Abstract
Mammalian cells require growth-factor-receptor-initiated signaling to proliferate. Signal transduction not only initiates entry into the cell cycle, but also reprograms cellular metabolism. This instructional metabolic reprogramming is critical if the cell is to fulfill the anabolic and energetic requirements that accompany cell growth and division. Growth factor signaling mediated by the PI3K/Akt pathway plays a major role in regulating the cellular uptake of glucose, as well as the incorporation of this glucose carbon into lipids for membrane synthesis. Tyrosine-kinase-based regulation of key glycolytic enzymes such as pyruvate kinase also plays a critical role directing glucose carbon into anabolic pathways. In addition, the Myc transcription factor and mTOR kinase regulate the uptake and utilization of amino acids for protein and nucleic acid synthesis, as well as for the supply of intermediates to the mitochondrial Krebs cycle. However, the relationship between cellular signaling and metabolism is not unidirectional. Cells, by sensing levels of intracellular metabolites and the status of key metabolic pathways, can exert feedback control on signal transduction networks through multiple types of metabolite-derived protein modifications. These mechanisms allow cells to coordinate growth and division with their metabolic activity.
Receptor-mediated signal transduction, initiated by external growth factors, drives cell proliferation. It also reprograms cell metabolism to fulfill the biosynthetic needs of cell growth and division.
1. INTRODUCTION
Unicellular organisms have evolved to grow and divide rapidly when nutrients are abundant, and they take up nutrients in a cell-autonomous manner. The macromolecular precursors and free energy derived from metabolism of these nutrients are used to synthesize the new biomass required for cell growth and division. When the nutrient supply dwindles, anabolic metabolism in these organisms decreases. The cells then shift to catabolic pathways that maximize the efficiency of energy production to survive periods of nutrient limitation (Vander Heiden et al. 2009).
In multicellular organisms, cells are generally surrounded by sufficient nutrients to engage in continuous cell growth and proliferation. However, organismal integrity requires that proliferation not be a cell-autonomous process dictated by available nutrients. Mammalian cells require receptor-mediated signal transduction initiated by extracellular growth factors to leave the quiescent state and enter the cell cycle. The onset of cell growth and division introduces a metabolic requirement for sufficient carbon, nitrogen, and free energy to support synthesis of the new proteins, lipids, and nucleic acids needed by a proliferating cell. Recent studies have shown that this additional uptake of nutrients is regulated by signal transduction pathways (Fig. 1). This growth-factor-directed uptake of nutrients is critical to supporting a rate of macromolecular synthesis sufficient for growth (DeBerardinis et al. 2008; Vander Heiden et al. 2009).
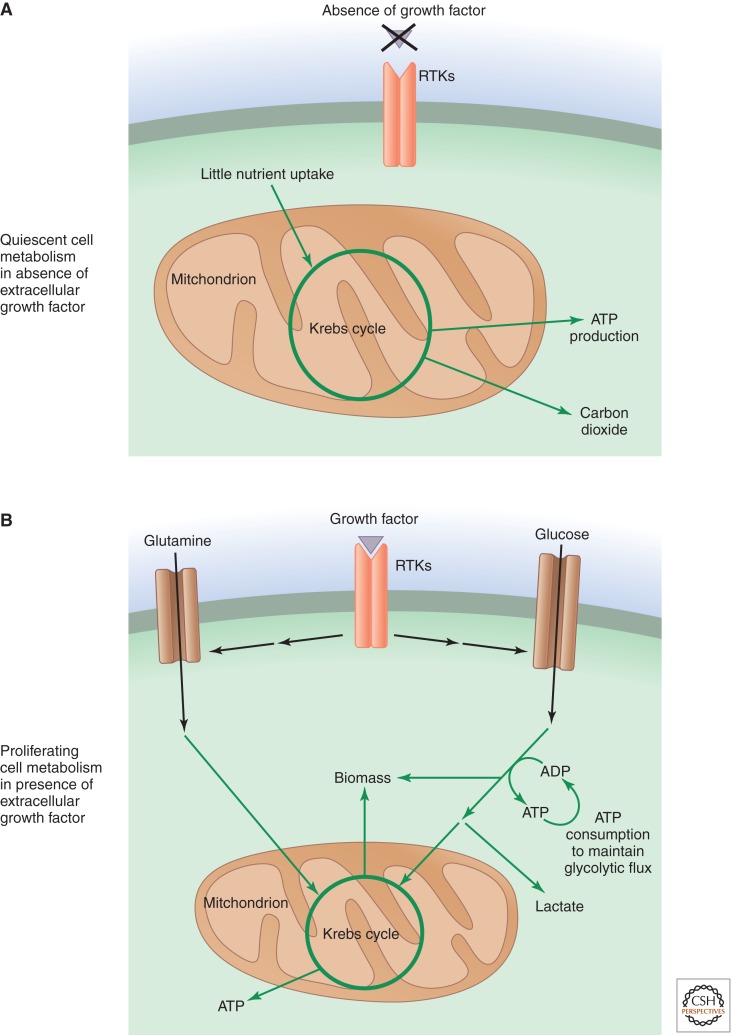
Figure 1.
Growth-factor-initiated signaling reprograms metabolism in proliferating cells. (A) In multicellular organisms, cells that are not instructed to proliferate by extracellular growth factors are generally quiescent. In these cells, glucose carbon is predominantly metabolized to carbon dioxide in the mitochondrial Krebs cycle when oxygen is available. This mitochondrial oxidation maximizes free-energy generation in the form of ATP. (B) When cells are instructed to proliferate by growth factor signaling, they increase their nutrient uptake, particularly that of glucose and glutamine. Much of this increased nutrient uptake is used to fulfill the lipid, protein, and nucleotide synthesis (biomass) required for cell growth, and the excess carbon is secreted as lactate. Proliferating cells also may adopt strategies to increase their ATP consumption to maintain glycolytic flux. Metabolic pathways are indicated by green arrows.
Mammalian cells instructed to proliferate via signal transduction are generally successful at avoiding metabolic collapse. Assuming that extracellular nutrients are abundant, these signaling-instructed cells will increase both uptake of nutrients and nutrient flux through anabolic pathways. However, if the regulation of cell growth by signaling pathways goes unchecked, problems can rapidly develop. The availability of a key extracellular nutrient could be limited in a particular context, or an important enzyme in a critical anabolic pathway may, for some reason, be deficient. Thus, to ensure that cell growth is properly coordinated with both the availability of key nutrients and with the cellular capacity to use them effectively, cells need a way to slow their own growth if their metabolic state cannot support biomass production. Such a brake on anabolic metabolism must be able to function even in the presence of growth-factor-initiated signaling. Metabolically sensitive posttranslational modifications, including acetylation and glycosylation, of signaling proteins provide an important mechanism by which cellular metabolism can exert feedback control on the output of signal transduction cascades. The relationship between cell signaling and metabolism is thus bidirectional.
2. PI3K/AKT SIGNALING CONTROLS GLUCOSE METABOLISM AND THE INCORPORATION OF CARBON INTO MACROMOLECULES
A highly conserved signal transduction pathway initiated by extracellular growth factors is the phosphoinositide 3-kinase (PI3K)/Akt pathway, whose components are conserved throughout metazoan species (Hemmings and Restuccia 2012). In mammals, the pathway plays a particularly critical role downstream from insulin signaling to facilitate glucose uptake in insulin-dependent tissues such as fat and muscle. In these tissues, the PI3K/Akt pathway promotes the trafficking of the glucose transporter GLUT4 to the cell surface (Kohn et al. 1996; Hardie 2012). However, this pathway plays multiple other roles in glucose metabolism, and its activity is not limited to those tissues classically described as insulin dependent.
In the normal, non-cancerous, setting, PI3K is activated in cells when cell membrane receptor tyrosine kinases (RTKs), as well as G-protein-coupled receptors (GPCRs) and cytokine receptors, are stimulated by extracellular growth factors. Following activation, PI3K phosphorylates membrane phosphatidylinositol lipids, which, in turn, leads to the recruitment and activation of additional kinases, most notably Akt (Fig. 2). One of the major effects of Akt on glucose metabolism is at the level of glucose uptake through increased expression of GLUT1 on the cell surface (Rathmell et al. 2003). GLUT1 is the major glucose transporter in most cell types. The translation of GLUT1 mRNA is also increased by Akt signaling through mTORC1 and 4EBP (Taha et al. 1999). In addition, Akt signaling stimulates the activity of several glycolytic enzymes. Hexokinase, which performs the first enzymatic reaction of glycolysis—glucose → glucose 6-phosphate (G6P)—is more active when associated with mitochondria, and this association is promoted by Akt (Gottlob et al. 2001). Akt also directly phosphorylates phosphofructokinase 2, and the resulting increase in fructose 2,6-bisphosphate levels enhances the activity of the glycolytic enzyme phosphofructokinase 1 (PFK1) (Deprez et al. 1997).
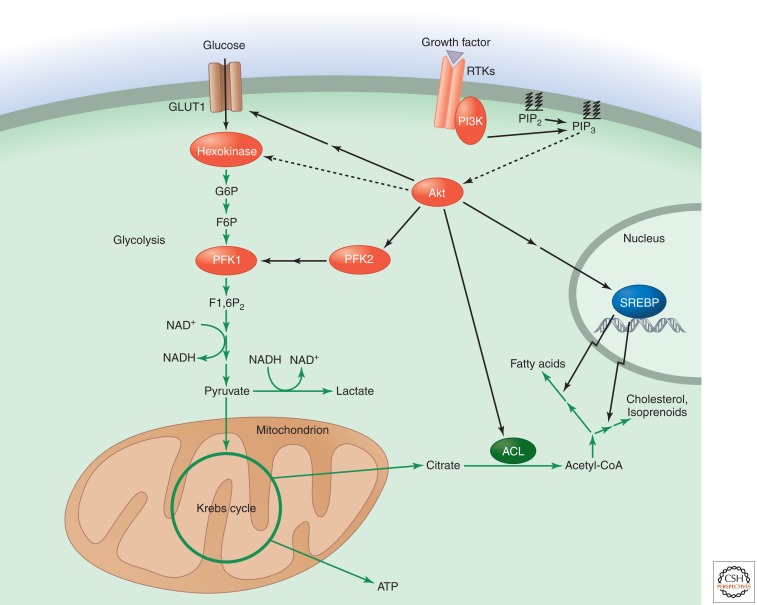
Figure 2.
Glucose carbon flux is regulated by PI3K/Akt signaling. PI3K generates phosphatidylinositol 3,4,5-trisphosphate (PIP3) from phosphatidylinositol 4,5-bisphosphate (PIP2). PIP3 recruits Akt to the cell membrane, permitting its activation by upstream kinases. Akt can then activate a variety of proteins that increase glycolytic metabolism and direct glucose carbon flux toward biosynthesis. Akt enhances glucose transporter 1 (GLUT1) translation and localization to the cell surface to increase glucose uptake and increases the activity of the glycolytic enzymes hexokinase and phosphofructokinase 1 (PFK1). Akt also promotes lipid synthesis from glucose carbon by multiple mechanisms. It can activate ATP-citrate lyase (ACL) to increase the availability of cytosolic acetyl-CoA. Akt activation can also indirectly enhance the proteolytic release of the sterol regulatory element-binding protein transcription factors (SREBPs), thereby promoting the expression of genes involved in the pathways for fatty acid and cholesterol/isoprenoid synthesis. Abbreviations: G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6P2, fructose 1,6-bisphosphate.
Akt signaling may also increase glycolytic flux indirectly by transcriptionally up-regulating the endoplasmic reticulum enzyme ENTPD5 (Fang et al. 2010). ENTPD5 promotes proper protein glycosylation and folding and does so through a cycle of reactions coupled to ATP hydrolysis. Thus, Akt-mediated up-regulation of ENTPD5 can increase the cellular ADP:ATP ratio. Increasing the ADP:ATP ratio can relieve allosteric inhibition of PFK1 by ATP. It can also lead to increased activity of downstream glycolytic enzymes that require ADP as a cofactor. Enhanced ATP consumption caused by PI3K/Akt signaling may therefore enhance the proliferating cell’s ability to rapidly metabolize glucose carbon.
Through these multiple effects on glycolytic gene expression and enzyme activity, activated Akt is able to increase the overall glycolytic rate in the cell. Even under aerobic conditions, cells with activated Akt display a greatly increased rate of glycolysis and secrete a high proportion of glucose carbon from the cell in the form of lactate (Elstrom et al. 2004). Originally described by Otto Warburg as a phenomenon specific to tumor cells, “aerobic glycolysis” is now known to be a general characteristic of rapidly proliferating cells, both cancerous and non-cancerous (Bauer et al. 2004).
The PI3K/Akt pathway also regulates the fate of glucose carbon beyond glycolysis for biosynthetic purposes. When glycolysis metabolizes glucose to pyruvate, the pyruvate can enter mitochondria and be converted to acetyl-CoA. This acetyl-CoA can then condense with oxaloacetate to form citrate. The best-understood fate of mitochondrial citrate is oxidation within the mitochondrial matrix through the Krebs citric acid cycle for ATP production. Indeed, this fate predominates in tissues such as skeletal muscle and heart, where the demand for free energy in the form of ATP is high. However, proliferating cells with high levels of glucose uptake do not oxidize all of their mitochondrial citrate, but instead transport some to the cytosol. This cytosolic citrate can be converted to cytosolic acetyl-CoA by ATP-citrate lyase (ACL), another enzyme stimulated by Akt (Berwick et al. 2002; Bauer et al. 2005).
Cytosolic acetyl CoA can initiate and/or elongate fatty acid chains as part of de novo lipid synthesis. By activating ACL, Akt signaling thus increases the availability of acetyl-CoA precursors for lipid synthesis. It also induces lipogenic genes involved in isoprenoid, cholesterol, and fatty acid biosynthesis. These actions depend on mTOR-mediated activation of protein synthesis. Enhanced synthesis of ER-exported proteins leads to depletion of ER-Golgi lipids. This, in turn, promotes ER-to-Golgi transport and proteolytic release of the sterol regulatory element-binding protein (SREBP) transcription factors (Porstmann et al. 2005; Bobrovnikova-Marjon et al. 2008; Hardie 2012). The additional lipids synthesized as a consequence of PI3K/Akt activation play a critical role as components of the plasma and organelle membranes that must be synthesized if cells are to grow and proliferate. Some of them obviously also play important signaling roles within the cell.
3. HIF1 SIGNALING PROVIDES ADDITIONAL REGULATION OF GLUCOSE METABOLISM IN RESPONSE TO BOTH OXYGEN AVAILABILITY AND NUTRIENT STATUS
The HIF1 (hypoxia inducible factor 1) signaling pathway also plays a central role in the regulation of cellular glucose metabolism. HIF1 is a transcription factor that was initially identified through its role in the adaptive cellular response to hypoxia (low oxygen tension). When oxygen is limited, the Krebs cycle in the mitochondria will cause mitochondrial redox stress if ATP continues to be produced solely by oxidative phosphorylation. Under these conditions, HIF1 promotes the expression of genes whose products are involved in anaerobic glycolysis (Fig. 3). This leads to increased generation of ATP in the cytosol from the conversion of glucose to pyruvate (Semenza et al. 1994).
Figure 3.
HIF-1 signaling responds to both hypoxia and growth factors to regulate glucose metabolism. The stability of the transcription factor HIF1 is regulated through hydroxylation of proline residues on the HIF1 α-subunit by prolyl hydroxylase enzymes (PHDs). Molecular oxygen (O2) is a substrate for PHD activity, which targets HIF1α for recognition by the Von Hippel–Lindau (VHL) ubiquitin E3 ligase and subsequent degradation by the proteasome. Inhibition of PHD activity and HIF1 degradation occurs in the presence of elevated levels of reactive oxygen species (ROS) or the Krebs cycle intermediates succinate and fumarate. HIF1 can also be positively regulated transcriptionally and translationally downstream from growth factor signaling. When not degraded, HIF1 can promote the expression of genes encoding glucose transporters and most enzymes of glycolysis. It also inhibits glucose carbon flow into the mitochondria by promoting the expression of pyruvate dehydrogenase kinase (PDK), which acts to inhibit pyruvate dehydrogenase (PDH). Furthermore, HIF1 up-regulates lactate dehydrogenase A (LDH-A) expression to increase the conversion of pyruvate to lactate. This conversion regenerates the ratio of NAD+/NADH necessary for continued flux through glycolysis.
To maintain a high rate of glycolysis, cells must also have a high rate of pyruvate → lactate conversion, which regenerates NAD+ from NADH. This is important for maintaining a sufficiently high cytoplasmic NAD+:NADH ratio to facilitate flux through the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase. HIF1 promotes lactate production and NAD+ regeneration by positively regulating the expression of the enzyme lactate dehydrogenase A. HIF1 also reduces the rate of pyruvate entry into the mitochondria by promoting the expression of pyruvate dehydrogenase kinase, an enzyme that phosphorylates and inactivates the mitochondrial pyruvate dehydrogenase complex (PDH) responsible for the formation of mitochondrial acetyl CoA from pyruvate (Kim et al. 2006; Papandreou et al. 2006). HIF1-mediated regulation of PDH flux serves to limit the flow of glucose-derived carbon into the mitochondrial, oxidative, Krebs cycle when excessive levels of pyruvate are generated. If pyruvate is oxidized in the Krebs cycle and increases electron transport beyond the cell’s ability to assimilate the resulting electron flux using molecular oxygen and/or the electrochemical potential of hydrogen in ATP generation, a considerable increase in the levels of potentially damaging reactive oxygen species (ROS) will result. HIF1-mediated regulation of PDH also limits the flow of glucose carbon into biosynthetic pathways that emanate from mitochondrial Krebs cycle intermediates (Lum et al. 2007).
HIF1 is a heterodimer composed of a labile α-subunit and a stable β-subunit (Majmundar et al. 2010). At normal oxygen levels, the α-subunit of HIF1 (HIF1α) is posttranslationally modified by hydroxylation of specific proline residues by a prolyl hydroxylase (PHD). This proline hydroxylation reaction requires both α-ketoglutarate and O2 as reactants, and generates succinate and CO2 as products. Proline hydroxylation of HIF1α promotes its association with a ubiquitin E3 ligase complex that is composed of the von Hippel–Lindau (VHL) tumor suppressor as the substrate specificity subunit together with a cullin-Rbx1 ligase (CRL). Ubiquitylation of HIF1α by the CRL–VHL complex then targets it for degradation by the proteasome. At low oxygen levels, the prolyl hydroxylation of HIF1α does not occur, because the PHD reaction requires molecular oxygen (O2) as a substrate. This permits stabilization of HIF1α during hypoxia and enhanced expression of HIF1 target genes.
In proliferating cells, several mechanisms increase HIF1 signaling even at normal oxygen tensions. Activated Akt, through mTORC1, enhances translation of HIF1α mRNA (Zhong et al. 2000; Hudson et al. 2002; Laplante and Sabatini 2011). Increased cellular nutrient uptake can also lead to inhibition of the proline hydroxylation reactions that would otherwise target HIF-1α for degradation. Excess nutrient metabolism can lead to increased cellular levels of reactive oxygen species (ROS) if reactions in the Krebs cycle occur at a rate exceeding the capacity of the electron transport chain to capture the electrons generated (Wellen and Thompson 2010). ROS are potent inhibitors of the HIF-targeting PHDs (Shatrov et al. 2003). Levels of succinate and fumarate may also increase with signaling-induced elevations in nutrient metabolism; both of these substrates can also inhibit HIF-targeting PHDs (Isaacs et al. 2005; Selak et al. 2005). PHD inhibition by succinate is an example of product inhibition. The result of PHD inhibition by ROS, succinate, and/or fumarate is a feedback mechanism that decreases the flow of glucose carbon into the mitochondria and thus inhibits further generation of ROS through mitochondrial metabolism.
4. TYROSINE KINASE SIGNALING AND PYRUVATE KINASE: REGULATION OF A METABOLIC SWITCH IN PROLIFERATING CELLS
A unique aspect of glycolysis in proliferating cells is the isoform specificity of pyruvate kinase, the enzyme that converts phosphoenolpyruvate (PEP) to pyruvate with the concomitant generation of ATP. Several alternatively spliced isoforms of pyruvate kinase exist, but most cells in the adult predominantly express the M1 isoform (PKM1). However, in early development, embryonic tissues predominantly express an alternatively spliced M2 isoform (PKM2). Furthermore, in all cancer cells examined to date, PKM2 is the predominant isoform found (Christofk et al. 2008a). The splicing factors PTB, hnRNPA1, and hnRNPA2 that regulate the alternative splicing of PKM mRNA to favor PKM2 over PKM1 expression are themselves up-regulated by the Myc transcription factor, a proto-oncogene product (Clower et al. 2010; David et al. 2010). Myc-regulated transcription of these splicing factors thereby ensures a high PKM2/PKM1 ratio in cells with activated growth factor signaling. Myc also regulates the expression of several glycolytic enzymes (Shim et al. 1997; Osthus et al. 2000).
Unlike the constitutively active PKM1 and the other pyruvate kinase isoforms, PKM2 is uniquely sensitive to regulation by tyrosine kinase signaling pathways downstream from growth factor receptors (Fig. 4). PKM2 can bind to the phosphorylated tyrosine residues in tyrosine-phosphorylated proteins and is also a target for tyrosine phosphorylation on itself (Christofk et al. 2008b; Hitosugi et al. 2009). These events lead to inhibition of PKM2 enzymatic activity, at least partly by promoting the release of PKM2’s allosteric activator fructose 1,6-bisphosphate. Binding of the latter to PKM2 causes it to form an active tetramer, but tyrosine-phosphorylated peptides displace fructose 1,6-bisphosphate and cause the PKM2 tetramer to dissociate, resulting in enzyme inactivation. The sensitivity of PKM2 to inhibition by tyrosine kinase signaling allows it to act as a gatekeeper for the metabolic fate of glucose carbon. When growth factors are present, PKM2 is inhibited, which can promote the channeling of upstream glycolytic intermediates into anabolic pathways such as nucleotide biosynthesis. When growth factors are absent, PKM2 is active and can generate pyruvate for subsequent catabolism in the Krebs cycle (Vander Heiden et al. 2009). All of this is consistent with the growth advantage that PKM2-expressing cells show in vivo compared with cells exclusively expressing PKM1 (Christofk et al. 2008a).
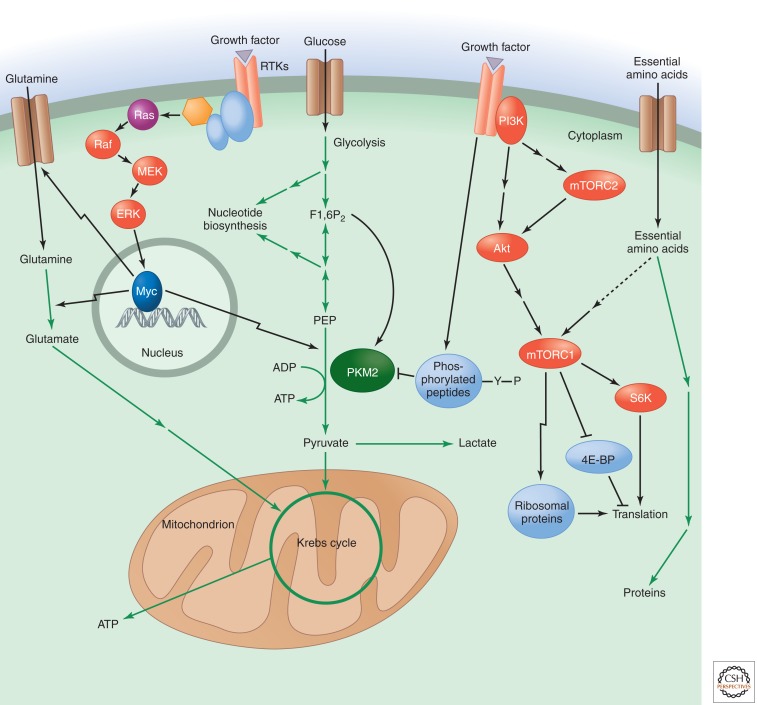
Figure 4.
Pyruvate kinase as a glycolytic switch, and mTOR and Myc regulate amino acid uptake and metabolism. Pyruvate kinase catalyzes the late step of glycolysis that converts phosphoenolpyruvate (PEP) to pyruvate, with the concomitant generation of ATP. The M2 isoform of pyruvate kinase (PKM2) is specific to proliferating cells. Its activity is allosterically activated by the upstream glycolytic intermediate fructose 1,6-bisphosphate (F1,6P2). Tyrosine kinase signaling downstream from growth factor receptors causes the release of F1,6P2 from PKM2, which results in decreased pyruvate kinase enzyme activity. This enzymatic inhibition can promote the redistribution of upstream glycolytic intermediates into anabolic pathways like nucleotide biosynthesis. The transcription factor Myc positively regulates the expression of PKM2 versus the constitutively active PKM1 isoform by up-regulating several RNA splicing factors. Glutamine uptake and metabolism are also regulated by the Myc transcription factor, which can be activated downstream from growth factor signaling pathways, such as those involving Ras. Myc positively regulates the expression of glutamine transporters as well as the enzyme glutaminase. Glutamate, the product of glutaminase activity, can be further metabolized to α-ketoglutarate to supply intermediates for the mitochondrial Krebs cycle. Another major regulator of amino acid metabolism is mTOR complex 1 (mTORC1), which can be activated downstream from PI3K/Akt signaling as well as through the sensing of essential amino acids. mTORC1 positively regulates protein synthesis in response to these inputs by activating S6 kinase (S6K), inhibiting eukaryotic initiation factor 4E binding protein (4E-BP), and promoting the expression of ribosomal proteins. A distinct mTOR complex, mTORC2, lies upstream of Akt and is positively regulated by PI3K signaling.
The full range of effects that PKM2 may have on cell growth and metabolism, however, remains incompletely characterized. Recently, an alternative glycolytic pathway has been proposed to be present in PKM2-expressing cells (Vander Heiden et al. 2010). When inactive PKM2 cannot effectively convert PEP to pyruvate, PEP can donate its high-energy phosphate to H11 of the upstream glycolytic enzyme phosphoglycerate mutase. This pathway may allow the generation of pyruvate from PEP in a step that is independent of ATP generation. Although further work is needed to characterize and purify an enzyme that can catalyze this proposed activity, the pathway may serve as an additional mechanism, besides Akt-mediated up-regulation of ENTPD5, by which proliferating cells can decrease their ATP:ADP ratio. The potential importance of PKM2 as a binding partner for a variety of signaling kinases and transcription factors has also received attention. For example, PKM2 translocates to the nucleus and potentiates the transcriptional activity of the Oct4 transcription factor involved in maintaining pluripotency (Lee et al. 2008). Many additional PKM2-interacting proteins have been reported, most recently HIF-1 and β-catenin, but the significance of each of these interactions for facilitating cell growth and proliferation requires further study.
The regulation of PKM2 activity by metabolites is also continuing to be investigated. For example, ROS have recently been proposed to cause oxidation, dissociation, and inactivation of the PKM2 tetramer (Anastasiou et al. 2011). Conversely, the non-essential amino acid serine allosterically activates PKM2 (Eigenbrodt et al. 1983). Ongoing work is addressing whether inactivation of PKM2 and the resultant accumulation of glycolytic intermediates can lead to enhanced flux through the serine synthetic pathway and thereby promote a feedback loop through serine synthesis that can modulate the flux of glycolytic intermediates to support cellular biosynthetic requirements.
5. AMINO ACID METABOLISM IS ALSO REGULATED BY CELLULAR SIGNALING CASCADES
Proliferating cells require a nitrogen source for protein and nucleotide biosynthesis. Mammalian cells acquire nitrogen through the uptake and metabolism of amino acids, using mechanisms that are also highly regulated by growth factor signaling. Myc, which is downstream from several signaling pathways, including those involving Ras and Hedgehog (see Ingham 2012; Morrison 2012), plays an especially important role in regulating metabolism of the amino acid glutamine (Yuneva et al. 2007; Wise et al. 2008; Gao et al. 2009). It is perhaps surprising that glutamine metabolism is highly regulated by growth factor signaling, because it is traditionally considered a non-essential amino acid that can be synthesized from other amino acids through enzymes such as glutamine synthetase. However, observations made by Eagle and others revealed more than 50 years ago that glutamine is uniquely critical for the proliferation of mammalian cells in culture (Wise and Thompson 2010). Myc stimulates glutamine uptake by promoting the expression of glutamine transporter proteins. It can also promote the intracellular metabolism of glutamine by increasing the expression of the enzyme glutaminase, which converts glutamine to glutamate and ammonia (Fig. 4).
Glutamine’s carbon backbone is important for replenishing the supply of mitochondrial Krebs cycle intermediates (anaplerosis). This is accomplished by the conversion of glutamine to glutamate and then to the Krebs cycle intermediate α-ketoglutarate (DeBerardinis et al. 2007). Importantly, treating cells that are glutamine-addicted because they express oncogenic Myc with a cell-permeable form of α-ketoglutarate can maintain cell viability in the absence of glutamine. However, for the cells to proliferate, the complete glutamine molecule that includes nitrogenous groups is required (Wise et al. 2008). Myc-induced glutamine catabolism can provide both the amide and amino groups needed for non-essential amino acid synthesis. It can also generate precursors for the biosynthesis of nucleotides and nicotinamide. Myc provides additional stimulation of nucleotide biosynthesis by promoting the expression of several key enzymes including TS, IMPDH2, and PRPS2 (Tong et al. 2009).
Growth factor signaling also regulates amino acid metabolism through the mTOR kinase. mTORC1, one of two protein complexes containing mTOR (see Laplante and Sabatini 2011), is stimulated by PI3K/Akt, which phosphorylates and consequently disrupts the tuberous sclerosis complex (TSC). When disrupted, the TSC complex can no longer hydrolyze the GTP-binding protein Rheb to its GDP-bound form, thus leaving the GTP-bound Rheb free to interact with and activate mTORC1 (Sengupta et al. 2010). mTORC1 inhibition can also be relieved by Akt phosphorylation of PRAS40 at T246 (Sancak et al. 2007).
mTORC1 promotes protein synthesis by phosphorylating and inhibiting the activity of 4E-BPs, which normally sequester eIF4E and thus block cap-dependent mRNA translation (Hara et al. 1997). mTORC1 can also phosphorylate and activate S6Ks to enhance translational elongation by relieving the suppression of eEF2 (Wang et al. 2001). In addition, mTORC1 regulates the translation of the 5′TOP mRNAs, an abundant class of mRNAs that includes many encoding components of the translational apparatus (Tang et al. 2001). mTORC1 also increases ribosome synthesis by activating RNA polymerase I to transcribe rRNAs (Mayer et al. 2004).
Through its responsiveness to PI3K/Akt signaling, mTORC1 activity enhances protein synthesis when a cell is directed to grow. However, mTORC1 is also highly sensitive to inputs that reflect the cell’s metabolic state. For example, upon depletion of cellular ATP levels, AMP-activated protein kinase (AMPK) can inhibit mTORC1 through multiple mechanisms, including activation of the TSC complex and inhibition of the mTOR binding partner Raptor (Corradetti et al. 2004; Gwinn et al. 2008). This provides a means for the cell to down-regulate the energetically costly process of protein synthesis when ATP is limiting. In contrast, mTORC1 can be activated by the availability of essential amino acids including leucine. Essential amino acids induce the Rag GTPase complex to assume an active conformation on lysosomal membranes. Active Rag can then recruit mTORC1 to the lysosomal surface, where it may be more amenable to activation by Rheb (Sancak et al. 2008; Sancak et al. 2010). Interestingly, recent work shows that cellular leucine uptake is coupled with glutamine efflux (Nicklin et al. 2009), linking Myc-regulated glutamine uptake with regulation of mTORC1 by essential amino acid availability.
The much less well understood mTORC2 complex also has important roles in growth factor signaling and metabolic regulation. Like mTORC1, it is activated by growth factors such as insulin. But, in contrast to mTORC1, mTORC2 lies upstream of Akt. Recent data indicate that growth-factor-stimulated activation of mTORC2 involves a direct association with ribosomes, which may ensure that mTORC2 is active only in cells that are growing and undergoing protein synthesis (Oh et al. 2010; Zinzalla et al. 2011). Once active, mTORC2 can phosphorylate Akt at S473, which is considered important both for enhancing the strength of Akt activation downstream from PI3K and for widening the range of effective Akt substrates.
6. METABOLICALLY SENSITIVE PROTEIN MODIFICATIONS LINK GROWTH FACTOR SIGNALING TO CELLULAR RESPONSES
Although many metabolic pathways that facilitate cell growth and proliferation are up-regulated in response to growth-factor-initiated signaling, the cell is not merely a passive recipient of instructions from growth factors. Rather, intracellular metabolites can exert feedback control on signaling initiated by growth factors through posttranslational modifications of critical signaling proteins (Metallo and Vander Heiden 2010).
One area where this regulation occurs is at the level of the growth factor receptor. Recent studies of cells whose ability to die by apoptosis upon nutrient withdrawal has been eliminated have shown that cells normally directed to take up nutrients upon stimulation by the growth factor interleukin 3 (IL3) can no longer take up glutamine when glucose is withdrawn from the medium (Wellen et al. 2010). This deficiency is due to the cell’s inability to continue displaying the IL3 receptor at the cell surface, necessary for IL3-dependent regulation of glutamine uptake and metabolism. Metabolism of glucose to UDP-N-acetylglucosamine (UDP-GlcNac) via the hexosamine biosynthetic pathway, which branches off glycolysis, is necessary for the proper N-linked glycosylation of the IL3 receptor α-subunit. This glycosylation is critical for the proper folding of the IL3 receptor and its localization to the cell surface. The production of UDP-GlcNac through the hexosamine pathway also requires glutamine as a nitrogen donor. Thus, the dependency of IL3 receptor signaling on receptor glycosylation ensures that this pathway remains active only when there are adequate sources of both glucose and glutamine, as well as intact enzymatic pathways for their metabolism (Fig. 5). Appearance of the TGFβ, epidermal growth factor (EGF), insulin-like growth factor, and Her2 receptors at the cell surface also responds to glucose availability and/or N-glycosylation (Wu and Derynck 2009; Fang et al. 2010).
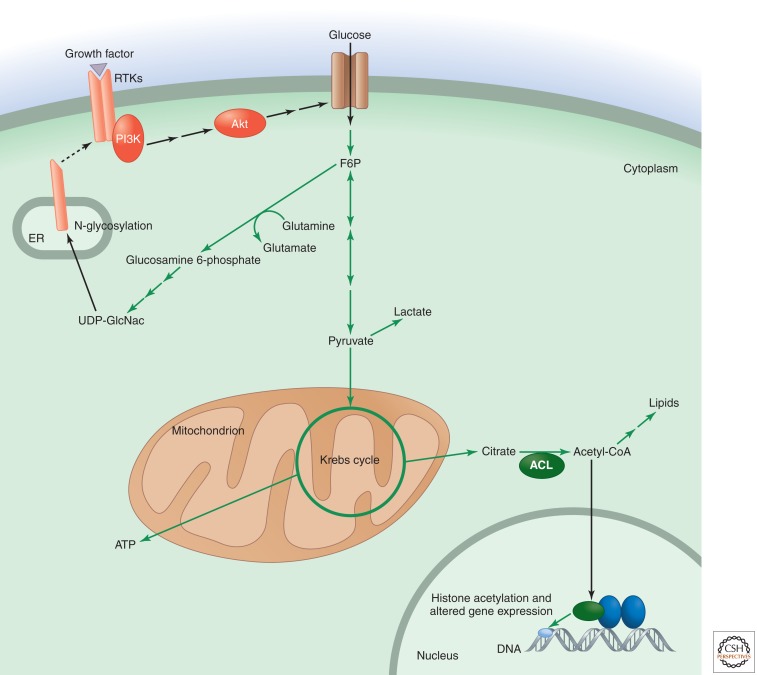
Figure 5.
Hexosamine and acetyl-CoA metabolite levels integrate signaling with cell growth and proliferation. In addition to being metabolized through glycolysis, fructose 6-phosphate can act as a nitrogen acceptor from glutamine and be converted to glucosamine 6-phosphate. Further metabolism through the hexosamine biosynthetic pathway results in UDP-N-acetylglucosamine (UDP-GlcNac). UDP-GlcNac can be used for N-linked-glycosylation reactions, including that involving growth factor receptor subunits as they are being processed in the ER. This glycosylation promotes the expression and localization of receptor subunits at the cell membrane, where they can respond to growth factor. Another metabolically sensitive protein modification occurs when acetyl-CoA is used as a substrate for the acetylation of histones in the nucleus, a modification that alters gene expression. The supply of acetyl-CoA for acetylation reactions depends on the activity of ATP-citrate lyase (ACL).
Histone acetylation is another metabolically sensitive protein modification that influences signaling outputs. Acetylation of histone lysine residues promotes open chromatin and increased gene expression (Li et al. 2007). In mammalian cells, the acetyl donor for histone acetylation, acetyl-CoA, is predominantly generated from glucose-derived citrate through the enzyme ACL (Wellen et al. 2009). Through ACL activity, increased levels of glucose availability and metabolism are thus able to facilitate open chromatin formation via increased histone acetylation. Further evidence for the concept that physiological variations in acetyl-CoA levels can alter the degree to which histones are acetylated has been found in yeast, where the transcription of cell growth genes is linked to increased histone acetylation that occurs following a surge in acetyl-CoA production upon entry into a growth phase (Cai and Tu 2011). In yeast, the acetylation of all proteins is not regulated by variations in acetyl-CoA levels; this regulation appears limited to substrates of the histone acetyltransferase Gcn5. Gcn5 has an in vitro Kd of 8.5 µm and Km of 2.5 µm, within the range of yeast intracellular acetyl-CoA concentrations that have been estimated to vary from 3 to 30 µm over the metabolic cycle (Cai et al. 2011). As in yeast, in mammalian cells, not all protein acetylation varies in response to the acetyl-CoA concentration. Although the addition of acetyl groups to histone proteins has been shown to vary with acetyl-CoA availability, the acetylation of tubulin remains relatively constant (Wellen et al. 2009). The removal of acetylation marks from histones can also be metabolically responsive. When glucose availability and metabolism are diminished, cellular NAD+ levels and the NAD+:NADH ratio are increased. This activates the sirtuin enzymes, class III histone deacetylases that remove the acetylation mark from histone lysine residues (Haigis and Sinclair 2010).
Note that cells use multiple other mechanisms to modify signaling pathways and outputs based on the metabolic state of the cell. As previously discussed, mTORC1 activity is regulated in part by the availability of essential amino acids, and cells also sense their metabolic state through AMPK (Hardie 2012). AMPK generally functions to activate/up-regulate ATP-producing pathways and to down-regulate ATP-consuming activities when cellular free-energy levels are low. Other targets of AMPK activity, in addition to mTORC1, include key enzymes for fatty acid synthesis and cholesterol synthesis: acetyl-CoA carboxylase and HMG-CoA reductase.
7. PERTURBATIONS OF CELLULAR METABOLISM IN DISEASE
In disease, particularly in cancer, the control of cellular metabolism by growth-factor-receptor-initiated signaling pathways often becomes dysregulated. As discussed previously, the preferential expression of PKM2 over PKM1 has been found in all cancer cells examined to date, and this selectivity is promoted by the Myc oncogene product. Loss-of-function mutations in the PTEN tumor suppressor that antagonizes PI3K are also common. These mutations impair negative regulation of PI3K/Akt signaling, thereby enhancing glucose uptake and the flux of glucose into lipid synthesis. Overexpression and/or constitutive activation of growth factor receptors, such as the EGF receptor and Her2, are also found frequently and can promote increased nutrient uptake and anabolic metabolism for cancer cells.
Not all disease-associated mutations affecting cellular metabolism are so clearly linked to enhanced anabolic pathways. For example, recent investigations have discovered that specific mutations in the active site of NADP+-linked cytosolic isocitrate dehdyrogenase 1 (IDH1), or in its mitochondrial relative IDH2, facilitate a neomorphic enzyme activity. This neomorphic activity converts the Krebs cycle intermediate α-ketoglutarate to a rare metabolite not found at high levels in mammalian cells under normal conditions, 2-hydroxyglutarate (2HG) (Dang et al. 2009; Ward et al. 2010, 2011). IDH1 and IDH2 mutations occur in cancers including glioma, acute myeloid leukemia, and chondrosarcoma, and in a large percentage of patients with the inborn error of metabolism 2HG aciduria (Mardis et al. 2009; Yan et al. 2009; Kranendijk et al. 2010; Ward et al. 2010; Amary et al. 2011). Although the function of the “oncometabolite” 2HG in the context of various cell types and tissues remains under active investigation, current evidence suggests that its major role is to competitively inhibit α-ketoglutarate-dependent enzymes that modify chromatin, particularly the TET family of DNA 5-methylcytosine hydroxylases and Jumonji-C domain histone demethylases (Figueroa et al. 2010; Chowdhury et al. 2011). Unlike other cancer-associated mutations, direct evidence for IDH mutations promoting cell-autonomous growth and activation of anabolic pathways is lacking, which is consistent with IDH mutations predominantly arising in lower-grade, more indolent lesions. This has led to the alternative proposal that the effects of mutant IDH and 2HG in the tumor and its microenvironment ultimately lead to a block in cellular differentiation (Ward et al. 2010).
8. CONCLUDING REMARKS
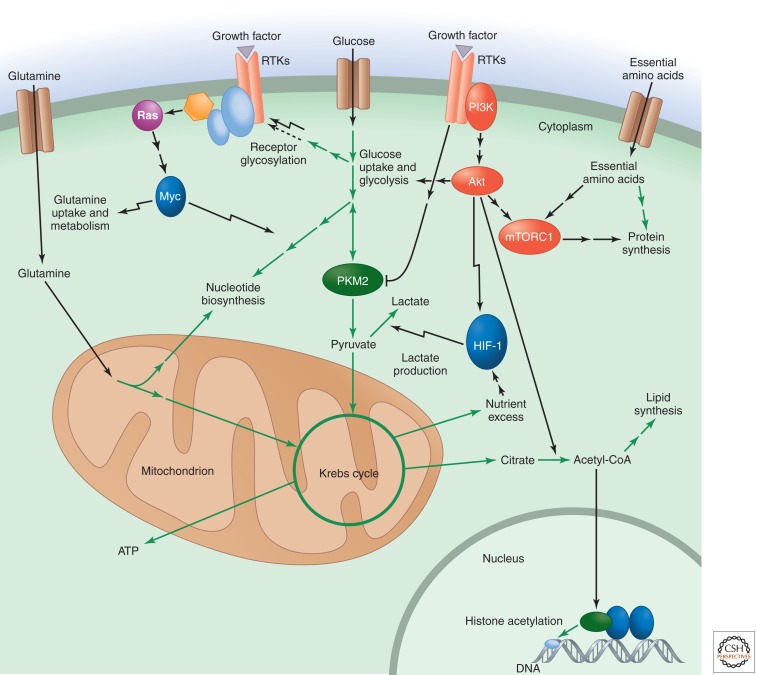
In multicellular organisms, cell growth and proliferation are normally not cell autonomous. Receptor-mediated signal transduction, initiated by extracellular growth factors, promotes entry into the cell cycle and reprograms cellular metabolism to fulfill the biosynthetic needs of cell growth and division (Fig. 6). However, despite having become highly dependent on instruction from extracellular growth factors, mammalian cells have retained the ability to sense their internal metabolic reserves and adjust their growth and biosynthetic activities accordingly. Much of this feedback control occurs at the level of posttranslational modifications of signal transduction proteins by key cellular metabolites. Moreover, intracellular metabolites can also regulate chromatin accessibility to control gene expression.
Figure 6.
Growth factor signaling reprograms cellular metabolism to promote biosynthesis but is sensitive to feedback control by metabolically sensitive protein modifications. Signaling downstream from PI3K/Akt enhances glucose uptake, glycolysis, and the flux of glucose carbon into cytosolic acetyl-CoA and lipids. HIF-1 signaling further promotes glycolysis while also enhancing the flux of pyruvate into lactate under conditions of O2 limitation or nutrient excess. The Myc transcription factor, activated downstream from Ras, enhances glutamine uptake and metabolism as well as nucleotide biosynthesis. mTORC1 signaling responds to both upstream PI3K/Akt signaling and the levels of essential amino acids to promote protein synthesis. PKM2 is a pyruvate kinase isoform specific to proliferating cells that is uniquely sensitive to inhibition by tyrosine kinase signaling downstream from growth factors. Metabolically sensitive protein modifications, such as receptor glycosylation and nuclear histone acetylation, provide a way for the cell to exert feedback control on the output of growth factor signaling.
The evolution of the ability to regulate chromatin accessibility by specific metabolites may have preceded the ability of growth factor signaling to reprogram metabolism in multicellular organisms. Nonetheless, many aspects of how variations in cellular metabolism influence chromatin accessibility remain to be fully characterized. In addition to intracellular glucose metabolism directly altering the acetylation state of histones, the methylation state of histone lysine and DNA cytosine may also be metabolically responsive. These methylation states were once thought to be irreversible, but recent work has described demethylating enzymes for both histones and DNA that are sustained by α-ketoglutarate production downstream from glutamine metabolism. As discussed above, these α-ketoglutarate-dependent enzymes can be inhibited by the 2HG produced by IDH1/IDH2 mutations.
Another area of continuing investigation is how minimizing ATP production and enhancing ATP consumption can facilitate the rapid nutrient metabolism of proliferating cells (Israelsen and Vander Heiden 2010). Minimizing cellular ATP accumulation runs counter to the metabolic strategy of quiescent cells, which completely oxidize the majority of glucose carbon in the mitochondria to maximize ATP production. But proliferating cells with activated growth factor signaling pathways usually take up nutrients far in excess of the level required to maintain ATP levels and avoid AMPK activation. Moreover, glycolysis in proliferating cells is limited by the rate of ATP consumption, not ATP production (Scholnick et al. 1973). Increasing cellular ATP consumption and the ADP:ATP ratio may be critical for relieving the inhibition of glycolytic enzymes that can occur when ATP levels are high, inhibition that would otherwise prevent high glycolytic flux and enhanced macromolecular biosynthesis from glycolytic intermediates.
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org.
REFERENCES
*Reference is also in this collection.
- Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T, et al. 2011. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 224: 334–343 [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. 2011. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to antioxidant responses. Science 334: 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Harris MH, Plas DR, Lum JJ, Hammerman PS, Rathmell JC, Riley JL, Thompson CB 2004. Cytokine stimulation of aerobic glycolysis in hematopoietic cells exceeds proliferative demand. FASEB J 18: 1303–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB 2005. ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24: 6314–6322 [DOI] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM 2002. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem 277: 33895–33900 [DOI] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA 2008. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci 105: 16314–16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Tu BP 2011. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb Symp Quant Biol 10.1101/sqb.2011.76.010769 [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP 2011. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008a. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC 2008b. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452: 181–186 [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci 107: 1894–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL 2004. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev 18: 1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB 2007. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci 104: 19345–19350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB 2008. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20 [DOI] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH 1997. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem 272: 17269–17275 [DOI] [PubMed] [Google Scholar]
- Eigenbrodt E, Leib S, Kramer W, Friis RR, Schoner W 1983. Structural and kinetic differences between the M2 type pyruvate kinases from lung and various tumors. Biomed Biochim Acta 42: S278–S282 [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. 2004. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64: 3892–3899 [DOI] [PubMed] [Google Scholar]
- Fang M, Shen Z, Huang S, Zhao L, Chen S, Mak TW, Wang X 2010. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell 143: 711–724 [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N 2001. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 15: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 30: 214–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA 2010. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol 5: 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J 1997. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 272: 26457–26463 [DOI] [PubMed] [Google Scholar]
- *.Hardie DG 2012. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hemmings BF, Restuccia DF 2012. The PI3K/Akt pathway. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. 2009. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal 2: ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT 2002. Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol Cell Biol 22: 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingham P 2012. Hedgehog signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, et al. 2005. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell 8: 143–153 [DOI] [PubMed] [Google Scholar]
- Israelsen WJ, Vander Heiden MG 2010. ATP consumption promotes cancer metabolism. Cell 143: 669–671 [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kohn AD, Summers SA, Birnbaum MJ, Roth RA 1996. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem 271: 31372–31378 [DOI] [PubMed] [Google Scholar]
- Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, Amiel J, Buist NR, Das AM, de Klerk JB, et al. 2010. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science 330: 336. [DOI] [PubMed] [Google Scholar]
- *.Laplante M, Sabatini DM 2011. mTOR signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim HK, Han YM, Kim J 2008. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol 40: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB 2007. The transcription factor HIF-1α plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev 21: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC 2010. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40: 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med 361: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Zhao J, Yuan X, Grummt I 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 18: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Vander Heiden MG 2010. Metabolism strikes back: Metabolic flux regulates cell signaling. Genes Dev 24: 2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrison D 2012. MAP kinase pathways. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E 2010. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J 29: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275: 21797–21800 [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC 2006. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197 [DOI] [PubMed] [Google Scholar]
- Porstmann T, Griffiths B, Chung YL, Delpuech O, Griffiths JR, Downward J, Schulze A 2005. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 24: 6465–6481 [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB 2003. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol 23: 7315–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM 2010. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholnick P, Lang D, Racker E 1973. Regulatory mechanisms in carbohydrate metabolism. IX. Stimulation of aerobic glycolysis by energy-linked ion transport and inhibition by dextran sulfate. J Biol Chem 248: 5175–5182 [PubMed] [Google Scholar]
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E 2005. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7: 77–85 [DOI] [PubMed] [Google Scholar]
- Semenza GL, Roth PH, Fang HM, Wang GL 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem 269: 23757–23763 [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM 2010. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatrov VA, Sumbayev VV, Zhou J, Brune B 2003. Oxidized low-density lipoprotein (oxLDL) triggers hypoxia-inducible factor-1α (HIF-1α) accumulation via redox-dependent mechanisms. Blood 101: 4847–4849 [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV 1997. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha C, Liu Z, Jin J, Al-Hasani H, Sonenberg N, Klip A 1999. Opposite translational control of GLUT1 and GLUT4 glucose transporter mRNAs in response to insulin. Role of mammalian target of rapamycin, protein kinase b, and phosphatidylinositol 3-kinase in GLUT1 mRNA translation. J Biol Chem 274: 33085–33091 [DOI] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O 2001. Amino acid–induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Zhao F, Thompson CB 2009. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev 19: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. 2010. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329: 1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG 2001. Regulation of elongation factor 2 kinase by p90 (RSK1) and p70 S6 kinase. EMBO J 20: 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Cross JR, Lu C, Weigert O, Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA, Thompson CB 2011. Identification of additional IDH mutations associated with oncometabolite R(−)-2-hydroxyglutarate production. Oncogene 10.1038/onc.2011.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Thompson CB 2010. Cellular metabolic stress: Considering how cells respond to nutrient excess. Mol Cell 40: 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24: 2784–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB 2010. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem Sci 35: 427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Derynck R 2009. Essential role of TGF-β signaling in glucose-induced cell hypertrophy. Dev Cell 17: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y 2007. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol 178: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Chiles K, Feldser D, Laughner E, Hanrahan C, Georgescu MM, Simons JW, Semenza GL 2000. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res 60: 1541–1545 [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN 2011. Activation of mTORC2 by association with the ribosome. Cell 144: 757–768 [DOI] [PubMed] [Google Scholar]