Abstract
Wnt genes are important regulators of embryogenesis and cell differentiation in vertebrates and insects. New data revealed by comparative genomics have now shown that members of the Wnt signaling pathway can be found in all clades of metazoans, but not in fungi, plants, or unicellular eukaryotes. This article focuses on new data from recent genomic analyses of several basal metazoan organisms, providing evidence that the Wnt pathway was a primordial signaling pathway during evolution. The formation of a Wnt signaling center at the site of gastrulation was instrumental for the formation of a primary, anterior–posterior body axis, which can be traced throughout animal evolution.
The Wnt pathway originated in pre-bilaterian metazoans. It served as a signaling center during gastrulation and was instrumental for the formation of a primary, anterior–posterior body axis.
Wnt genes are specific for metazoans, a monophyletic group of eukaryotes with a common origin in protozoans (Ruiz-Trillo et al. 2008; Schierwater et al. 2009). So far, no Wnt genes have been described from any unicellular eukaryotes, neither from choanoflagellates, which are presumed to represent the common ancestor of animals (King et al. 2008), nor from other protists or fungi (Sebe-Pedros et al. 2011).
Fossil records from 580-million-year (Myr)-old Ediacaran assemblages indicate that the first metazoans were diploblastic organisms similar to modern sponges and cnidarians (Xiao and Laflamme 2009). These organisms are primarily radially symmetric (Radiata) and can be distinguished from the rest of animals showing two body axes (Bilateria). Paleontological records of the Burgess Shale assemblages document that this bilaterian diversification occurred within a short period in the lower Cambrium (Conway Morris 2000). However, major questions concerning the origin of the major phyla remain unsettled, indicating that the deep nodes in metazoan evolution are difficult to resolve (Rokas et al. 2005).
Bilaterian animals are patterned along two major body axes (Niehrs 2010): the anterior–posterior (AP) axis, which is oriented parallel to the gut and, in a perpendicular orientation, the dorsal–ventral (DV) axis. Increasing evidence suggests that the primary body axis is the AP axis, whereas the secondary body axis is the DV axis (Niehrs 2010; Holstein et al. 2011). A localized Wnt/β-catenin signaling center determines the orientation of the primary body axis in both bilaterians and non-bilaterians, indicating that the conserved Wnt signaling pathway is at the base of polarized development in all metazoans (Petersen and Reddien 2009). The secondary DV axis, which is patterned by the antagonistic BMP-Chordin network, has been conserved between vertebrate and invertebrate phyla in patterning the DV axis (De Robertis 2010). Pre-bilaterian groups like sponges and cnidarians lack a DV axis. The antagonistic BMP-Chordin network seems absent in sponges, whereas in cnidarians it is initially expressed along the primary body axis (Rentzsch et al. 2008).
Bilaterians are further split into protostomes and deuterostomes based on the site of origin of their mouth (Brusca and Brusca 2002). The protostomes comprise two major phyla designated as Lophotrochozoa and Ecdysozoa, the latter including two prominent model organisms, Drosophila melanogaster and Caenorhabditis elegans. Deuterostomes include chordates (i.e., vertebrates, urochordates, and the cephalochordate Amphioxus), hemichordates, and echinoderms. The transition from radial to bilateral symmetry and the origin of the two major bilaterian groups are a matter of ongoing debate with controversial hypotheses (De Robertis and Sasai 1996; Arendt et al. 2001; Martindale and Hejnol 2009; Schierwater et al. 2009). Based primarily on the expression of Wnts and anterior marker genes, even the existence of a common ur-bilaterian ancestor has been questioned, positing that protostome and deuterostome lineages evolved independently from a radially symmetric cnidarian-, gastrula-like organism (Meinhardt 2002, 2004, 2006). The evolution of Wnt genes has therefore not only an impact for our understanding of stem cell self-renewal, cell differentiation, and apoptosis, but also for the evolution and diversification of the metazoan body plans.
ROOTS OF WNT SIGNALING IN PRE-METAZOANS
The Wnt signaling pathway is an invention of the first multicellular animals (Metazoa), because in no single-cell organism (Protozoa) has a complete Wnt signaling pathway been discovered. Nevertheless, it is possible to identify modules of this signaling pathway in several protozoans. This suggests that a successful assembly of these modules was a driving force for the formation of tissues and a signaling center at the transition of single-cell to multicellular organisms. Figure 1 summarizes the evolutionary origin of genes involved in the Wnt signaling pathway(s).
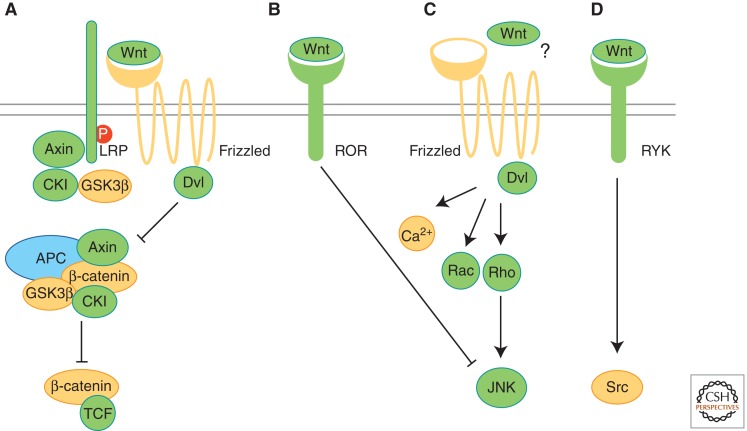
Figure 1.
Evolution of Wnt pathways. Proteins involved in different Wnt pathways (A–D) are shown and labeled according to their occurrence in protozoans (yellow), pre-bilaterians (green), and bilaterians (blue). (A) Binding of Wnt to Frizzled receptor and LRP activates β-catenin/TCF signaling. The β-catenin destruction complex, including APC, Axin, GSK3β, and CKI, sequesters and phosphorylates β-catenin. Phosphorylation of LRP is crucial for Wnt/β-catenin signaling. (B) Binding of Wnt to the receptor tyrosine kinase (RTK) Ror2 activates Jnk and inhibits β-catenin/TCF signaling. (C) Frizzled receptors in planar cell polarity (PCP) of vertebrates act via downstream messengers including Dishevelled (Dvl), small Rho GTPases, Jnk, or Ca2+. In flies, PCP can be activated independently of Wnt-Frizzled binding (?). (D) Binding of Wnt proteins to RYK RTKs results in the activation of Src proteins. (Adapted from van Amerongen and Nusse 2009; reprinted, with permission, from the authors.)
Wnt Ligands
No genes encoding for Wnt ligands have been found in any protozoan. This is different from other metazoan-specific signaling pathways, for example, Hedgehog signaling, where proteins containing a Hedge and/or Hog domain of the Hedgehog ligand can be traced back to a variety of single-cell organisms, including red algae and dinoflagellates (Ingham et al. 2011). No members of the Wnt secretion machinery like Wntless (Banziger et al. 2006; Bartscherer et al. 2006) and Porcupine (Kadowaki et al. 1996) have been found outside of the Metazoa. The invention of Wnt ligands must therefore be tightly coupled with the secretory control of Wnt proteins.
Wnt Receptors and Mediators
Wnt ligands bind to the cysteine-rich domain (CRD) of Frizzled (Fzd) receptors (Dann et al. 2001) in a ternary complex with low-density lipoprotein receptor–related proteins (LRP5/6) (Tamai et al. 2000), recruiting Dishevelled (Dsh) to the membrane. GSK3 and CK1γ phosphorylate the cytoplasmic domain of LRP5/6 (Heuberger and Birchmeier 2010), which, in turn, recruits Axin to the membrane and stabilizes β-catenin.
Wnt-related receptors have been found in the cellular slime mold Dictyostelium discoideum. In these social amoebae, several Fzd-related genes have been described, but no LRP5/6 orthologs (Eichinger et al. 2005; Prabhu and Eichinger 2006). Sixteen Fzd-related genes encode for a putative cysteine-rich domain (CRD) and have been denoted Frizzled/Smoothened-like (fsl) (Harwood 2008). Two members (fslJ and fslK) contain an additional KTXXXW motif, which is required for activation of the canonical pathway (Harwood 2008).
Three main components of the Wnt signal transduction cascade, that is, GSK3, CK1, and an ortholog related to β-catenin called Aardvark, have also been found in Dictyostelium (Grimson et al. 2000; Coates et al. 2002), but other components of the Wnt signal transduction cascade, including Dsh, Axin, and APC proteins, have not been identified (Harwood 2008). A GSK3 ortholog (GSKA) acts in cell growth and differentiation processes of Dictyostelium (Harwood 2008), for example, in the homeostasis of prestalk and prespore cells (Harwood et al. 1995; Schilde et al. 2004). Although GSK3 was also identified in other eukaryotes (Srivastava et al. 2008), the Axin-binding site present in GSK3 and other non-metazoan GSK3-related kinases can be considered as a pre-adaptation for the function of GSK3 in Wnt signaling.
The most notable member of the Wnt pathway present in Dictyostelium is the β-catenin-related molecule Aardvark (Aar) (Grimson et al. 2000). Aardvark is essential for the cAMP-mediated induction of pre-spore cells, where it acts together with GSKA. However, in contrast to the negative regulation of canonical Wnt signaling by GSK3, that is, phosphorylation of β-catenin by GSK3, Dictyostelium GSKA acts positively on Aar to induce pspA expression (Harwood 2008). As pointed out by Harwood (2008), this mechanism is reminiscent to the regulation of noncanonical Wnts in C. elegans (Hardin and King 2008).
Recently, an α-catenin ortholog has been discovered that, together with the β-catenin ortholog Aar, plays a role in cell adhesion (Dickinson et al. 2011). Mutations of Aar result in a loss of cell junctions (Coates and Harwood 2001; Hardin and King 2008). This is remarkable because cadherin-like molecules have not been found in Dictyostelium and were only identified in the genome of the choanoflagellate Monosiga brevicollis (Abedin and King 2008). This could indicate that β-catenin has an ancient function in cell adhesion that predates the evolution of its function in Wnt signaling (Dickinson et al. 2011).
Wnt Transcription Factors and Target Genes
No orthologs to the TCF/Lef transcription factors have been described outside metazoans, suggesting that these are metazoan-specific innovations (Srivastava et al. 2010). However, a Brachyury-related T-box gene was found in Capsaspora owczarzaki, a protist that is closely related to choanoflagellates and metazoans (Sebe-Pedros et al. 2011). Brachyury is a direct downstream target of Wnt signaling during posterior growth of vertebrates (Yamaguchi et al. 1999), where it directly regulates Wnt signaling (Martin and Kimelman 2008, 2009). At which point in animal evolution this positive Brachyury–Wnt loop was established is unknown so far (Martin and Kimelman 2008), but the coexpression of Wnt and Brachyury in basal metazoans (Technau et al. 2000) suggests that it was established during early metazoans evolution.
ORIGIN OF DIFFERENT WNT PATHWAYS IN PRE-BILATERIANS
Wnt signaling controls a variety of different cellular behaviors including cell proliferation, stem cell maintenance and differentiation, coordinated cell movement, and the establishment of tissue polarity (Croce and McClay 2008; van Amerongen and Nusse 2009). Based on their ability to induce an ectopic axis in Xenopus embryos (McMahon and Moon 1989) or planar cell polarity and convergent extension movements (Heisenberg et al. 2000), the different Wnts have frequently been classified as “canonical” or “noncanonical” Wnts. However, this is an oversimplification because how different Wnts signal is also determined by the receptors with which they interact. For example, the “noncanonical” Wnt5a can also signal in the canonical β-catenin pathway, if it is exposed to the right receptor on the cell, that is, Fz4 (van Amerongen et al. 2008; van Amerongen and Nusse 2009). As shown in Figure 1, in canonical β-catenin signaling, Wnts can bind to Frizzled and LRP and thereby activate β-catenin/TCF. Wnts can also interact in a β-catenin-independent manner with RYK receptors in Src signaling or with ROR in JNK signaling (via small Rho GTPases and c-Jun amino-terminal kinase) (van Amerongen and Nusse 2009). In some cases, multiple Wnts are required to induce a specific cellular response (van Amerongen and Nusse 2009).
It is difficult to judge which of the Wnt functions might represent the ancestral form—the β-catenin dependent or independent—or how this might be related to the specific structure of Wnt ligands. There are hints that canonical and noncanonical functions of Wnt proteins may be conserved during evolution (Rigo-Watermeier et al. 2011), but the distinction between both groups of ligands is not sharp (see above). It could therefore easily be that the first Wnt ligands had multiple functions in different pathways. Because the repertoire of Wnt receptors was smaller in basal metazoans compared with higher bilaterians, it is tempting to speculate that Wnt receptors in basal metazoans are more promiscuous than in vertebrates and that the evolution of Wnt receptors resulted in a larger and more refined Wnt-receptor repertoire.
Considering the core function of β-catenin in cell adhesion, an ancient role of β-catenin in cell adhesion might have further evolved to control local changes in cell signaling by Wnt ligands by creating a collective of cells with similar signaling properties (Fig. 2). These positional cues were probably the origin of the canonical pathway, fostered by the evolution of multidomain scaffolding proteins.
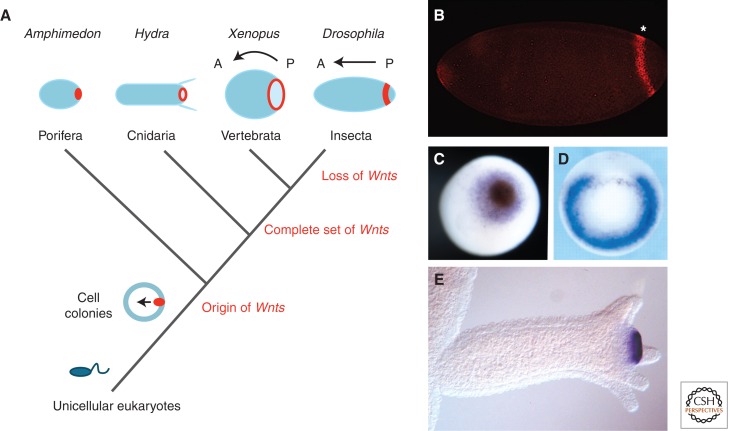
Figure 2.
Evolution of Wnt signaling and axis formation during metazoan evolution. (A) Wnt signaling centers (red) evolved at the transition from unicellular (e.g., Monosiga) to multicellular eukaryotes. In all studied metazoans (sponges, cnidarians, deuterostomes, and protostomes), a posterior Wnt signaling center defines the posterior pole of the body axis. The number of Wnt gene subfamilies increased from the first metazoans to cnidarians, whose complete set of Wnt genes is only retained in basal deuterostomes and protostomes. Protostomes are characterized by frequent loss of Wnt gene subfamilies. (B) The posterior Wg (Wnt1) stripe in Drosophila. (Figure kindly provided by Drs. P. Vorwald and E. De Robertis, Howard Hughes Medical Institute, UC Los Angeles.) (C) Wnt expression at the blastopore of a Amphimedon gastrulae (AmqWntA), at the blastopore of an early Xenopus gastrulae (Wnt8) (figure kindly provided by Dr. Maja Adamska, SARS Center, Bergen), and (D) and (E) at the hypostome of Hydra polyps (Wnt3). (Fig. 2D from Steiner et al. 2006; reprinted, with permission, from The Company of Biologists © 2006; Fig. 2E from Hobmayer et al. 2000; reprinted, with permission, from the author.)
Porifera, Placozoa, and Ctenophora
Porifera (sponges), Placozoa, and Ctenophora (comb jellies) are simple metazoans for which a basal set of Wnt ligands, receptors, and cytoplasmic transducers has been identified. Their Wnt gene repertoire is less complex than that of any other metazoans. Although the phylogenetic position of placozoans and ctenophores is still under debate (Ruiz-Trillo et al. 2008), it is likely that they represent a metazoan clade that branched off early in metazoan evolution.
Placozoans, disc-shaped creeping creatures with unknown embryology, have two epithelial layers lacking any gastric cavity (Srivastava et al. 2008). The genome contains 11,514 protein-coding genes that are closely related to those of cnidarians (Srivastava et al. 2008). The only known species, Trichoplax adhaerens, has three unclassified Wnt genes and major components of canonical Wnt signaling (Dsh, Frz, GSK3, Axin, β-catenin TCF), but no Wnt antagonists like Dkk or secreted Frizzled-related proteins (Srivastava et al. 2008). The expression patterns of the Wnt genes and β-catenin are unknown.
Sponges have a simple body plan that develops from a blastula, producing a tube-shaped diploblastic larva that is similar to the planula larva of cnidarians. This diploblastic tube invaginates at multiple positions and develops a branched filtering channel system (Brusca and Brusca 2002). The filtering cells (choanocytes) are similar in morphology to the protozoan choanoflagellate Monosiga (King et al. 2008). The genome of the demosponge Amphimedon queenslandica shows significant conservation of gene families with those of cnidarians and bilaterians (Srivastava et al. 2008). Besides β-catenin, Fzd, and GSK3, the main components of the canonical Wnt/β-catenin pathway (SFRP, Lrp5/6, Dvl, Axin, APC, TCF, and Groucho) are present, but those of the noncanonical pathways are missing (Adamska et al. 2007, 2010). Amphimedon contains three, and the homoscleromorph sponge Oscarella two, Wnt genes, which have proven difficult to classify (Lapebie et al. 2009; Adamska et al. 2010; Srivastava et al. 2010). WntA expression is restricted to the posterior pole of the Amphimedon larva, which is also marked by a pigment ring (Fig. 2C). In Oscarella, Wnt-I is also expressed at the canal openings (ostia), and β-catenin activation induces ectopic ostia (Lapebie et al. 2009). This is reminiscent of the blastoporal organizer in cnidarians (Windsor and Leys 2010), and future work must reveal to what extent these Wnt genes also contribute to non-canonical Wnt function.
In the ctenophore Mnemiopsis leidyi, orthologs of the main components of Wnt signaling have been found (β-catenin, Dishevelled, 2Fz, sFrp, TCF, Pygopus, Porcupine, LRP5/6, GSK3, APC, CK1, Groucho, Wntless), including four Wnt ligands (WntA, Wnt6, Wnt9, Wntx) (Pang et al. 2010). β-catenin is expressed at the oral pole during gastrulation, whereas the four Wnt ligands are expressed only late at the aboral side of the larva, when tentacles and sensory organs form (Pang et al. 2010).
Cnidaria
Cnidarians show an archetypal gastrula-shaped body plan. The genomes of the sea anemone Nematostella (Putnam et al. 2007) and the freshwater polyp Hydra (Chapman et al. 2010) reveal a gene repertoire with about 18,000 bona fide protein-coding genes, illustrating the high genomic complexity of the common bilaterian–cnidarian ancestor (Putnam et al. 2007; Chapman et al. 2010). All bilaterian Wnt gene subfamilies are present in these cnidarian genomes (Kusserow et al. 2005; Lee et al. 2006; Lengfeld et al. 2009). In Nematostella, Wnt-9 has not been found (Kusserow et al. 2005; Lee et al. 2006), whereas in Hydra, Wnt4, -6, and -A are missing. Three Hydra Wnt genes were classified as HyWnt9/10-a, -b, and -c (Lengfeld et al. 2009). This completeness of Wnt gene subfamilies in cnidarians suggests that the common (eumetazoan) ancestor of cnidarians and bilaterians already possessed a complete repertoire of Wnt gene ligands (Kusserow et al. 2005).
Cnidarian Wnts act in the canonical and the PCP pathways. All core components of the Wnt receptor and the β-catenin destruction complex are present. The number of Frizzled (Fzd) receptors is lower than that of the ligands (four Fzd in Hydra, and six in Nematostella), suggesting that the radiation of Wnt genes was followed by diversification of the receptors. Similarly to sponges, Nematostella Axin is lacking a clear β-catenin interaction domain, and APC shows only clear Armadillo repeats and a PDZ-domain (Adamska et al. 2010), raising questions as to whether the assembly of the cnidarian β-catenin destruction complex is as complete as in bilaterians. Orthologs of PCP signaling like Strabismus/Van Gogh, RhoA, Rock-2, and RAC1 are present, and all secreted Wnt antagonists have been identified, that is, sFrp, Wnt inhibitory factor (WIF), Cerberus, and Dkk proteins (Augustin et al. 2006; Guder et al. 2006a; Lee et al. 2006; Holstein et al. 2011; Technau and Steele 2011). Components of Ca2+ signaling (PLC, PKC, CaMKII, and Calcineurin) are also present, but no functional data exist on the putative role of Wnt5 or any other cnidarian Wnt ligand in Ca2+ signaling.
Wnt/β-catenin signaling has a major function in gastrulation and anterior–posterior patterning throughout metazoan development, because most Wnt genes are expressed at the site of the blastopore (Fig. 2E). During gastrulation, β-catenin is activated at the site of the future blastopore, and blocking GSK3 causes extended gastrulation movements in Nematostella (Wikramanayake et al. 2003). Dsh is necessary for this blastoporal activation (Lee et al. 2007). In the hydrozoan Clytia, Fz1 and maternal Wnt3 are required for oral β-catenin activation (Momose and Houliston 2007; Momose et al. 2008). In Nematostella gastrulation and planula larvae, Wnts are expressed in overlapping domains along the oral–aboral axis (Kusserow et al. 2005).
Most Wnt genes are also expressed in a similar cascade during head regeneration and budding of Hydra (Lengfeld et al. 2009). Here, canonical Wnt signaling acts upstream of non-canonical Wnt signaling, which is required for mass-tissue movements during tissue evaginations. Noncanonical processes are mediated by Wnt5, Wnt8, Fzd2, and Dsh together with JNK (Philipp et al. 2009). A role for Nematostella Wnts in Xenopus Wnt-5a/Ror2 and Wnt-11 PCP signaling was shown by different morphant phenotypes that differ in PAPC regulation, cell polarization, cell protrusion formation, and microtubule orientation (Rigo-Watermeier et al. 2011). NvWnt-5 rescued XWnt-11 and NvWnt-11 rescued XWnt-5a, suggesting that specific structures in Wnt ligands are important for receptor complex recognition in Wnt signaling.
PATTERNS OF GENE GAIN AND LOSS IN BILATERIANS
An intriguing feature in metazoan evolution is the frequent loss of Wnt gene subfamilies in protostomes (Table 1). All protostome lineages lack the Wnt3 ortholog, whereas Wnt1, Wnt5, and WntA are well conserved in most protostomes. The other Wnt gene subfamilies have been significantly reduced in various groups (Table 1). Gene loss of Wnts is also evident in some deuterostomes, but to a much lesser extent.
Table 1.
Distribution of metazoan Wnt genes
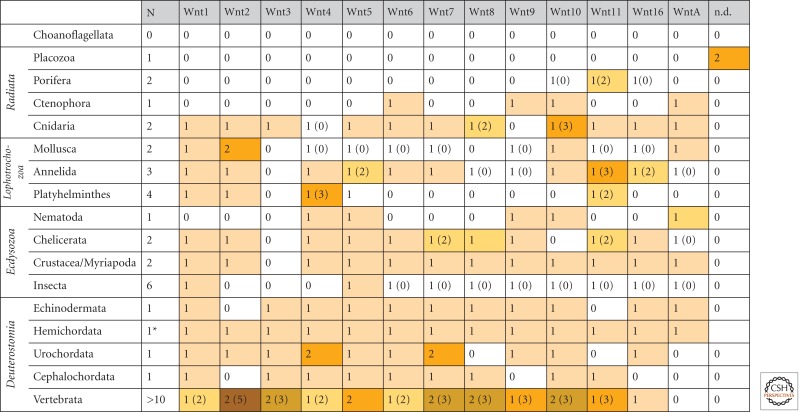 |
Notes: N indicates the number of species analyzed in a clade; numbers in parentheses indicate deviations of the number of Wnt orthologs found in species of a given clade. Colors in filled boxes indicate the abundance of Wnt gene subfamily members present in a given clade (gene duplication or gene loss). Note the loss of Wnt3 in protostomes and the loss of WntA in chordates. Data are based on molecular Wnt phylogenies by Kusserow et al. (2005), Lee et al. (2006), Croce and McClay (2008), Srivastava et al. (2008), Lengfeld et al. (2009), Lapebie et al (2009), Adamska et al (2010), Gurley et al. (2010), Janssen et al (2010), and Riddiford and Olson (2011).
(*) The Wnts from the hemichordate Saccoglossus kowalevskii correspond to Gl: 259013321 (Wnt1), Gl: 259013323 (Wnt2), Gl: 259013327 (Wnt3), Gl: 291234645 (Wnt4), Gl: 269785077 (Wnt5), Gl: 268054405 (Wnt6), Gl: 269785051 (Wnt7), Gl: 269785053 (Wnt8), Gl: 269785055 (Wnt9), Gl: 291244845 (Wnt10), Gl: 269785075 (Wnt11), Gl: 197320537 (Wnt16), Gl: 284005183 (WntA).
Lophotrochozoa
The lophotrochozoans were identified as a monophyletic group by molecular phylogenies (Halanych et al. 1995) and constitute forms with spiral cleavage and a trochophora larva (annelids and mollusks) and flatworms. Twelve Wnt subfamilies have been reported for annelids (Capitella teleta and Platynereis dumerlii), a major lophotrochozoan group. This indicates that basal protostomes also had an almost complete set of Wnt genes (Cho et al. 2010). Other annelids and mollusks have lost several Wnt gene subfamilies, that is, Wnt8, -9, and -A in Helobdella robusta or Wnt4, -5, -6, -7, -8, -9, -11, and -16 in Patella vulgata (Prud’homme et al. 2002; Cho et al. 2010; Janssen et al. 2010; Riddiford and Olson 2011). The Wnt genes of P. dumerilii show blastoporal and posterior expression in the growth zone (segment addition zone) of trochophoran larvae, and at the posterior side of each segment (Janssen et al. 2010), reminiscent to arthropods (see below).
It has been suggested that ciliated acoel and turbellarian flatworms are the most ancient bilaterians (Ruiz-Trillo et al. 1999; Baguna and Riutort 2004). Their shape is similar to cnidarian planulae, but they have an elaborated gut with one pharyngeal opening derived from the blastopore. A survey of Wnt genes in platyhelminths, including Schmidtea mediterranea (turbellarian planarian), the parasitic Schistosoma mansoni (cestode), Echinococcus granulosis (trematode), and Hymenolepis microstoma (trematode) revealed a strong tendency toward gene loss (Riddiford and Olson 2011). Planarians possess nine Wnt genes, classified as members of the Wnt1, -2, -5, and -11 subfamilies, with putative gene duplications in the Wnt11 group (Gurley et al. 2010; Riddiford and Olson 2011). Because most platyhelminths are highly diverged, it will be essential for future studies to unravel the Wnt gene repertoire of acoel flatworms, the putative bilaterian stem group.
In S. mediterranea, all Wnt genes are predominantly expressed with sFrp genes in discrete, complex, overlapping domains along the A/P axis, in a manner that suggests a possible steady-state posterior-to-anterior gradient of β-catenin activity (Gurley et al. 2008, 2010; Petersen and Reddien 2008). This is reminiscent of the overlapping Wnt expression patterns observed in the radially symmetric cnidarian body plan, the main axis of which is determined by β-catenin signaling (Gurley et al. 2010). Wnt and sFrp genes play an important role in flatworm regeneration (Reddien and Sanchez Alvarado 2004; Gurley et al. 2008, 2010; Petersen and Reddien 2008; Iglesias et al. 2011). Down-regulation of β-catenin in regenerating planarians produces an enhancement of anterior head structures (Gurley et al. 2008; Iglesias et al. 2008; Petersen and Reddien 2008). Inhibition of APC or silencing of Axin results in the regeneration of tails from anterior-facing wounds (Gurley et al. 2008) or in two-tailed planarians without a brain (Iglesias et al. 2008). In line with these data, inhibition of the Wnt inhibitor notum induces the regeneration of anterior-facing tails instead of a head, and double-RNAi experiments indicate that notum inhibits Wnt signaling in a feedback inhibition to promote head regeneration (Petersen and Reddien 2011). Thus, Wnt/β-catenin signaling clearly controls the tail-versus-head (i.e., posterior–anterior) axis in intact and regenerating planarians.
Ecdysozoa
Gene loss has also occurred in various ecdysozoan groups (Table 1), the most prominent having occurred in Caenorhabditis elegans (nematodes), which is lacking eight Wnt genes subfamilies (Wnt1–3, Wnt6–8, Wnt11, and Wnt16). The other major ecdysozoan groups are arthropods. Basal arthropods show a rich Wnt gene repertoire, indicating that the common ancestor of arthropods only lacked Wnt-3 (Janssen et al. 2010). Members of 12 Wnt gene subfamilies were characterized at the transcriptional level (Janssen et al. 2010). A relatively complete set of protostome Wnt genes was found in the genomes of the crustacean Daphnia pulex (12 Wnt subfamilies), and in chelicerates and myriapods, where Wnt10 has been lost (Janssen et al. 2010). In comparison, insects show significant gene loss (Table 1). Only Wnt1 (wingless) and Wnt5 are present in all insects investigated so far. The other Wnt genes are absent in several insects, and there seems to be a tendency for increasing loss of Wnt genes, for example, Wnt2 and Wnt4 are lacking in all insects, Wnt16 in all holometabolous insects, Wnt11 in all dipterans, and WntA in Drosophila (Janssen et al. 2010).
The expression patterns of arthropod Wnt genes reveal that most overlap in a segment addition or growth zone, and in segmentally reiterated patterns defining the parasegments (Janssen et al. 2010).
These expression patterns are remarkably similar to annelids’, suggesting similar Wnt regulation mechanisms during segment and parasegment formation in annelids and arthropods, respectively (Janssen et al. 2010). Wnt1/Wg is a prototypical Wnt gene for the posterior growth zone in annelids and arthropods. It is notable that even in Drosophila it shows an early posterior expression domain (Fig. 2B). In Drosophila, all segments are formed simultaneously (long germ band formation) and the posterior band of Wg is expressed in the early blastoderm before the segment-polarity stripes of Wg are formed (Vorwald-Denholtz and De Robertis 2011). This posterior Wg band might represent a remnant of a posterior signaling center (Vorwald-Denholtz and De Robertis 2011).
Of particular interest is the fate of Wnt8 in arthropods. In the spider Achaearanea sp. Wnt8 is required for posterior development and the maintenance of the growth zone (McGregor et al. 2008). It is expressed in a solid circular domain at the posterior end of a radial symmetric embryo before its transition to axial symmetry (Fig. 1A in McGregor et al. 2008). In Drosophila, Wnt8 has a completely different function and acts as a feedback inhibitor of the NF-κB homolog Dorsal (the gene was therefore named Wnt inhibitor of Dorsal, WntD) in DV signaling and immunity (Gordon et al. 2005; Gordon and Nusse 2006). WntD expression is under the control of Toll/Dorsal signaling, and increased levels of WntD block the nuclear accumulation of Dorsal. WntD signals independently of the β-catenin homolog Armadillo (Ganguly et al. 2005; Gordon et al. 2005).
Deuterostomia
In deuterostomes, data from more than 10 completely sequenced vertebrate genomes show unambiguously that all Wnt gene subfamilies except WntA were present at the base of vertebrate radiation (Table 1). Because WntA was identified in the genome of echinoderms (Sodergren et al. 2006; Croce and McClay 2008), the common ancestor of deuterostomes must have had a complete set of Wnt genes. Genome data suggest some Wnt gene loss in the cephalochordate amphioxus (Wnt2, -9, -16, and -A) (Putnam et al. 2007) and the urochordate ascidian Ciona (Wnt8 and -11) (Dehal et al. 2002). According to NCBI annotations, the only deuterostome with a complete set of Wnt gene subfamilies is the hemichordate Saccoglossus kowalevskii (Table 1). Although the Saccoglossus data must be confirmed by a specific phylogenetic study, it is likely that the deuterostome radiation started with a complete set of Wnt gene subfamilies.
The function and localization of Wnt and β-catenin signaling in deuterostome AP-axis formation has been recently reviewed in detail (De Robertis 2010; Niehrs 2010; Holstein et al. 2011). The general picture that emerges is that β-catenin plays an important function in the early polarization of the animal–vegetal axis of the embryo (Martin and Kimelman 2009). Similar to pre-bilaterian animals, β-catenin is vegetally localized in many basal deuterostomes. In sea urchins (echinoderms), development starts with clear polarity from the animal to the vegetal pole that resembles the oral–aboral body axis in cnidarians. β-catenin signaling marks the vegetal pole of the sea urchin embryo (Wikramanayake et al. 1998, 2004; Logan et al. 1999) and is required for the formation of the endoderm and mesoderm. Overactivation of β-catenin with LiCl results in vegetalized embryos with ectopic endoderm (Darras et al. 2011). Furthermore, in many chordates, a peak expression of β-catenin at the vegetal and blastoporal side defines a gradient along the AP axis. In both ascidians and amphioxus, nuclear-localized β-catenin marks the blastopore, which, in turn, defines the future posterior end of the embryo (Imai et al. 2000). Activation of β-catenin signaling by LiCl posteriorizes the amphioxus embryo, which is evident by loss of the neural plate (Yu et al. 2007; Onai et al. 2009). Similarly to the posterior growth zone in amphioxus tail buds (Schubert et al. 2001), nuclear β-catenin also forms a posterior-to-anterior gradient in vertebrates. Wnt signaling is involved in tail development in zebrafish (Agathon et al. 2003; Shimizu et al. 2005; Thorpe et al. 2005), where Wnt3a determines the anterior–posterior position of the somite determination front (Aulehla et al. 2003; Dunty et al. 2008; De Robertis 2010; Niehrs 2010).
SYNTENY AND EVOLUTION OF WNT GENES
Wnt genes show intriguing axial expression patterns along the anterior–posterior axis in cnidarians (e.g., in Nematostella). However, this is also true in many other bilaterians with complete sets of Wnts, where expression during embryogenesis or regeneration indicates an underlying order in transcriptional regulation. The pattern of overlapping Wnt ligands that is used in cell fate specification and patterning was characterized as the “Wnt code” (Guder et al. 2006a) or the “Wnt landscape” (Janssen et al. 2010).
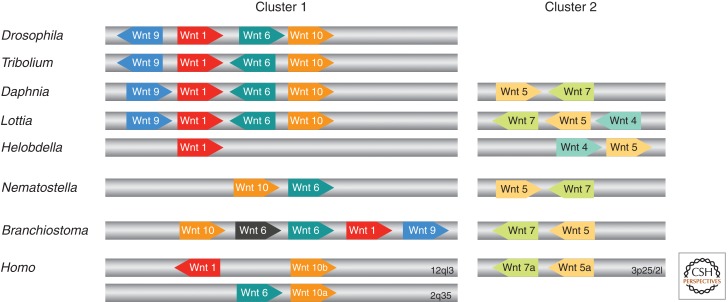
Although Wnt genes do not show a proper Hox-type collinearity on the genomic level (Sullivan et al. 2007), there is a certain degree of Wnt clustering and synteny (see also Fig. 3). A primordial cluster of Wnt genes (Wnt1, -6, and -10) was first postulated by Nusse (2001) for the Drosophila and human genomes. Accordingly, the human genome contains duplicated copies of this cluster, but Wnt6 was deleted from 12q13 and Wnt1 was deleted from 2q35 (Fig. 3). The clustering of Wnt1, -6, and -9/10 in a molecular phylogeny with Wnts from Nematostella (Kusserow et al. 2005) and Hydra (Lengfeld et al. 2009) supports this hypothesis. A second conserved cluster was found for Wnt5 and Wnt7 (Sullivan et al. 2007). New data on the arrangement of Wnt genes in the Daphnia genome revealed two syntenic clusters of these genes: Wnt9–Wnt1–Wnt6–Wnt10 and Wnt5–Wnt7 (Janssen et al. 2010). Lottia gigantea shows a similar organization of these Wnt genes and therefore reflects the ancient cluster of Wnt genes in eumetazoans (Nusse 2001; Sullivan et al. 2007; Cho et al. 2010; Janssen et al. 2010). These two Wnt clusters are also supported by recent molecular phylogenies on Wnt genes (Cho et al. 2010; Janssen et al. 2010). How the interacting Wnt ligands are regulated is rather unknown so far, but it is tempting to speculate that there is a regulative hierarchy of Wnt gene expression similar to that described for Hydra (Nakamura et al. 2011) or Drosophila (van de Wetering et al. 1997; Lessing and Nusse 1998).
Figure 3.
Synteny of metazoan Wnt genes. The relative position and orientation of Wnt genes with common scaffolds/chromosomes are shown for D. melanogaster, Tribolium castaneum, D. pulex, L. gigantea, H. robusta, Nematostella vectensis, Branchiostoma floridae, and Homo sapiens (not to scale). Numbers indicate specific chromosomes in the case of H. sapiens. (Figure created from data from Nusse [2001], Sullivan et al. [2007], Cho et al. [2010], and Janssen et al. [2010].) Note that there are more Wnts for a given species that are not clustered (see Table 1).
THE ORGANIZER: EVOLUTION OF THE POSTERIOR WNT SIGNALING CENTER
Cnidarians are the only pre-bilaterian animals with a complete set of Wnt genes and a blastoporal signaling center that is similar to that in deuterostomes and protostomes. This signaling center has been investigated in detail in the freshwater polyp Hydra, where it was shown to exhibit inductive properties that are reminiscent of the classical vertebrate organizers.
The Hydra Organizer
The Hydra organizer corresponds to the oral end of an adult polyp, which has retained the signaling properties of the blastoporal signaling center. It is known as the Hydra “head” organizer. The naming is misleading because the Hydra “head,” being a blastoporal region, represents the posterior pole of the body axis, and the Hydra “foot” is therefore the anterior pole (Meinhardt 2002; Niehrs 2010; Holstein et al. 2011). We therefore name it the “Hydra Organizer (HO).” A group of 10–50 ectodermal and endodermal epithelial cells constitutes this signaling center (Technau et al. 2000), and when transplanted to an ectopic site, the HO induces a secondary body axis by recruiting host tissue (Browne 1909; Broun and Bode 2002).
Short- and Long-Range Wnt Signaling
Activation of Wnt signaling stimulates the expression of all Wnt genes as well as of the Wnt target genes Tcf and Brachyury, resulting in numerous spot-like Wnt expression domains along the body column (Broun et al. 2005; Guder et al. 2006b; Nakamura et al. 2011). It is generally accepted that each of these Wnt signaling centers creates a gradient of diffusible Wnt, although thus far the distribution of none of the cnidarian Wnts has been visualized by using either antibodies or reporter constructs.
The parameters of the HO were determined experimentally under conditions of de novo pattern formation (Technau et al. 2000) in reaggregates from dissociated single-cell suspensions (Gierer et al. 1972). Clusters with a minimal size of five to 15 epithelial Wnt3-expressing cells are necessary and sufficient to form de novo a HO in an aggregate, generating a lateral inhibition with a diffusion range of up to 1000 µm (Technau et al. 2000).
These parameters fit with the reaction-diffusion model of Hydra pattern formation, assuming short-range autocatalytic activation and long-range inhibition (Gierer and Meinhardt 1972). In transgenic Hydra, an activator element of the Hydra Wnt3 promoter was identified that maintains Wnt3 transcription in the HO by recruiting TCF and β-catenin (Nakamura et al. 2011).
The molecular nature of long-range inhibition could also be explained by Wnt secretion. According to a model proposed by Bartscherer and Boutros (2008), Wnt ligands may have different diffusion ranges related to their mode of secretion into different vesicle populations (Bartscherer and Boutros 2008; Lorenowicz and Korswagen 2009). Wnt can be secreted with a short diffusion range at the apical side of an epithelial layer, whereas Wnt proteins that are associated with lipoprotein particles result in a longer range. We presume that both modes of Wnt secretion contribute to short- and long-range signaling in Hydra (Fig. 4B). Because β-catenin and Wnt3 act in an autoregulatory loop, a gradient of Wnt3 will result in a graded distribution of nuclear β-catenin along the body column (Guder et al. 2006a). A long-range Wnt3 signal can activate both a transcriptional repressor (see below) and the Wnt antagonist Dkk1/2/4 in the body column (Fig. 4B). Fields of lateral inhibition can be visualized by the suppression of Dkk1/2/4 expression in Hydra polyps after activation of Wnt signaling by using alsterpaullone treatment (Guder et al. 2006a,b). The double function of Wnt3 as a short-range activator and long-range inhibitor was recently proposed and elaborated in a model by Meinhardt (2012).
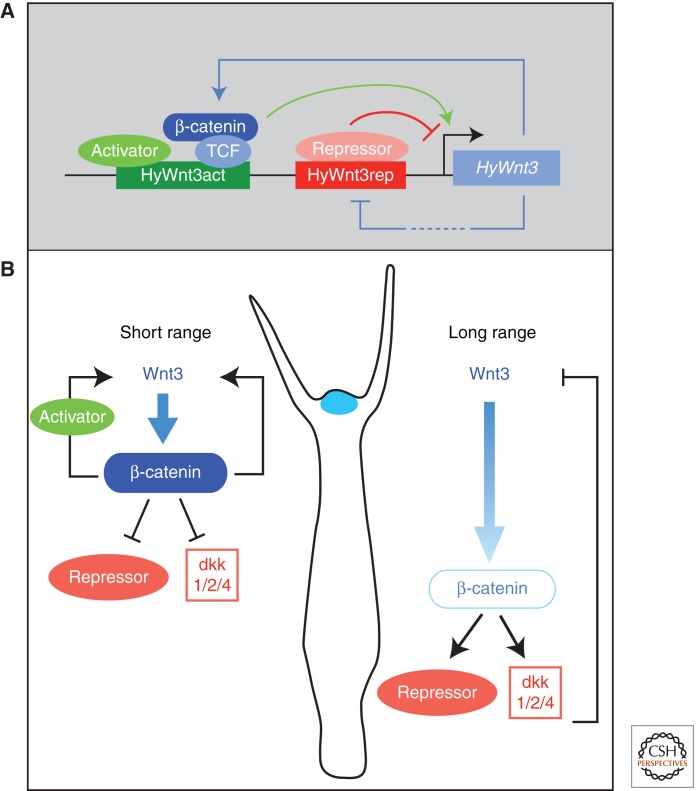
Figure 4.
Transcriptional control of Wnt signaling in Hydra. (A) Model showing the transcriptional regulation of head organizer-specific HyWnt3 expression. HyWnt3 expression is controlled by two distinct cis-regulatory elements; the activator (HyWnt3act, green) and the repressor (HyWnt3rep, red) are positively and negatively regulated by Wnt/β-catenin signaling, respectively. The β-catenin/TCF complex and putative activators (light green) bind to HyWnt3act, and their combinatorial inputs act in HyWnt3 transcription (green arrow). Potential repressors (red) bind to a repressor element (red) and inhibit HyWnt3 expression. (Figure adapted from Nakamura et al. 2011; reprinted, with permission, from the author.) (B) Presumed distribution or activity of β-catenin depends on the diffusion range of Wnt3. Following short-range diffusion, the concentrations of Wnt3 and β-catenin are high, thereby repressing putative repressors and dkk1/2/4. Following long-range diffusion, the level of nuclear β-catenin is lower, inducing putative repressors including dkk1/2/4. Positive and negative regulation restricts HyWnt3 expression (blue) to the head organizer region.
An alternative explanation for the long-range signaling could be that a cascade of sequential Wnt gene activation and expression could explain the “field” behavior of Wnt signaling, thereby extending its range. The cascade of consecutive Wnt activation during gastrulation in Nematostella (Kusserow et al. 2005) and during regeneration and bud formation in Hydra (Lengfeld et al. 2009) would support this alternative model of long-range Wnt signaling.
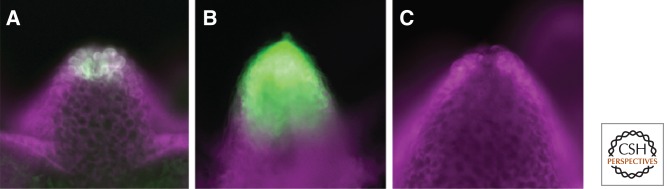
Local Restriction of the Organizer
In line with these new hypotheses, we found that Wnt signaling can also be effectively suppressed at the transcriptional level (Nakamura et al. 2011). The removal of a repressor element in the regulatory region of Wnt3 resulted in an expansion of the Wnt3 gene expression domain toward the gastric region (Fig. 5) (Nakamura et al. 2011). This shows that transcriptional regulation is essential to restrict Wnt expression to the site of the signaling center. Transcriptional regulation of Wnt3 expression is probably at the core of Wnt/β-catenin signaling in Hydra, and only complemented by Dkk1/2/4. The creation of local sources for the secretion of Wnt ligands might have been an important step in the evolution of Wnt signaling centers. We propose that cis-regulatory control mechanisms combining short-range autoactivation and long-range repression resulted in a locally defined Wnt signaling center. This signaling center may be independent of the function of extracellular Wnt antagonists, which could explain why Wnt antagonists are absent from several metazoan lineages (Nakamura et al. 2011).
Figure 5.
Expression of Wnt3 in the Hydra organizer. Reporter constructs for HyWnt3-EGFP (green) were used with an independent transformation marker (HyActin–RFP reporter gene, magenta) to ensure that all cells carry the EGFP reporter gene under examination. (A) The transgenic Hydra strain shows the localization of Wnt3 in epithelial cells at the tip of the hypostome with a complete Wnt3 promoter (HyWnt3FL). (B) Transgenic Hydra with a reporter construct lacking the HyWnt3 repressor element show dramatic expansion of expression. (C) Transgenic Hydra with a reporter construct lacking the HyWnt3 activator sequence including TCF binding sites show no expression. (From Nakamura et al. 2011; reprinted, with permission, from the author.)
CONCLUDING REMARKS
The broad variety in animal forms appears to be founded on astonishingly few signaling pathways that are shared by all major metazoan phyla. The Wnt signaling pathways had a critical function in setting up an ancient blastoporal signaling center. Molecular components of this signaling network can only be traced in part to pre-metazoan origins, indicating that major novelties were linked to the emergence of the metazoan stem. A core set of Wnt genes was already present in sponges, but only in cnidarians is a complete functional repertoire of Wnt ligands present. All Wnt genes act in these archetypical animals during gastrulation, pattern formation, and regeneration, providing a basic tool set to understand the twist of canonical and noncanonical functions in Wnt signaling. Future work should focus on unraveling the cis-regulatory control mechanisms (Nakamura et al. 2011) that link the coordinated action of Wnt genes (van Amerongen and Nusse 2009).
ACKNOWLEDGMENTS
Thanks to Ildiko Somorjai (Heidelberg) and Suat Özbek (Heidelberg) for critically reading the manuscript, to Michael Boutros (Heidelberg) and our Wnt research group for discussions, to Hans Meinhardt (Tübingen) for sharing unpublished data, to Peggy Vorwald and Edward De Robertis (Los Angeles) for providing Fig. 2B, and to Maja Adamska (Bergen) for providing Fig. 2C. This work is supported by the DFG (FOR 1036/A1).
Footnotes
Editors: Roel Nusse, Xi He, and Renee van Amerongen
Additional Perspectives on Wnt Signaling available at www.cshperspectives.org
REFERENCES
- Abedin M, King N 2008. The premetazoan ancestry of cadherins. Science 319: 946–948 [DOI] [PubMed] [Google Scholar]
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM 2007. Wnt and TGF-β expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS ONE 2: e1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamska M, Larroux C, Adamski M, Green K, Lovas E, Koop D, Richards GS, Zwafink C, Degnan BM 2010. Structure and expression of conserved Wnt pathway components in the demosponge Amphimedon queenslandica. Evol Dev 12: 494–518 [DOI] [PubMed] [Google Scholar]
- Agathon A, Thisse C, Thisse B 2003. The molecular nature of the zebrafish tail organizer. Nature 424: 448–452 [DOI] [PubMed] [Google Scholar]
- Arendt D, Technau U, Wittbrodt J 2001. Evolution of the bilaterian larval foregut. Nature 409: 81–85 [DOI] [PubMed] [Google Scholar]
- Augustin R, Franke A, Khalturin K, Kiko R, Siebert S, Hemmrich G, Bosch TC 2006. Dickkopf related genes are components of the positional value gradient in Hydra. Dev Biol 296: 62–79 [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG 2003. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev Cell 4: 395–406 [DOI] [PubMed] [Google Scholar]
- Baguna J, Riutort M 2004. The dawn of bilaterian animals: The case of acoelomorph flatworms. Bioessays 26: 1046–1057 [DOI] [PubMed] [Google Scholar]
- Banziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K 2006. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125: 509–522 [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Boutros M 2008. Regulation of Wnt protein secretion and its role in gradient formation. EMBO Rep 9: 977–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M 2006. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125: 523–533 [DOI] [PubMed] [Google Scholar]
- Broun M, Bode HR 2002. Characterization of the head organizer in hydra. Development 129: 875–884 [DOI] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR 2005. Formation of the head organizer in Hydra involves the canonical Wnt pathway. Development 132: 2907–2916 [DOI] [PubMed] [Google Scholar]
- Browne EN 1909. The production of new hydranths in Hydra by the insertion of small grafts. J Exp Zool 7: 1–37 [Google Scholar]
- Brusca RC, Brusca GJ 2002. Invertebrates. Sinauer, Sunderland, MA [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature 464: 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Valles Y, Giani VC Jr, Seaver EC, Weisblat DA 2010. Evolutionary dynamics of the wnt gene family: A lophotrochozoan perspective. Mol Biol Evol 27: 1645–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates JC, Harwood AJ 2001. Cell–cell adhesion and signal transduction during Dictyostelium development. J Cell Sci 114: 4349–4358 [DOI] [PubMed] [Google Scholar]
- Coates JC, Grimson MJ, Williams RS, Bergman W, Blanton RL, Harwood AJ 2002. Loss of the β-catenin homologue Aardvark causes ectopic stalk formation in Dictyostelium. Mech Dev 116: 117–127 [DOI] [PubMed] [Google Scholar]
- Conway Morris S 2000. The Cambrian “explosion”: Slow-fuse or megatonnage? Proc Natl Acad Sci 97: 4426–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce JC, McClay DR 2008. Evolution of the Wnt pathways. Methods Mol Biol 469: 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ 2011. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412: 86–90 [DOI] [PubMed] [Google Scholar]
- Darras S, Gerhart J, Terasaki M, Kirschner M, Lowe CJ 2011. β-Catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development 138: 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298: 2157–2167 [DOI] [PubMed] [Google Scholar]
- De Robertis EM 2010. Wnt signaling in axial patterning and regeneration: Lessons from planaria. Sci Signal 3: pe21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Sasai Y 1996. A common plan for dorsoventral patterning in Bilateria. Nature 380: 37–40 [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI 2011. A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331: 1336–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunty WC Jr, Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP 2008. Wnt3a/β-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135: 85–94 [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Jiang J, Ip YT 2005. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development 132: 3419–3429 [DOI] [PubMed] [Google Scholar]
- Gierer A, Meinhardt H 1972. A theory of biological pattern formation. Kybernetik 12: 30–39 [DOI] [PubMed] [Google Scholar]
- Gierer A, Berking S, Bode H, David CN, Flick K, Hansmann G, Schaller H, Trenkner E 1972. Regeneration of Hydra from reaggregated cells. Nat New Biol 239: 98–101 [DOI] [PubMed] [Google Scholar]
- Gordon MD, Nusse R 2006. Wnt signaling: Multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem 281: 22429–22433 [DOI] [PubMed] [Google Scholar]
- Gordon MD, Dionne MS, Schneider DS, Nusse R 2005. WntD is a feedback inhibitor of Dorsal/NF-κB in Drosophila development and immunity. Nature 437: 746–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ 2000. Adherens junctions and β-catenin-mediated cell signalling in a non-metazoan organism. Nature 408: 727–731 [DOI] [PubMed] [Google Scholar]
- Guder C, Philipp I, Lengfeld T, Watanabe H, Hobmayer B, Holstein TW 2006a. The Wnt code: Cnidarians signal the way. Oncogene 25: 7450–7460 [DOI] [PubMed] [Google Scholar]
- Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, Niehrs C, Holstein TW 2006b. An ancient Wnt–Dickkopf antagonism in Hydra. Development 133: 901–911 [DOI] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sanchez Alvarado A 2008. β-Catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sanchez Alvarado A 2010. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347: 24–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halanych KM, Bacheller JD, Aguinaldo AM, Liva SM, Hillis DM, Lake JA 1995. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science 267: 1641–1643 [DOI] [PubMed] [Google Scholar]
- Hardin J, King RS 2008. The long and the short of Wnt signaling in C. elegans. Curr Opin Genet Dev 18: 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ 2008. Dictyostelium development: A prototypic Wnt pathway? Methods Mol Biol 469: 21–32 [DOI] [PubMed] [Google Scholar]
- Harwood AJ, Plyte SE, Woodgett J, Strutt H, Kay RR 1995. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell 80: 139–148 [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW 2000. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405: 76–81 [DOI] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W 2010. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2: a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbächer U, Holstein TW 2000. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407: 186–189 [DOI] [PubMed] [Google Scholar]
- Holstein TW, Watanabe H, Ozbek S 2011. Signaling pathways and axis formation in the lower metazoa. Curr Top Dev Biol 97: 137–177 [DOI] [PubMed] [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Salo E, Adell T 2008. Silencing of Smed-β-catenin1 generates radial-like hypercephalized planarians. Development 135: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Iglesias M, Almuedo-Castillo M, Aboobaker AA, Salo E 2011. Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/β-catenin pathway. Dev Biol 358: 68–78 [DOI] [PubMed] [Google Scholar]
- Imai K, Takada N, Satoh N, Satou Y 2000. β-Catenin mediates the specification of endoderm cells in ascidian embryos. Development 127: 3009–3020 [DOI] [PubMed] [Google Scholar]
- Ingham PW, Nakano Y, Seger C 2011. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet 12: 393–406 [DOI] [PubMed] [Google Scholar]
- Janssen R, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, Schwager EE, Hopfen C, Colbourne JK, Budd GE, Brown SJ, et al. 2010. Conservation, loss, and redeployment of Wnt ligands in protostomes: Implications for understanding the evolution of segment formation. BMC Evol Biol 10: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N 1996. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev 10: 3116–3128 [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451: 783–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, et al. 2005. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433: 156–160 [DOI] [PubMed] [Google Scholar]
- Lapebie P, Gazave E, Ereskovsky A, Derelle R, Bezac C, Renard E, Houliston E, Borchiellini C 2009. WNT/β-catenin signalling and epithelial patterning in the homoscleromorph sponge Oscarella. PLoS ONE 4: e5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PN, Pang K, Matus DQ, Martindale MQ 2006. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin Cell Dev Biol 17: 157–167 [DOI] [PubMed] [Google Scholar]
- Lee PN, Kumburegama S, Marlow HQ, Martindale MQ, Wikramanayake AH 2007. Asymmetric developmental potential along the animal–vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev Biol 310: 169–186 [DOI] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Ozbek S, Bode H, Holstein TW 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330: 186–199 [DOI] [PubMed] [Google Scholar]
- Lessing D, Nusse R 1998. Expression of wingless in the Drosophila embryo: A conserved cis-acting element lacking conserved Ci-binding sites is required for patched-mediated repression. Development 125: 1469–1476 [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR 1999. Nuclear β-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126: 345–357 [DOI] [PubMed] [Google Scholar]
- Lorenowicz MJ, Korswagen HC 2009. Sailing with the Wnt: Charting the Wnt processing and secretion route. Exp Cell Res 315: 2683–2689 [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D 2008. Regulation of canonical Wnt signaling by Brachyury is essential for posterior mesoderm formation. Developmental cell 15: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Kimelman D 2009. Wnt signaling and the evolution of embryonic posterior development. Curr Biol 19: R215–R219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale MQ, Hejnol A 2009. A developmental perspective: Changes in the position of the blastopore during bilaterian evolution. Dev Cell 17: 162–174 [DOI] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WG 2008. Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr Biol 18: 1619–1623 [DOI] [PubMed] [Google Scholar]
- McMahon AP, Moon RT 1989. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell 58: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Meinhardt H 2002. The radial-symmetric Hydra and the evolution of the bilateral body plan: An old body became a young brain. Bioessays 24: 185–191 [DOI] [PubMed] [Google Scholar]
- Meinhardt H 2004. Different strategies for midline formation in bilaterians. Nat Rev Neurosci 5: 502–510 [DOI] [PubMed] [Google Scholar]
- Meinhardt H 2006. Primary body axes of vertebrates: Generation of a near-Cartesian coordinate system and the role of Spemann-type organizer. Dev Dyn 235: 2907–2919 [DOI] [PubMed] [Google Scholar]
- Meinhardt H 2012. Modeling pattern formation in Hydra—a route to understand essential steps in development. Int J Dev Biol 10.1387/ijdb.113483hm [DOI] [PubMed] [Google Scholar]
- Momose T, Houliston E 2007. Two oppositely localised Frizzled RNAs as axis determinants in a cnidarian embryo. PLoS Biol 5: e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T, Derelle R, Houliston E 2008. A maternally localised Wnt ligand required for axial patterning in the cnidarian Clytia hemisphaerica. Development 135: 2105–2113 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Tsiairis CD, Ozbek S, Holstein TW 2011. Autoregulatory and repressive inputs localize Hydra Wnt3 to the head organizer. Proc Natl Acad Sci 108: 9137–9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C 2010. On growth and form: A Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137: 845–857 [DOI] [PubMed] [Google Scholar]
- Nusse R 2001. An ancient cluster of Wnt paralogues. Trends Genet 17: 443. [DOI] [PubMed] [Google Scholar]
- Onai T, Lin HC, Schubert M, Koop D, Osborne PW, Alvarez S, Alvarez R, Holland ND, Holland LZ 2009. Retinoic acid and Wnt/β-catenin have complementary roles in anterior/posterior patterning embryos of the basal chordate amphioxus. Dev Biol 332: 223–233 [DOI] [PubMed] [Google Scholar]
- Pang K, Ryan JF, Mullikin JC, Baxevanis AD, Martindale MQ 2010. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. Evodevo 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2008. Smed-β-catenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319: 327–330 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2009. Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1068 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2011. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332: 852–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp I, Aufschnaiter R, Ozbek S, Pontasch S, Jenewein M, Watanabe H, Rentzsch F, Holstein TW, Hobmayer B 2009. Wnt/β-catenin and noncanonical Wnt signaling interact in tissue evagination in the simple eumetazoan Hydra. Proc Natl Acad Sci 106: 4290–4295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu Y, Eichinger L 2006. The Dictyostelium repertoire of seven transmembrane domain receptors. Eur J Cell Biol 85: 937–946 [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Lartillot N, Balavoine G, Adoutte A, Vervoort M 2002. Phylogenetic analysis of the Wnt gene family. Insights from lophotrochozoan members. Curr Biol 12: 1395. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sanchez Alvarado A 2004. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757 [DOI] [PubMed] [Google Scholar]
- Rentzsch F, Fritzenwanker JH, Scholz CB, Technau U 2008. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 135: 1761–1769 [DOI] [PubMed] [Google Scholar]
- Riddiford N, Olson PD 2011. Wnt gene loss in flatworms. Dev Genes Evol 221: 187–197 [DOI] [PubMed] [Google Scholar]
- Rigo-Watermeier T, Kraft B, Ritthaler M, Wallkamm V, Holstein TW, Wedlich D 2011. Functional conservation of Nematostella Wnts in canonical and noncanonical Wnt-signaling. Biol Open 10.1242/bio.2011021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Kruger D, Carroll SB 2005. Animal evolution and the molecular signature of radiations compressed in time. Science 310: 1933–1938 [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Riutort M, Littlewood DT, Herniou EA, Baguna J 1999. Acoel flatworms: Earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science 283: 1919–1923 [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF 2008. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol 25: 664–672 [DOI] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R 2009. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol 7: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C, Araki T, Williams H, Harwood A, Williams JG 2004. GSK3 is a multifunctional regulator of Dictyostelium development. Development 131: 4555–4565 [DOI] [PubMed] [Google Scholar]
- Schubert M, Holland LZ, Stokes MD, Holland ND 2001. Three amphioxus Wnt genes (AmphiWnt3, AmphiWnt5, and AmphiWnt6) associated with the tail bud: The evolution of somitogenesis in chordates. Dev Biol 240: 262–273 [DOI] [PubMed] [Google Scholar]
- Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I 2011. Unexpected repertoire of Metazoan transcription factors in the unicellular Holozoan Capsaspora owczarzaki. Mol Biol Evol 28: 1241–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Bae YK, Muraoka O, Hibi M 2005. Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev Biol 279: 125–141 [DOI] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. 2006. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454: 955–960 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner AB, Engleka MJ, Lu Q, Piwarzyk EC, Yaklichkin S, Lefebvre JL, Walters JW, Pineda-Salgado L, Labosky PA, Kessler DS 2006. FoxD3 regulation of Nodal in the Spemann organizer is essential for Xenopus dorsal mesoderm development. Development 133: 4827–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JC, Ryan JF, Mullikin JC, Finnerty JR 2007. Conserved and novel Wnt clusters in the basal eumetazoan Nematostella vectensis. Dev Genes Evol 217: 235–239 [DOI] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X 2000. LDL-receptor-related proteins in Wnt signal transduction. Nature 407: 530–535 [DOI] [PubMed] [Google Scholar]
- Technau U, Steele RE 2011. Evolutionary crossroads in developmental biology: Cnidaria. Development 138: 1447–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Cramer von Laue C, Rentzsch F, Luft S, Hobmayer B, Bode HR, Holstein TW 2000. Parameters of self-organization in Hydra aggregates. Proc Natl Acad Sci 97: 12127–12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CJ, Weidinger G, Moon RT 2005. Wnt/β-catenin regulation of the Sp1-related transcription factor sp5l promotes tail development in zebrafish. Development 132: 1763–1772 [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R 2009. Towards an integrated view of Wnt signaling in development. Development 136: 3205–3214 [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R 2008. Alternative wnt singnaling is initiated by distinct receptors. Sci Signal 1: re9. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- Vorwald-Denholtz PP, De Robertis EM 2011. Temporal pattern of the posterior expression of Wingless in Drosophila blastoderm. Gene Expr Patterns 11: 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Huang L, Klein WH 1998. β-Catenin is essential for patterning the maternally specified animal–vegetal axis in the sea urchin embryo. Proc Natl Acad Sci 95: 9343–9348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, Xu R, Martindale MQ 2003. An ancient role for nuclear β-catenin in the evolution of axial polarity and germ layer segregation. Nature 426: 446–450 [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH 2004. Nuclear β-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis 39: 194–205 [DOI] [PubMed] [Google Scholar]
- Windsor PJ, Leys SP 2010. Wnt signaling and induction in the sponge aquiferous system: Evidence for an ancient origin of the organizer. Evol Dev 12: 484–493 [DOI] [PubMed] [Google Scholar]
- Xiao S, Laflamme M 2009. On the eve of animal radiation: Phylogeny, ecology and evolution of the Ediacara biota. Trends Ecol Evol 24: 31–40 [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev 13: 3185–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JK, Satou Y, Holland ND, Shin IT, Kohara Y, Satoh N, Bronner-Fraser M, Holland LZ 2007. Axial patterning in cephalochordates and the evolution of the organizer. Nature 445: 613–617 [DOI] [PubMed] [Google Scholar]