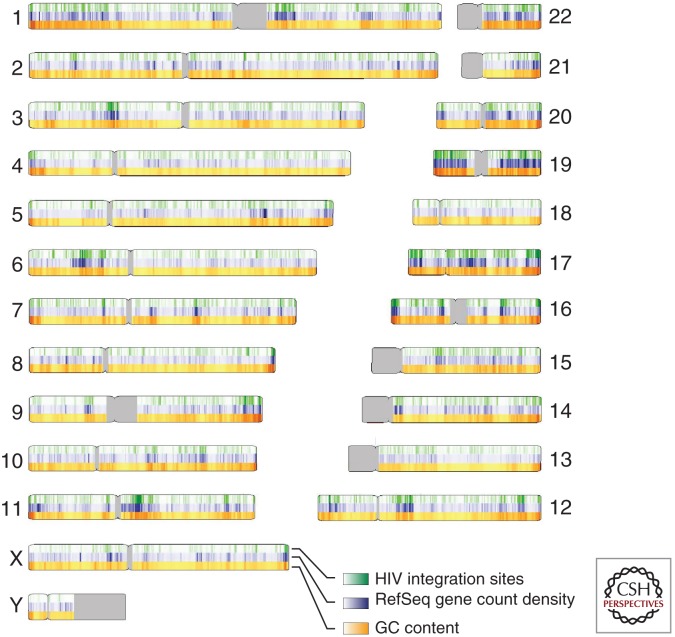
With the completion of the draft human genome sequence in 2001 (Lander 2001; Venter 2001), it became possible to study integration target site selection genome wide using high-throughput DNA sequencing. In the first such study, 524 sites of HIV integration were mapped after acute infection of the T-cell line SupT1 and the relationship with genomic annotation analyzed (Schroder et al. 2002). This experiment revealed that HIV favored integration within transcription units quite strongly. The relationship with gene activity was then probed by transcriptional profiling analysis of the SupT1 cells, revealing that active transcription units were particularly strongly favored for integration. In the human genome, many genomic features are correlated with each other, and this complicates identifying the primary determinants of integration targeting. Active transcription units are associated with regions of high G/C content, high gene density, high CpG island density, short introns, high frequencies of Alu repeats, low frequencies of LINE repeats, and characteristic epigenetic modifications. These features are also associated with high frequencies of HIV integration (Fig. 4). Subsequently, HIV integration site selection has been studied after acute infection in many cell types (Wu et al. 2003; Mitchell et al. 2004; Barr et al. 2006; Berry et al. 2006; Ciuffi et al. 2006; Wang et al. 2007; Brady et al. 2009a), and the favoring of integration in active transcription units has been seen in all cases except in the presence of artificially engineered tethering factors (described below).
Figure 4.
HIV integration site distributions on the human chromosomes (Wang et al. 2007). The human chromosomes are numbered at the sides of the diagram. HIV integration sites (20,000 total) are shown in green, gene density is shown in blue (measured as the count of RefSeq genes in a 500-kb interval), and the G/C content is shown in orange (measured in 500-kb intervals). The gray coloring indicates regions of centromeric repeats that have not been sequenced.
Why did HIV evolve to favor integration in active transcription units? Studies of many types of integrating genomic parasites show that their targeting preferences have evolved to optimize their ability to persist in their hosts and leave progeny. The yeast Ty retrotransposons, for example, must coexist with their hosts indefinitely, and they have evolved to integrate into benign genomic locations that do not harm the yeast cell (reviewed in Bushman 2001; Craig et al. 2002). For HIV, infected T cells typically have a half-life of only a day or two before cells are killed by the cellular immune system or by the toxicity of infection (Perelson et al. 1996). Thus HIV has only a limited time for the production of progeny. Recent studies show that integration within transcription units is usually favorable for efficient transcription (Jordan et al. 2001; Lewinski et al. 2005), potentially explaining the targeting preference.
Other retroviruses, however, show different favored target sites in chromosomes. The gammaretroviruses strongly favor integration at transcription start sites (Wu et al. 2003). Other retroviral genera show no strong preferences, with integration only slightly favored in transcription units or gene 5′ ends (Mitchell et al. 2004; Narezkina et al. 2004; Brady et al. 2009b).
These observations argue against the simplest version of the open chromatin model for targeting—if relative exposure of the target DNA was the only determinant of integration site selection, then how could the retroviruses be so different from each other? Considerable evidence now indicates that a simple tethering model explains HIV integration targeting. Another type of model could invoke the different modes of nuclear entry. The gammaretroviruses require mitosis for efficient infection (Roe et al. 1993), whereas HIV can infect nondividing cells, raising the possibility that PICs from the two types of retroviruses are encountering chromatin at different points in the cell cycle. If the chromatin is in different states at these stages, differences in integration targeting could result. Studies of HIV integration in two models of cells arrested at G1 showed favored integration in active transcription units for both (Barr et al. 2006; Ciuffi et al. 2006), but more data on integration at other points in the cell cycle is needed to explore this model more fully.
An IN “swap” experiment implicated IN as a dominant determinant of integration targeting. In this study, the IN region of the gammaretrovirus MLV was substituted for that of HIV, and integration target site selection monitored (Lewinski et al. 2006). The chimeric virus showed favored integration near transcription start sites and reduced integration along the length of transcription units, thus resembling MLV and differing from HIV. Thus MLV IN was a dominant determinant of MLV-like integration in this context.
With the development of “next-generation” sequencing methods, it has become possible to generate much larger collections of integration sites for analysis. In parallel, new kinds of genome-wide annotation have become available thanks to “ChIP-seq” and other methods based on massively parallel sequencing for genome-wide mapping of bound proteins, sites of histone modification, and DNA methylation. Analysis of 40,000 sites of HIV integration in the Jurkat T-cell line has indicated that integration commonly occurs on the outer surface of DNA wrapped on nucleosomes in vivo (Wang et al. 2007), as was suggested from earlier experiments in vitro. The distributions of a wide variety of histone posttranslational modifications have been associated with distributions of HIV integration sites, indicating that marks characteristic of active transcription units are favorable (e.g., H3K4me1 and me2, H3K27me1, and H3K36me3), whereas those characteristic of intergenic regions or inactive genes are unfavorable (e.g., H3K9me2 and me3, and H3K27me2 and me3).
Extensive mapping of both HIV integration sites and genomic annotation allows extraction of the genomic features most strongly directing integration targeting. Logistic regression or machine-learning methods can be used to compare integration site distributions to random control models, and then variable selection schemes can be used to identify the most strongly associated forms of annotation. For HIV, the genomic features that best allow discrimination of HIV integration sites from random sites included the local sequence at the point of integration, G/C content, gene density, and DNAse I cleavage site density (Berry et al. 2006; Wang et al. 2007), the latter three being associated with transcriptionally active regions of the genome.
LEDGF/p75, a Host Cell Factor Affecting HIV Integration
HIV takes advantage of host cell factors to allow efficient integration and optimize integration target site selection. Of particular importance is the host cell protein LEDGF/p75 (product of the PSIP1 gene), which both boosts the efficiency of integration and mediates targeting to active transcription units.
LEDGF/p75 was first identified as a transcriptional mediator protein that promoted activator-dependent transcription in vitro (Ge et al. 1998). Subsequently, several groups used affinity-based screens to identify cellular proteins that bound to HIV IN and thereby identified LEDGF/p75 as a tight binder (Cherepanov et al. 2003; Turlure et al. 2004; Emiliani et al. 2005). Analysis of the LEDGF/p75 protein showed that it contains a PWWP chromatin-binding domain at the amino terminus, an A/T hook domain likely involved in DNA binding, a nuclear localization signal, and a carboxy-terminal domain that bound tightly to IN. Imaging studies showed that LEDGF/p75 could be visualized bound to condensed chromosomes at mitosis, and in the presence of LEDGF/p75, IN would accumulate on chromatin as well, suggesting that LEDGF/p75 could tether IN to chromatin (Maertens et al. 2003).
Initially it was unclear whether LEDGF/p75 was important for efficient HIV infection, but experiments eventually showed that even trace amounts of LEDGF/p75 were sufficient to promote HIV replication. Once LEDGF/p75 was efficiently depleted or knocked out, a substantial reduction in infectivity was detected. Furthermore, mapping the level of the block showed inhibition selectively at the integration step (Llano et al. 2006b; Shun et al. 2007). Quantitative polymerase chain reaction (PCR) methods, in addition to documenting reduced provirus formation, also showed that the formation of 2-LTR circles was actually increased by LEDGF/p75 depletion (Llano et al. 2006b). An increase in 2-LTR circles is also seen during infection in the presence of IN inhibitors (Arts and Hazuda 2011), likely because inhibition of integration within the nucleus provides more viral cDNA substrate to the circularization reaction, boosting circle formation. Further studies showed that still greater reductions in HIV infection efficiency could be accomplished by overexpression of the LEDFGF/p75 IN binding domain fragment by itself (De Rijck et al. 2006; Llano et al. 2006a), further supporting the importance of the LEDGF/p75-IN interaction in vivo.
Mapping of integration site distributions in the presence of LEDGF/p75 knockdowns showed that much of the targeting to transcription units was lost when LEDGF/p75 was depleted, and restoration of LEDGF/p75 rescued proper targeting, directly implicating LEDGF/p75 in the targeting mechanism (Ciuffi et al. 2005; Marshall et al. 2007; Shun et al. 2007). Binding sites for LEDGF/p75 have been mapped on chromosomes experimentally for the ENCODE regions, which comprises about 1% of the human genome. Well-supported LEDGF/p75 binding sites (“LEDGF islands”) were found to lie preferentially in transcription units, paralleling the observed preference of these locations for HIV integration (De Rijck et al. 2010). Several additional aspects of HIV target site selection were also affected by the LEDGF/p75 knockdown. Integration was more favored in regions of higher G/C content in the knockdown, and integration near CpG islands was increased. Overall, the data support a simple tethering model, in which LEDGF/p75 binds to HIV IN and simultaneously to chromatin at active transcription units, thereby directing integration to these locations.
Subsequent studies of HIV integration targeting in different cell types showed that the proportion of integration sites in transcription units differed among cell types, providing another angle for studying LEDGF/p75 function (Marshall et al. 2007). LEDGF expression levels were compared over these same cell types, which revealed that higher LEDGF/p75 expression correlated with a greater proportion of integration sites in transcription units. These findings support the importance of LEDGF/p75 for integration targeting in primary cells that were not subjected to harsh manipulations such as siRNA treatment or gene deletion.
The tethering model has received strong support from studies of artificial derivatives of LEDGF/p75, in which substitution of the LEDGF/p75 chromatin-binding region, comprised of the PWWP domain and a pair of A/T hooks (Llano et al. 2006b; Turlure et al. 2006) for alternative chromatin-binding domains allowed clear-cut retargeting of integration (Ferris et al. 2010; Gijsbers et al. 2010; Silvers et al. 2010). One dramatic example took advantage of the HP1/Cbx protein, which directs binding to sites of histone H3K9 di- and trimethylation. Substitution of the HP1/Cbx binding unit for the LEDGF/p75 chromatin-binding domain yielded a fusion protein that, when expressed in cells depleted for wild-type LEDGF/p75, retargeted integration to sites of H3K9 di- and trimethylation. Because this histone modification is enriched outside of transcription units, integration was reprogrammed to favor regions outside of genes. This provided strong support for the idea that LEDGF/p75 tethers integration complexes to target sites in vivo. Use of such fusions provides a possible means of controlling integration targeting for use in gene therapy applications, where it is desirable to target integration away from cancer-related genes to avoid insertional activation.
Might the IN-LEDGF/p75 interaction provide a target for small molecule therapy to inhibit HIV infection? X-ray crystallography studies have defined the interaction surface between the IN catalytic domain dimer and the LEDGF/p75 IN binding domain, defining the target for potential inhibitors (Cherepanov et al. 2005). An early study of small molecules binding to IN identified a binding site at the dimer interface in the catalytic domain (Molteni et al. 2001), and this later turned out to be the interaction site for LEDGF/p75 (Cherepanov et al. 2005). A more recent structure-based design effort yielded potent inhibitors that bind this site, block the IN-LEDGF/p75 interaction in vitro, and inhibit HIV replication in vivo (Christ et al. 2010). Thus the IN-LEDGF/p75 interaction appears to be a promising target for therapeutic inhibitors of HIV replication.
Why Does HIV Infection Not Cause Insertional Activation of Proto-Oncogenes?
Many studies document that retroviral infection in animal models can be associated with activation of proto-oncogenes and cancer, but remarkably this is never seen with HIV. HIV infection and AIDS are associated with elevated risks for several cancers, but in no case do the transformed cells harbor integrated proviruses, ruling out the known mechanisms of insertional activation. This issue is of considerable importance because HIV-based vectors are increasingly used in gene therapy, in part because of this observation. Possible explanations include (1) HIV vpr arrests the cell cycle, (2) HIV env expression is cytotoxic, (3) HIV infects terminally differentiated cells that consequently have limited proliferative potential, (4) cells expressing HIV proteins are quickly killed off by cytotoxic T cells, and (5) integration targeting is not optimal for transformation. However, in a recent lentiviral gene therapy trial that successfully treated β thalassemia major in one human subject, a cell clone expanded that contained an integrated vector in the proto-oncogene HMGA2 (Cavazzana-Calvo et al. 2010). This finding raises questions about possible effects of HIV integration on cell growth that may be more subtle than overt transformation.
Other Host Factors Affecting HIV Integration
A variety of additional host proteins are implicated in HIV integration. Following the completion of reverse transcription, the viral cDNA is likely to become coated by DNA- binding proteins, some of which may be contributed by the host cell. Product formation in integration reactions in vitro can be increased by the addition of proteins that assist assembly of protein/DNA complexes by altering DNA conformation (“architectural” DNA-binding proteins). However, multiple proteins show such activity in integration reactions in vitro, and it is unclear which are most important biologically. Proteins with reported stimulatory activity include LEDGF/p75, BAF, HMGA, HMGB, Ini-1, YY1, and the viral NC protein. Some of these proteins might contribute to PIC function by coating and condensing the viral DNA, thereby assisting the assembly of the viral nucleoprotein complexes. Several cellular chromatin proteins have also been suggested to influence integration including Ini-1, EED, SUV39H1, HP1γ, and others (for reviews of some of this work see Greene and Peterlin 2002; Bushman et al. 2005; Suzuki and Craigie 2007).
Cellular DNA repair proteins likely support the final steps of integration, in which DNA gaps at host-virus DNA junctions are processed and repaired. Collections of well-known host cell DNA repair enzymes can process model DNA substrates containing such gaps in vitro, but so far the enzymes most relevant in vivo during infection have not been identified (Yoder and Bushman 2000). The cellular double-strand break repair proteins Ku, ligase IV, and XRCC4, responsible for forming 2-LTR circles, have also been proposed to be important in supporting infection (Daniel et al. 1999), although this has been controversial (Baekelandt et al. 2000; Li et al. 2001). In several viral and transposon systems, DNA repair enzymes have also been suggested to inhibit replication, and additional DNA repair pathways are reportedly inhibitory for HIV-1 replication (Yoder et al. 2006).
Additional Roles for IN in HIV Biology
Studies of IN mutants have implicated IN protein in viral replication functions in addition to catalyzing DNA cutting and joining reactions (see Engelman et al. 1999 for review and references). Most amino acid substitutions within the IN active site residues selectively eliminate its catalytic activities without affecting other steps of the replication cycle—these have been termed “class I” mutants. However, amino acid substitutions in other parts of the IN protein, termed “class II” mutants, can have more pleiotropic effects, including disruption of correct core assembly in viral particles and impairment of reverse transcription, implicating IN in particle assembly and structure. Most deletions of IN result in a class II phenotype, suggesting that IN may be important for proper assembly of the viral core (Bukovsky and Gottlinger 1996; Dar et al. 2009).
CONCLUSIONS AND NEW RESEARCH AREAS
Progress in the integration field provides a classic example of how science ought to work. Early studies clarified the steps of the retroviral replication cycle and identified the IN protein. After the discovery of HIV, the field established assays for purified IN protein, which then provided the basis for small molecule screens to identify lead inhibitors active against virus. An enormous effort within the pharmaceutical industry then succeeded in turning early-stage inhibitors into pharmaceutical products. Full FDA approval of the first IN inhibitor was obtained in 2007. Now efforts turn to (1) developing inhibitors active against drug-resistant viruses and obligate IN cofactors, (2) understanding structures in more detail, and (3) understanding the integration system in its full cellular context.
ACKNOWLEDGMENTS
We thank Peter Cherepanov, Alan Engelman, and members of the Bushman and Craigie laboratories for helpful comments on the manuscript.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- *.Arts EJ, Hazuda DJ 2011. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt V, Claeys A, Cherepanov P, De Clercq E, De Strooper B, Nuttin B, Debyser Z 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J Virol 74: 11278–11285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D 1970. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature 226: 1209–1211 [DOI] [PubMed] [Google Scholar]
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD 2006. HIV integration site selection: Targeting in macrophages and the effects of different routes of viral entry. Mol Ther 14: 218–225 [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD 2006. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol 2: e157 10.1371/journal.pcbi.0020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bor YC, Bushman F, Orgel L 1995. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc Natl Acad Sci 92: 10334–10338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Brown PO, Bishop JM, Varmus HE 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev 3: 469–478 [DOI] [PubMed] [Google Scholar]
- Brady T, Agosto LM, Malani N, Berry CC, O’Doherty U, Bushman F 2009a. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 23: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T, Lee YN, Ronen K, Malani N, Berry CC, Bieniasz PD, Bushman FD 2009b. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev 23: 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319: 921–926 [DOI] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM 1987. Correct integration of retroviral DNA in vitro. Cell 49: 347–356 [DOI] [PubMed] [Google Scholar]
- Brown PO, Bowerman B, Varmus HE, Bishop JM 1989. Retroviral integration: Structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci 86: 2525–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz RA, Skalka AM 1995. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol 253: 333–346 [DOI] [PubMed] [Google Scholar]
- Bukovsky A, Gottlinger H 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol 70: 6820–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley GW, Stevenson M 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci 90: 6125–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD 2001. Lateral DNA transfer: Mechanisms and consequences. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Bushman FD, Craigie R 1992. Integration of human immunodeficiency virus DNA: Adduct interference analysis of required DNA sites. Proc Natl Acad Sci 89: 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Fujiwara T, Craigie R 1990. Retroviral DNA integration directed by HIV integration protein in vitro. Science 249: 1555–1558 [DOI] [PubMed] [Google Scholar]
- Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci 90: 3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C 2005. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol 3: 848–858 [DOI] [PubMed] [Google Scholar]
- Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol 4: 567–577 [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. 2010. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278: 372–381 [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator 75. Proc Natl Acad Sci 102: 17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu TK, Davies DR 2004. Structure and function of HIV-1 integrase. Curr Top Med Chem 4: 965–977 [DOI] [PubMed] [Google Scholar]
- Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, et al. 2010. Rational design of small-molecule inhibitors of the LEDGF/p75–integrase interaction and HIV replication. Nat Chem Biol 6: 442–448 [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Bushman FD 2006. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet 22: 388–395 [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med 11: 1287–1289 [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD 2006. Integration site selection by HIV-based vectors in dividing and growth-arrested IMR-90 lung fibroblasts. Mol Ther 13: 366–373 [DOI] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz AM 2002. Mobile DNA II. ASM Press, Washington, DC [Google Scholar]
- Craigie R, Fujiwara T, Bushman F 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62: 829–837 [DOI] [PubMed] [Google Scholar]
- Daniel R, Katz RA, Skalka AM 1999. A role for DNA-PK in retroviral DNA integration. Science 284: 644–647 [DOI] [PubMed] [Google Scholar]
- Dar MJ, Monel B, Krishnan L, Shun MC, Di Nunzio F, Helland DE, Engelman A 2009. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 6: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289: 77–85 [DOI] [PubMed] [Google Scholar]
- De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z 2006. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol 80: 11498–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, Bartholomeeusen K, Ceulemans H, Debyser Z, Gijsbers R 2010. High-resolution profiling of the LEDGF/p75 chromatin interaction in the ENCODE region. Nucleic Acids Res 38: 6135–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Varmus HE 1984. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci 81: 6461–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Ilyinskii PO, Lally K, Desrosiers RC, Engelman A 1997. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol 71: 8124–8132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR 1994. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science 266: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Eijkelenboom APAM, Puras Lutzke RA, Boelens R, Plasterk RHA, Kaptein R, Hard K 1995. The DNA binding domain of HIV-1 integrase has an SH3-like fold. Nature Struct Biol 2: 807–810 [DOI] [PubMed] [Google Scholar]
- Eijkelenboom APAM, van den Ent FMI, Vos A, Doreleijers JF, Hard K, Tullius T, Plasterk RHA, Kaptein R, Boelens R 1997. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase; A three-helix bundle stabilized by zinc. Cur Biol 1: 739–746 [DOI] [PubMed] [Google Scholar]
- Ellison VH, Abrams H, Roe T, Lifson J, Brown PO 1990. Human immunodeficiency virus integration in a cell-free system. J Virol 64: 2711–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, et al. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem 280: 25517–25523 [DOI] [PubMed] [Google Scholar]
- Engelman A, Craigie R 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol 66: 6361–6369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Mizuuchi K, Craigie R 1991. HIV-1 DNA integration: Mechanism of viral DNA cleavage and DNA strand transfer. Cell 67: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Engelman A, Maramorosch K, Murphy FA, Shatkin AJ 1999. In vivo analysis of retroviral integrase structure and function. In Advances in virus research. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc Natl Acad Sci 87: 4164–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA 1991a. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol 65: 6942–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA 1991b. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol 65: 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A 2006. HIV infection of non-dividing cells: A divisive problem. Retrovirology 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. 2010. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci 107: 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Craigie R 1989. Integration of mini-retroviral DNA: A cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci 86: 3065–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mizuuchi K 1988. Retroviral DNA integration: Structure of an integration intermediate. Cell 54: 497–504 [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Roeder RG 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J 17: 6723–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z 2010. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther 18: 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett DP, Vora AC, Schiff RD 1978. A 32,000-dalton nucleic acid-binding protein from avian retrovirus cores possesses DNA endonuclease activity. Virology 89: 119–132 [DOI] [PubMed] [Google Scholar]
- Greene WC, Peterlin BM 2002. Charting HIV’s remarkable voyage through the cell: Basic science as a passport to future therapy. Nat Med 8: 673–680 [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, Martinache C, Rieux-Laucat F, Latour S, Belohradsky BH, et al. 2010. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 363: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P 2009. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. Plos Pathogens 5 10.1371/journal.ppat.1000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J 1999. HMG protein family members stimulate human immunodeficiency virus type 1 avian sarcoma virus concerted DNA integration in vitro. J Virol 73: 2994–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman AG, Coffin JM 2005. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci 102: 6103–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, et al. 2008. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest 118: 3143–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hu W-S, Hughes SH 2011. HIV reverse transcription: Virally encoded enzyme. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A 2009. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J 276: 2926–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanson L, Subra F, Vaganay S, Hervy M, Marangoni E, Bourhis J, Mouscadet JF 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300: 100–108 [DOI] [PubMed] [Google Scholar]
- Jenkins TM, Hickman AB, Dyda F, Ghirlando R, Davies DR, Craigie R 1995. Catalytic domain of human immunodeficiency virus type 1 integrase: Identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci 92: 6057–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20: 1726–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju G, Skalka AM 1980. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: Structural similarities with transposable elements. Cell 22: 379–386 [DOI] [PubMed] [Google Scholar]
- Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63: 87–95 [DOI] [PubMed] [Google Scholar]
- Katz RA, Gravuer K, Skalka AM 1998. A preferred target DNA structure for retroviral integrase in vitro. J Biol Chem 273: 24190–24195 [DOI] [PubMed] [Google Scholar]
- Katzman M, Katz RA, Skalka AM, Leis J 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol 63: 5319–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilzer JM, Stracker TH, Beitzel B, Meek K, Weitzman MD, Bushman FD 2003. Roles of host cell factors in circularization of retroviral DNA. Virology 314: 460–467 [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Li XA, Naraharisetty HL, Hare S, Cherepanov P, Engelman A 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci 107: 15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J, Jones KS, Katz RA, Mack JPG, Skalka AM 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol 12: 2331–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Leavitt AD, Shiue L, Varmus HE 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem 268: 2113–2119 [PubMed] [Google Scholar]
- Lee MS, Craigie R 1998. Protection of retroviral DNA from autointegration: Involvement of a cellular factor. Proc Natl Acad Sci 95: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, et al. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski M, Bisgrove D, Shinn P, Chen H, Verdin E, Berry CC, Ecker JR, Bushman FD 2005. Genome-wide analysis of chromosomal features repressing HIV transcription. J Virol 79: 6610–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, et al. 2006. Retroviral DNA integration: Viral and cellular determinants of target-site selection. PLoS Pathog 2: e60 10.1371/journal.ppat.0020060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Craigie R 2005. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem 280: 29334–29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olvera JM, Yoder K, Mitchell RS, Butler SL, Lieber MR, Martin SL, Bushman FD 2001. Role of the non-homologous DNA end joining pathway in retroviral infection. EMBO J 20: 3272–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR Jr, Craigie R 2006. Retroviral DNA integration: Reaction pathway and critical intermediates. EMBO J 25: 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM 2006a. An essential role for LEDGF/p75 in HIV integration. Science 314: 461–464 [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM 2006b. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol 360: 760–773 [DOI] [PubMed] [Google Scholar]
- Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM 1995. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry 34: 9826–9833 [DOI] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278: 33528–33539 [DOI] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468: 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman F 2007. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One 2: e1340 10.1371/journal.pone.0001340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2: pE234 10.1371/journal.pbio.0020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K 1992. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem 287: 21273–21276 [PubMed] [Google Scholar]
- Mizuuchi K, Adzuma K 1991. Inversion of the phosphate chirality at the target site of Mu-DNA strand transfer—Evidence for a one-step transesterification mechanism. Cell 66: 129–140 [DOI] [PubMed] [Google Scholar]
- Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S 2001. Identification of a small molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr D 57: 536–544 [DOI] [PubMed] [Google Scholar]
- Murphy JE, Goff SP 1992. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol 66: 5092–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, De Los Santos T, Goff SP 1993. Mutational analysis of the sequences at the termini of the Moloney murine leukemia virus DNA required for integration. Virology 195: 432–440 [DOI] [PubMed] [Google Scholar]
- Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA 2004. Genome-wide analyses of avian sarcoma virus integration sites. J Virol 78: 11656–11663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban AT, Temin HM 1983. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature 306: 155–160 [DOI] [PubMed] [Google Scholar]
- Panganiban AT, Temin HM 1984. The retrovirus pol gene encodes a product required for DNA integration: Identification of a retrovirus int locus. Proc Natl Acad Sci 81: 7885–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD 1996. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science 271: 1582–1586 [DOI] [PubMed] [Google Scholar]
- Pruss D, Bushman FD, Wolffe AP 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci 91: 5913–5917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak PM, Varmus HE 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69: 769–780 [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Sil A, Varmus HE 1992. Retroviral integration into minichromosomes in vitro. EMBO J 11: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Craigie R, Davies DR 1996. Retroviral integrases and their cousins. Curr Opin Struct Biol 6: 76–83 [DOI] [PubMed] [Google Scholar]
- Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD 2009. Molecular architecture of the Mos1 paired-end complex: The structural basis of DNA transposition in a eukaryote. Cell 138: 1096–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J 12: 2099–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110: 521–529 [DOI] [PubMed] [Google Scholar]
- Schwartzberg P, Colecilli J, Goff SP 1984. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: A new viral function required for productive infection. Cell 37: 1043–1052 [DOI] [PubMed] [Google Scholar]
- Shimotohno K, Mizutani S, Temin HM 1980. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature 285: 550–554 [DOI] [PubMed] [Google Scholar]
- Shoemaker CS, Goff S, Giboa E, Paskind M, Mitra SW, Baltimore D 1980. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: Implications for retrovirus integration. Proc Natl Acad Sci 77: 3932–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21: 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers RM, Smith JA, Schowalter M, Litwin S, Liang ZH, Geary K, Daniel R 2010. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther 21: 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Pursley MH, Grandgenett DP 2002. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J Virol 76: 3105–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Grandgenett DP 2005. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol 79: 8208–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Griffith JD 1994. Human immunodeficiency virus type 1 may preferentially integrate into chromatin occupied by L1Hs repetitive elements. Proc Natl Acad Sci 91: 5557–5561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Craigie R 2007. The road to chromatin—Nuclear entry of retroviruses. Nat Rev Microbiol 5: 187–196 [DOI] [PubMed] [Google Scholar]
- Temin HM 1976. The DNA provirus hypothesis. Science 192: 1075–1080 [DOI] [PubMed] [Google Scholar]
- Temin H, Mizutani S 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226: 1211–1213 [DOI] [PubMed] [Google Scholar]
- Turlure F, Devroe E, Silver PA, Engelman A 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front Biosci 9: 3187–3208 [DOI] [PubMed] [Google Scholar]
- Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res 34: 1653–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Oude Groeneger AAM, Plasterk RHA 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci 89: 9598–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus HE, Brown PO, Berg DE, Howe MM 1989. Retroviruses. In Mobile DNA, pp. 53–108 American Society for Microbiology, Washington, DC [Google Scholar]
- Venter JC 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Wang JY, Ling H, Yang W, Craigie R 2001. Structure of a two-domain fragment of HIV-1 integrase: Implications for domain organization in the intact protein. EMBO J 20: 7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD 2007. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17: 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R, Teich N, Varmus H, Coffin J 1984. RNA tumor viruses. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300: 1749–1751 [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM, Munroe DJ 2005. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J Virol 79: 5211–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder K, Bushman FD 2000. Repair of gaps in retroviral DNA integration intermediates. J Virol 74: 11191–11200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R 2006. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc Natl Acad Sci 103: 4622–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4: 495–504 [DOI] [PubMed] [Google Scholar]