Abstract
In Drosophila and other Dipterans, homologous chromosomes are in close contact in virtually all nuclei, a phenomenon known as somatic homolog pairing. Although homolog pairing has been recognized for over a century, relatively little is known about its regulation. We performed a genome-wide RNAi-based screen that monitored the X-specific localization of the male-specific lethal (MSL) complex, and we identified 59 candidate genes whose knockdown via RNAi causes a change in the pattern of MSL staining that is consistent with a disruption of X-chromosomal homolog pairing. Using DNA fluorescent in situ hybridization (FISH), we confirmed that knockdown of 17 of these genes has a dramatic effect on pairing of the 359 bp repeat at the base of the X. Furthermore, dsRNAs targeting Pr-set7, which encodes an H4K20 methyltransferase, cause a modest disruption in somatic homolog pairing. Consistent with our results in cultured cells, a classical mutation in one of the strongest candidate genes, pebble (pbl), causes a decrease in somatic homolog pairing in developing embryos. Interestingly, many of the genes identified by our screen have known roles in diverse cell-cycle events, suggesting an important link between somatic homolog pairing and the choreography of chromosomes during the cell cycle.
Keywords: RNAi, homolog pairing, male-specific lethal, cell cycle, interchromosomal interaction
The eukaryotic nucleus is organized and dynamic. During interphase, the genome is packaged to permit expression of active genes and facilitate the silencing of inactive genes, all while maintaining plasticity to allow for variation in gene expression in response to the environment (reviewed in Branco and Pombo 2007; Heard and Bickmore 2007). As cells divide, chromosomes undergo radical changes in their conformations, most often through large-scale condensation followed by the coordinated events of mitosis, after which interphase organization is reestablished in daughter nuclei (Essers et al. 2005; Gerlich et al. 2003; Thomson et al. 2004; Walter et al. 2003). Although we have gained considerable understanding of these chromosomal dynamics, the genes responsible for establishing and maintaining order in the interphase nucleus remain poorly understood.
Drosophila melanogaster provides an excellent model for the study of nuclear organization. Drosophila has a relatively small genome, and high throughput genetic and genomic tools have been used to generate genome-wide maps of chromatin modifications (Filion et al. 2010; Kharchenko et al. 2011) and three-dimensional chromosomal interactions (Sexton et al. 2012). The Drosophila nucleus follows a simple higher-order organizational principle, namely, that homologous chromosomes are paired together from end to end in the majority of somatic cells (reviewed by McKee 2004). Notably, somatic homolog pairing can influence gene expression through interactions between regulatory elements on homologous chromosomes, a phenomenon known as transvection (reviewed by Duncan 2002; Kennison and Southworth 2002). Extensive and stable pairing between homologous chromosomes appears to be unique to Dipteran insects, but cytological analyses have demonstrated that interactions between homologous sequences can also occur in somatic nuclei of vertebrate species (Bacher et al. 2006; Koeman et al. 2008; Xu et al. 2006). Furthermore, widespread pairing of homologous chromosomes is critical for the proper execution of meiosis in many organisms (reviewed by Jordan 2006; McKee 2004; Sybenga 1999), suggesting that mechanisms that identify and align homologous sequences are important to diverse species.
How are homologous chromosomes paired in somatic nuclei of Drosophila? Previous analyses of developing Drosophila embryos suggest that contacts between homologs initiate independently along the chromosome (Fung et al. 1998) rather than spreading from fixed pairing centers as observed for meiotic pairing in C. elegans (MacQueen et al. 2005). In addition, somatic homolog pairing is dynamic and sensitive to events of the cell cycle, as analyses based on DNA FISH have shown that progression through either S phase (Csink and Henikoff 1998) or late stages of mitosis (Fung et al. 1998; also see Williams et al. 2007) can disrupt pairing. Several gene products have been shown to influence somatic homolog pairing in flies; for example, experiments using dsRNAs and chemical inhibitors targeting Topoisomerase 2 (Top2) have demonstrated its requirement for normal pairing of homologous euchromatic sequences in cultured Drosophila cells (Williams et al. 2007). Similarly, loss-of-function mutations in the zinc finger protein encoded by Suppressor of Hairy wing [Su(Hw)] cause a reduction in pairing of homologous euchromatic sequences in developing embryos (Fritsch et al. 2006). In contrast, genetic and cytological analyses suggest that Chromosome-associated protein H2 (Cap-H2), a component of the condensin II complex, antagonizes somatic homolog pairing (Hartl et al. 2008). Other genetic analyses have had the potential to uncover genes involved in somatic homolog pairing, but their reliance on phenotypes generated by transvection in vivo (e.g. Gelbart 1982; Lewis 1954; Su et al. 2001) or restriction to a specific developmental window (Bateman and Wu 2008) has limited their capacity to uncover novel pairing regulators. Thus, systematic approaches to identify Drosophila genes that affect somatic homolog pairing specifically, and nuclear organization in general, are as yet incomplete.
Here, we describe a genome-wide RNAi-based screen that uncovers factors affecting somatic homolog pairing in Drosophila cell culture. Our screen took advantage of the MSL complex, a key regulator of dosage compensation that specifically associates with the X chromosome (Belote and Lucchesi 1980). The MSL complex is a histone acetyltransferase complex that increases transcript levels of X-linked genes in males 2-fold to equalize transcript levels with females, which have two X chromosomes (Hamada et al. 2005; Smith et al. 1998). In male-derived S2 cultured cells (Schneider 1972), which are amenable to manipulation by RNAi (Clemens et al. 2000), the MSL complex maintains its specificity for X-chromosomal sequences; in this case, approximately two X chromosomes per nucleus are targeted due to tetraploidy/aneuploidy of the cell line (Hamada et al. 2005; Williams et al. 2007; Zhang et al. 2010). In cells where X chromosomes are paired, MSL staining appears as one large nuclear body, whereas separation of X chromosomes will increase the number of MSL staining bodies per nucleus. Thus, antibodies targeting the MSL complex can be used to monitor X-chromosomal pairing in S2 cell cultures.
Our genome-wide screen identified 59 candidate genes whose disruption causes an increase in the proportion of nuclei with more than one MSL staining body. We demonstrated that disruption of 17 of these genes has a strong effect on somatic homolog pairing as assayed by DNA-FISH targeting the 359 bp repeat on the X chromosome. Furthermore, we confirmed that mutation of one of these genes, pebble (pbl), causes a decrease in somatic homolog pairing in developing embryos. Finally, we showed that dsRNAs targeting Pr-set7, which encodes an H4K20 methyltransferase, cause a disruption of somatic homolog pairing in cultured cells. The genes identified by our screen, many of which have roles in diverse cell-cycle processes, provide novel avenues for exploration of mechanisms that regulate somatic homolog pairing.
Materials and Methods
Immunostaining for genome-wide RNAi screen
A genome-wide RNAi screen was performed at the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School using standard protocols (Ramadan et al. 2007) with some adaptations. First, each plate contained two wells with dsRNA targeting msl2 (amplicon DRSC00829) as positive controls and two wells with dsRNA targeting gfp (Gelbart et al. 2009) as negative controls. Second, all RNAi treatments were performed for five days. Third, screening was performed by immunostaining in 384-well plates with a rabbit anti-MSL1 antibody (Hamada et al. 2005). Following dsRNA incubation, cells were fixed with 2% formaldehyde for 30 min, then treated with cold (4°) methanol for two minutes as an additional fixation step. Next, cells were blocked in PBST/10% donkey serum (Jackson Immunoresearch) prior to incubation in primary antibody. MSL1 staining was visualized using an anti-rabbit secondary antibody conjugated to AlexaFluor 594 (Invitrogen), and nuclei were visualized with Hoechst dye. Imaging was performed on a Discovery automated microscope (Molecular Devices). Two single-plane images were taken per well, and both images were scored as described below.
Genome-wide screening was performed using the first generation RNAi library from the DRSC, which was provided in 62 384-well screening plates. Images generated from the approximately 40,000 wells were visually screened for the qualitative phenotypes “dim” or “supernumerary”; dim hits have reduced MSL1 staining relative to negative controls, and supernumerary hits were defined as those where a greater proportion of nuclei appeared to have two or more MSL1 staining bodies relative to control wells. Validation of the top candidates was performed with a second amplicon to reduce the likelihood of false positives due to off-target effects of the RNAi. GO terms were determined using the AmiGO term enrichment tool. For the selection of candidates that were scored visually for the number of MSL staining bodies per nucleus, only signals that were clearly separated were scored as distinct bodies. Due to the large size of the MSL staining signal, those separated by less than ∼1.5–2 µm between center points appeared as overlapping and were scored as a single focus.
FISH analysis of candidate genes
S2R+ cells were grown at 25° in Complete Schneider’s Medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum and penicillin-streptomycin according to standard procedures. Synthesis of dsRNAs and application of dsRNAs to cells were carried out as previously described (Ramadan et al. 2007). All treatments were performed in 96-well plates, and cells were incubated with dsRNAs for five days before fixation and analysis. Control cells were treated with sterile water rather than dsRNA.
DNA FISH was carried out as previously described (Williams et al. 2007) using a fluorescently labeled oligonucleotide probe complementary to the 359 bp repeat on the X chromosome (5′-ggg atc gtt agc act ggt aat tag ctg c-3′) or to the dodeca satellite on chromosome III (5′-AcG gGa CcA gTa CgG-3′, where uppercase letters indicate LNA-modified nucleotides; Silahtaroglu et al. 2003). Briefly, following incubation with dsRNAs, cells were added to 10-well glass slides (Erie Scientific) treated with 0.01% poly-L-lysine (Sigma-Aldrich), washed with PBS, fixed in 4% formaldehyde for 5 min, then stored in methanol at −20° for up to a week. Slides were then washed in 2X sodium-citrate buffer with 0.1% Tween-20 (SSCT) at 37° for 30 min, after which 250 ng/μl of labeled DNA probe in FISH hybridization buffer (FHB; 50% formamide, 2X SSC, 10% dextransulfate, and 0.05% salmon sperm DNA) was added to each well. The slides were then denatured at 91° for 4 min, incubated at 42° for 10 min, and washed in 2X SSCT at 42° for 30 min. Finally, nuclei were labeled with 10 nM Sytox Green (Invitrogen) in PBS with 0.1% Triton X-100 (PBST), washed in PBST, and mounted in Fluoromount-G (Southern Biotech).
Cells were visualized using a Zeiss Axioplan 2 microscope with a 510 Meta confocal laser scanning system. Optical sections were collected at 0.7 µm increments; prior to analysis, each z-stack was compressed into a single plane using ImageJ software. These files were then analyzed in Cellprofiler v2.0 image analysis software (Carpenter et al. 2006) using a pipeline designed to count the number of FISH signals per nucleus. Briefly, for each image, nuclei were identified by segmentation of the Sytox Green image using Otsu’s thresholding method (Otsu 1979). Next, the corresponding 359 or dodeca FISH image was masked based on the identified nuclei, and FISH signal intensity was enhanced to increase the contrast between staining and background. FISH signals were then segmented based on size and intensity relative to background levels, and each identified FISH signal was assigned to a parent nucleus based on its relative position in the image. Finally, the number of FISH signals was tallied for each nucleus. For the 359 bp repeat, pairing scores were calculated by the percentage of nuclei with a single signal under the assumption that there are two X chromosomes in most nuclei (Zhang et al. 2010). In control experiments, we rescored several images by eye, and we found pairing levels for the 359 bp repeat to be nearly identical to those calculated by our automated method (data not shown). The dodeca repeat is assumed to be carried on four copies of chromosome III per cell, making (4−1)! = 6 possible pairing interactions between dodeca loci in each nucleus. We generated dodeca pairing scores based on the method of Williams et al. (2007); nuclei with a single signal were scored as have 6/6 pairing interactions, those with four or more signals were scored as having 0/6 pairing interactions, those with three signals were scored as having 1/6 pairing interactions, and those with two signals, which could result from a 2+2 (2 pairs, 4 non-pairs) or a 3+1 (3 pairs, 3 non-pairs) configuration, were scored as having 2.5/6 pairing interactions. All statistical tests were performed in R.
To score colocalization of FISH signals targeting the 359 bp repeat and those targeting dodeca, nuclei and 359 FISH signals were identified in Cellprofiler software as outlined above. The corresponding dodeca image was then masked based on the identified 359 signals, and the remaining dodeca FISH signals were identified based on size and intensity. Each dodeca FISH signal was then assigned to a parent 359 FISH signal, and the percentage of 359 signals that overlapped at least one dodeca FISH signal was calculated.
Pr-set7 analysis
For three-dimensional FISH analysis in Pr-set7 knockdowns, FISH was performed as described above with the exception that slides were not treated with methanol; a probe complementary to a different portion of the 359 bp repeat was used (5′-Ttt Tcc Aaa Ttt Cgg Tca Tca Aat Aat Cat-3′; Bateman and Wu 2008), and the nuclear envelope was stained with fluorescently labeled wheat germ agglutinin (WGA; Invitrogen) prior to mounting in Vectashield mounting medium with DAPI (Vector Laboratories). Optical sections were collected at 0.5 µm increments using a Zeiss Axioplan 2 with a 510 Meta confocal system, and the number of distinct fluorescent signals in each nucleus was scored visually in three-dimensional z-stacks using ImageJ software. In general, neighboring FISH signals whose center points were separated by less than ∼0.6–0.7 µm appeared as overlapping and were scored as a single focus, whereas those separated by a greater distance were easily discernable as two or more foci and were scored as such. All scoring was performed blindly.
Embryo FISH
Embryos were collected from a stock of pbl2/TM3 mutants, aged for 2–4 hr after laying, and subjected to DNA FISH targeting the dodeca satellite as previously described (Bateman and Wu 2008). Early stage 9 embryos were selected based on the degree of germ band extension, and cells of zone 1 were imaged using a Deltavision imaging station and Softworx imaging software. Homozygous mutant embryos were identified by the presence of binucleate cells in zone 1. Scoring of pairing was carried out as previously described (Bateman and Wu 2008). For binucleate cells, each nucleus was scored separately.
Results
Our initial motivation in this study was to better understand the genes that regulate MSL complex targeting in Drosophila. We reasoned that dsRNA knockdown of genes required for localization of the MSL complex on the X chromosome would result in diminished or diffuse anti-MSL immunostaining. With this logic in mind, we performed a genome-wide screen in S2 cells using a library of 21,000 dsRNAs in 384-well plates. After a five-day incubation, we fixed and treated cells with antibodies to MSL1 to visualize the MSL complex and with Hoechst dye to visualize whole nuclei, and then imaged cells from each well using automated microscopy. Although the majority of the images produced were sufficient for subsequent analysis, there was considerable variation in image quality, which hampered automated scoring of the dataset at high stringency (see File S1 and Table S5 for an automated analysis of the image set). Thus, we began our analysis by screening visually for wells with noticeable changes in the pattern of MSL1 staining.
We expected that dsRNA knockdown of components of the MSL complex would result in diminished MSL1 staining (a “dim” phenotype), as each member of the complex contributes to achieve complex localization (reviewed by Gelbart and Kuroda 2009). Indeed, control experiments using dsRNAs to msl2 showed diminished MSL1 staining intensity (data not shown). By screening our genome-wide image set for wells with a similar dim phenotype, we identified knockdowns of each of the protein-coding components of the MSL complex, including msl1, msl2, msl3, mof, and mle (supporting information, Figure S1). Surprisingly, we identified only seven other dsRNAs with a similar dim phenotype (Table S1). Of these seven candidates, four had been previously identified in a genome-wide screen for cell growth and viability (Boutros et al. 2004), and six of the seven had a secondary phenotype of altered MSL localization (see below) that led us to believe that their effect on MSL1 staining intensity might be indirect. The seven dim candidates were not pursued further in the present analysis.
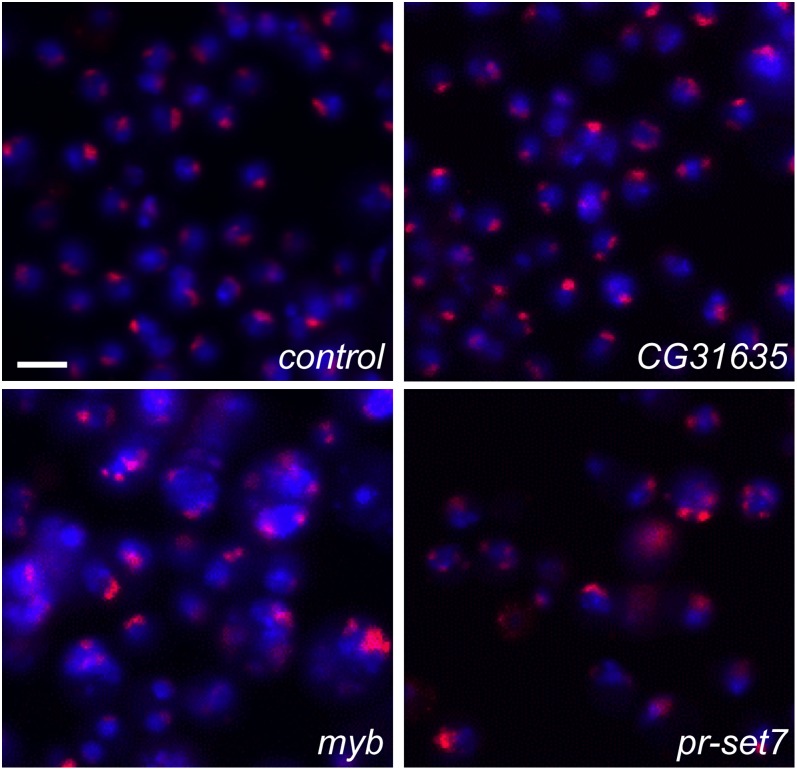
Our visual screen uncovered a second unanticipated change to the pattern of MSL1 staining in some wells. In wild-type cells and control cells treated with dsRNA to gfp, most nuclei showed a single MSL1-stained signal due to the tight pairing of the approximately two X chromosomes per nucleus. In contrast, we noted several dsRNAs that caused a change in this pattern, such that many nuclei showed two or more MSL1 signals, which we refer to as a “supernumerary” (SN) phenotype (Figure 1). Through our analysis of the genome-wide dataset and a secondary validation screen analyzing staining of MSL1 and MSL2, we identified 59 dsRNAs that caused a reproducible SN phenotype (Table S2). For a selection of the 59 candidates, we visually scored the number of MSL signals per nucleus using representative images from our screen (Table 1). In control cells treated with dsRNA to gfp, 88.1% of nuclei showed a single MSL signal, whereas knockdowns of candidate genes from our screen reduced this number to a range of 31.3% (pavarotti) to 82.9% (CG31635). Below, we refer to the genes targeted by these 59 dsRNAs as “SN hits.”
Figure 1.
Genome-wide screening identifies dsRNAs that increase the number of MSL staining bodies per nucleus (“supernumerary” phenotype). Cropped example images from our screen show DNA (blue) and MSL1 (red) from cells treated with a control dsRNA targeting gfp or with dsRNAs targeting candidate CG31635, myb, or Pr-set7. Scale bar represents 10 µm for all panels.
Table 1. Quantification of supernumerary MSL staining phenotype for select genome-wide screen hits.
| Gene Targeted | Screen Amplicon | % Nuclei With Single MSL Signal | Nuclei Scored | P |
|---|---|---|---|---|
| Control | (gfp) | 88.1 | 227 | — |
| Pr-set7 | DRSC15473 | 73.2 | 97 | 9 × 10−4 |
| incenp | DRSC21950 | 46.5 | 101 | <1 × 10−4 |
| CG31635 | DRSC02273 | 82.9 | 199 | 0.13 |
| Betatub56D | DRSC07583 | 71.6 | 81 | 5 × 10−4 |
| myb | DRSC20260 | 53.9 | 76 | <1 × 10−4 |
| pbl | DRSC11381 | 51.3 | 80 | <1 × 10−4 |
| skpA | DRSC18833 | 43.3 | 67 | <1 × 10−4 |
| feo | DRSC19398 | 35.9 | 39 | <1 × 10−4 |
| ial-1 | DRSC03548 | 56.6 | 76 | <1 × 10−4 |
| pav | DRSC08730 | 31.3 | 48 | <1 × 10−4 |
One representative image was scored for each selected screen hit. P values are derived from χ2 tests with expected values based on control cells treated with dsRNA targeting gfp. Although the modest effect of CG31635 knockdown was not significant by this analysis, it was qualitatively scored as having a supernumerary phenotype through multiple rounds of blind screening, and its effect on pairing was confirmed by FISH analysis (see text). See Table S2 for a complete list of all 59 SN hits.
DNA FISH shows diminished somatic homolog pairing in dsRNA-treated cells
The SN pattern of MSL localization could result from several changes to the biology of the nucleus, including aberrant aggregation of MSL complexes, incorrect targeting of the MSL complex to other chromosomes, or a loss of X-chromosomal homolog pairing. To more directly assess possible changes in somatic homolog pairing, we used DNA FISH targeting the 359 bp repeat in cells treated with dsRNAs for a subset of our SN hits. We expected that cells in which homologous 359 bp regions were paired would show a single FISH signal representing overlapping loci, whereas unpaired 359 bp regions would appear as separate FISH foci within one nucleus. Thus, any dsRNA treatment that adversely affects somatic homolog pairing would be expected to decrease the proportion of nuclei with a single FISH signal relative to untreated control cells. As an additional consideration in our FISH analysis, our genome-wide screen used a first-generation dsRNA library in which some dsRNAs had the potential to target more than one gene (see Table S2). We therefore chose new dsRNAs that were designed to target our SN hits with no known off-targets (Kulkarni et al. 2006).
We incubated S2R+ cells with dsRNAs for five days, subjected fixed cells to DNA FISH targeting the 359 bp repeat, and imaged cells in three dimensions using confocal microscopy. We then projected three-dimensional image stacks into a single plane for automated scoring of the number of FISH signals per nucleus. In cells treated with water in the place of dsRNA, 66.8 ± 3.4% of nuclei had a single FISH signal for the 359 bp repeat (4498 nuclei from 13 independently treated samples). From the list of 59 SN hits, we arbitrarily chose 47 for analysis using FISH (Table S3). In order to consider pairing of the 359 bp repeat to be significantly disrupted, we established a cutoff equivalent to 2.5 standard deviations below the mean percentage of nuclei with a single FISH signal in water-treated controls. While 0/13 water-treated control slides met this criterion, 17 of the dsRNA candidates tested had pairing scores below the threshold (Figure 2, Table 2). To determine whether these dsRNAs had a more general effect on pairing beyond the X chromosome, we also assessed the effect of dsRNA treatments on pairing of the dodeca satellite on the third chromosome, and we found that 16/17 caused an overall decrease in dodeca pairing relative to the mean of water-treated controls, with over half (9/17) decreasing pairing below a high stringency cutoff analogous to that used for the 359 bp repeat (Table S4).
Figure 2.
DNA FISH shows loss of somatic homolog pairing in dsRNA-treated S2R+ cells. Each row shows staining of DNA by DAPI, DNA FISH targeting the 359 bp repeat, and a merged image. All images represent a z-series that has been compressed into a single plane. In water-treated cells (wt), 66.8% of nuclei have a single FISH signal. Shown are representative nuclei from cells treated with dsRNAs targeting FISH hits, including chromosome bows (chb), Separase (Sse), CG31635, pebble (pbl), and fascetto (feo). Scale bar represents 10 µm for all panels. Inset, bottom right, is an example of a large nucleus (at equivalent scale) treated with dsRNA targeting feo where more than four FISH signals are apparent; such nuclei are assumed to be polyploid/aneuploid.
Table 2. FISH-validated genes required for normal somatic homolog pairing.
| Gene Targeted | Gene Function | Validation Amplicon | % Paired Nucleia | Nuclei Scored | Nuclear Sizeb | % Nuclei > 4 Signalsc |
|---|---|---|---|---|---|---|
| Control | — | (water) | 66.8 | 4498 | 2537 ± 892 | 0.002 |
| β-Tub56D | Cytoskeleton | DRSC33236 | 47.9 | 361 | 2943 ± 1330 | 1.1 |
| CG31635 | Unknown | DRSC31568 | 54 | 274 | 2573 ± 787 | 0.36 |
| CG5844 | Fatty acid metabolism | DRSC30977 | 52.3 | 151 | 2417 ± 925 | 0 |
| chb | Microtubule binding | DRSC36464 | 57.7 | 345 | 2643 ± 832 | 0.29 |
| feo | Cytokinesis | DRSC23773 | 53.1 | 241 | 2892 ± 1598 | 3.7 |
| geminin | Regulation of DNA replication | DRSC33311 | 56.3 | 190 | 3879 ± 1886 | 2.1 |
| His3 | Core histone | DRSC40904 | 57.6 | 184 | 2848 ± 1041 | 0.54 |
| ial-1 | Aurora B kinase; cytokinesis | DRSC36311 | 45.4 | 207 | 3360 ± 2048 | 6.3 |
| Incenp | Inner centromere protein | DRSC39343 | 46.6 | 161 | 3637 ± 2297 | 9.9 |
| Klp61F | Mitotic kinesin | DRSC30823 | 31.7 | 123 | 3727 ± 1967 | 13.9 |
| Myb | Oncogene | DRSC31099 | 39.7 | 209 | 3315 ± 1843 | 12 |
| pav | Kinesin; cytokinesis | DRSC33333 | 33.8 | 136 | 2972 ± 1903 | 8.8 |
| pbl | Cytokinesis | DRSC33335 | 41.5 | 299 | 2776 ± 1229 | 3.7 |
| polo | Kinetechore | DRSC34463 | 46.7 | 244 | 2628 ± 1180 | 2 |
| scra | Contractile ring | DRSC07679 | 41.4 | 280 | 3313 ± 1807 | 8.2 |
| Sse | Mitotic chromosome segregation | DRSC34476 | 53.6 | 308 | 2777 ± 1229 | 0.32 |
| tsr | Actin binding protein | DRSC31277 | 55.6 | 144 | 2289 ± 1256 | 4.1 |
Percentage of nuclei with a single FISH signal for the 359 bp repeat.
Nuclear sizes were determined from images representing flattened z-stacks and are presented as mean areas of DAPI staining plus or minus the standard deviation. Units are arbitrary.
Percentage of nuclei with greater than four FISH signals for the 359 bp repeat, which are assumed to be polyploid/aneuploid.
In addition to the influence of somatic homolog pairing, the nuclear localization of repetitive sequences such as the 359 bp repeat and the dodeca satellite can be impacted by mechanisms that aggregate heterochromatin irrespective of sequence homology (Francastel et al. 2000; Sage and Csink 2003). Thus, it is possible that the decreased pairing observed in our FISH analysis could reflect roles for some or all of our 17 candidate dsRNAs in clustering heterochromatin as opposed to somatic homolog pairing. Arguing against this, each of the candidate dsRNAs was originally identified by its SN phenotype for MSL complex localization; given that MSL complexes are enriched over transcribed genes in euchromatic regions across the X chromosome (Alekseyenko et al. 2006), it is likely that our 17 candidate dsRNAs decrease somatic homolog pairing. However, to address whether these dsRNAs affect heterochromatin clustering, we scored the frequency with which FISH signals from the 359 bp repeat overlap those from the dodeca satellite in cells treated with each dsRNA. In water-treated control slides, 14.8 ± 1.5% of 359 bp repeat FISH signals (n = 6272, 13 independent samples) overlap at least one dodeca signal. Notably, this level of colocalization is ∼10-fold greater than that of nonhomologous euchromatic loci (Bateman and Wu 2008; Williams et al. 2007), consistent with the tendency of heterochromatic sequences to cluster. Cells treated with each of the 17 candidate dsRNAs showed very similar levels of 359-dodeca overlap relative to water-treated controls, differing by only a few percentage points (Table S4). The largest change was seen for dsRNA targeting geminin, which increased the percentage of 359 signals that overlap dodeca signals to 20.6%, contrasting the decrease in pairing of homologous 359 loci caused by the same dsRNA. The strongest negative effect on heterochromatin clustering was observed for dsRNA targeting CG5844, in which overlap of 359 signals with dodeca signals was reduced by ∼3.5% relative to controls. Importantly, the minor perturbations of heterochromatin clustering observed for our dsRNA treatments are unlikely to account for the strong decreases in pairing of the 359 bp repeat observed in our FISH screen. On the basis of the combined data from our MSL staining and FISH analysis, we conclude that the genes targeted by these 17 dsRNAs are required for normal somatic homolog pairing in cultured Drosophila cells. We refer to these genes below as FISH hits.
The list of 17 FISH hits is enriched for several Gene Ontology (GO) terms related to the cytoskeleton and cell division, including the terms “cell cycle” (13/17, P = 1 × 10−10) and “cytokinesis” (9/17, P = 4.6 × 10−13) (Table S4). The GO term “cell cycle” was also highly enriched in the list of 59 SN hits from our genome-wide screen (34/59, P = 7 × 10−23) (Table S2), further highlighting a potential link between cell division and pairing. Indeed, 8 of the 17 FISH hits had been previously identified in a screen for genes required for cytokinesis (Echard et al. 2004; Eggert et al. 2004), and 12 of the 17 had been identified as important for normal cell morphology in cultured cells (Kiger et al. 2003) (Table S4).
We noticed that some of the dsRNAs targeting our FISH hits produced nuclei that appeared to be larger than those of control cells. We quantified nuclear areas based on DAPI staining in the images from our FISH analysis, and for many of the 17 dsRNAs, we found higher mean nuclear sizes and greater variation relative to water-treated control cells (Table 2). Changes in nuclear size could reflect alteration of chromatin structure, or given the importance of many of our FISH hits in cell-cycle processes, could result from changes in cell ploidy. Consistent with the latter notion, dsRNAs targeting 11/17 of our screen hits had been previously shown to increase the average DNA content of treated cells as assessed by FACS analysis (Bettencourt-Dias et al. 2004; Bjorklund et al. 2006; Somma et al. 2002) (Table S4). Furthermore, for some screen hits, we observed an increase in the percentage of nuclei with more than four FISH signals—the maximum number of X sister chromatids in a tetraploid S2 cell during G2 of the cell cycle—for the 359 bp repeat (Table 2), suggesting that some cells indeed had increased chromosome complements. Importantly, we did not find a significant relationship between average nuclear size and pairing of the 359 bp repeat among our 17 FISH hits (R2 = 0.18, P = 0.09), suggesting that changes in ploidy are not central to the observed disruption of homolog pairing. Indeed, pairing mechanisms are known to be tolerant of the altered ploidy of cultured cells, including that of the tetraploid S2 cells used in this analysis (Williams et al. 2007), implying that factors beyond simple changes in ploidy must contribute to the disruption of pairing that we observed (see Discussion).
Loss of Pr-set7 disrupts somatic homolog pairing
Of the SN hits identified by our genome-wide screen, we were particularly interested in the histone H4K20 methyltransferase encoded by Pr-set7 due to its proposed role in modifying chromatin, which we imagined could have a direct effect on somatic homolog pairing. Although Pr-set7 was not among the list of 17 FISH hits, our scoring of the SN phenotype showed that dsRNA targeting Pr-set7 significantly decreased the proportion of nuclei with a single MSL staining body (Table 1), and cells treated with dsRNA targeting Pr-set7 showed a moderate decrease in pairing of the 359 bp repeat relative to water-treated controls (Table S3). We reasoned that Pr-set7 could play a role in somatic homolog pairing that is difficult to substantiate via the high-stringency cutoff used in our FISH analysis, and that a more detailed study of Pr-set7 function in homolog pairing might uncover its significance.
To address a potential role for Pr-set7 in somatic homolog pairing, we treated S2R+ cells with the same dsRNA incubation conditions and FISH protocol as described for our screening approach. In this case, we visually scored pairing in three-dimensional image stacks. Using this scoring method, 77.7% (n = 300) of water-treated control cells were scored as having a single FISH signal for the 359 bp repeat. In contrast, two independent dsRNAs targeting Pr-set7 caused a modest but significant decrease in pairing of the 359 bp repeat, reducing the percentage of nuclei with a single focus to 63.5% (n = 274) and 58.9% (n = 263; χ2 test, P = 2 × 10−4 and <1 × 10−4, respectively) (Figure 3, Table 3). As an additional treatment, we also incubated cells with a dsRNA targeting zeste (z), a gene that impacts some transvection phenotypes but has previously been shown to have no effect on mean pairing levels in vivo (Gemkow et al. 1998), and we found the percentage of cells with a single FISH signal to be nearly identical to water-treated cells (78.5%, n = 107). Notably, quantitative RT-PCR showed that both Pr-set7 and z were knocked down to similar degrees, with roughly 10% of steady-state mRNA levels relative to water-treated cells (data not shown). Finally, we also assessed dsRNA targeting one of our strong FISH hits, pavarotti (pav), using the same scoring method. In this case, we observed a much stronger reduction in pairing of the 359 bp repeat relative to dsRNAs targeting Pr-set7 (Table 3), in line with our reasoning that loss of Pr-set7 has a modest effect on somatic homolog pairing in comparison with our 17 FISH hits.
Figure 3.
RNAi knockdown of Pr-set7 causes a reduction in somatic homolog pairing in S2R+ cells. Each row shows staining of the nuclear envelope by fluorescently labeled wheat germ agglutinin (WGA), DNA FISH targeting the 359 bp repeat, and a merged image. All images represent a z-series that has been compressed into a single plane. Top row, nuclei from representative water-treated cells (wt); bottom, nuclei from cells treated with dsRNA corresponding to amplicon DRSC27118 targeting Pr-set7. Scale bar represents 5 µm for all panels.
Table 3. dsRNA targeting Pr-set7 disrupts somatic pairing.
| Gene Targeted | dsRNA Amplicon | % Paired Nucleia | Nuclei Scored |
|---|---|---|---|
| Control | (water) | 77.7 | 300 |
| Pr-set7 | DRSC15473 | 63.5b | 274 |
| Pr-set7 | DRSC27118 | 58.9b | 263 |
| z | DRSC23134 | 78.5 | 107 |
| pav | DRSC08730 | 49.0b | 49 |
Percentage of nuclei with a single FISH signal for the 359 bp repeat.
P < 0.05 according to χ2 tests with expected values based on water-treated control cells.
Previous analysis has shown that loss of Pr-set7 does not alter cell ploidy in vivo (Sakaguchi and Steward 2007); consistent with this, we found zero nuclei with greater than four FISH signals for the 359 bp repeat in our FISH screen (n = 288) or in our visual scoring (n = 296). However, we found a small but significant increase in the average nuclear area of cells treated with dsRNA to Pr-set7 relative to water-treated controls (2537 ± 892 arbitrary units, n = 4488 for controls; 2783 ± 953, n = 288 for Pr-set7 knockdowns; Welch’s t-test, P = 2.4 × 10−4), which could reflect changes in chromatin compaction in the absence of Pr-set7 (reviewed by Brustel et al. 2011; Wu et al. 2010; see Discussion). Furthermore, scoring of colocalization between FISH signals in cells treated with dsRNA targeting Pr-set7 showed that 16.6% of 359 FISH signals (n = 434) overlapped at least one dodeca signal, representing a small increase relative to the mean for water-treated controls (14.8 ± 1.5%, n = 6272). Thus, it is unlikely that the loss of pairing between homologous 359 bp repeat loci in response to decreased Pr-set7 activity is related to changes in heterochromatin clustering. In sum, our data support a role for Pr-set7 in promoting somatic homolog pairing in cultured cells.
A mutation in pbl affects pairing in vivo
The results of our screens suggest a relationship between somatic homolog pairing and proper regulation of the cell cycle. As our screens were performed in cultured cells, we next tested whether this relationship exists in whole organisms. We focused on pbl, a well-characterized gene that encodes an activator of the Rho1 small GTPase and whose activity is required for the function of the contractile ring during cytokinesis (Prokopenko et al. 1999). Embryos lacking pbl activity carry out their first 13 mitoses with no significant differences from wild-type due to maternal loading of pbl-encoded RNA and protein (Hime and Saint 1992; Lehner 1992; Prokopenko et al. 2000). However, during mitosis 14, cells from pbl mutant embryos fail to complete cytokinesis, resulting in binucleate daughter cells.
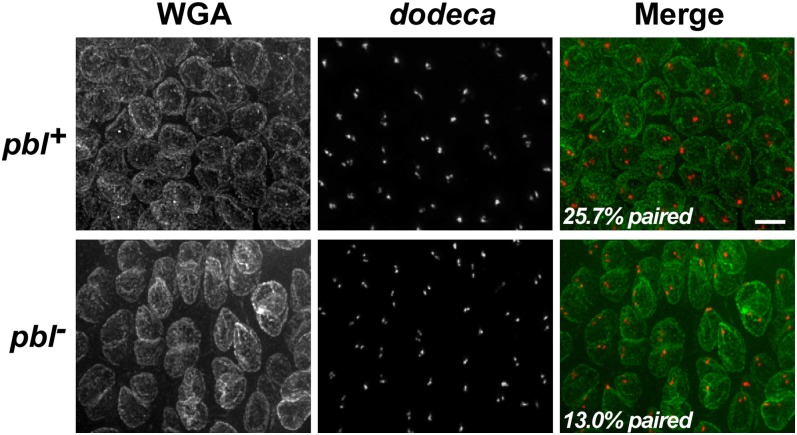
Using FISH targeting the dodeca repeat on chromosome III, we assessed somatic homolog pairing in homozygous pbl2 embryos and their balanced siblings during interphase 15, taking advantage of the binucleate phenotype of homozygous mutants to differentiate genotypes (Figure 4). We focused on a region of the embryo called zone 1, which contains the first group of cells to undergo mitosis 14 (Foe et al. 1993), during early stage 9 of embryogenesis. Consistent with previous analyses of dodeca pairing levels during early embryogenesis (Bateman and Wu 2008; Blumenstiel et al. 2008), 25.7% (n = 382) of nuclei from zone 1 of pbl+ embryos had a single FISH signal for the dodeca repeat. In contrast, nuclei from homozygous pbl2 embryos showed a significant reduction in dodeca pairing, with just 13.0% (n = 308; χ2 test, P < 1 × 10−4) of nuclei containing a single dodeca FISH signal. Notably, the decrease in pairing of the dodeca satellite in mutant embryos was unlikely to involve changes to ploidy, as embryos were aged only ∼40 min after mitosis 14 began, during which time the cells of zone 1 do not normally undergo another cell cycle (Foe et al. 1993). Thus, the loss of dodeca pairing associated with a classical mutation in the pbl gene in vivo is consistent with the reduction in somatic homolog pairing caused by dsRNA targeting pbl in cultured cells.
Figure 4.
A classical allele of pbl causes disruption of somatic homolog pairing in vivo. Each row shows staining of the nuclear envelope by fluorescently labeled wheat germ agglutinin (WGA), DNA FISH targeting the dodeca repeat on chromosome III, and a merged image. All images represent a z-series that has been compressed into a single plane. Nuclei are derived from zone 1 of early stage 9 pbl2/pbl2 embryos (pbl; identified by their binucleate phenotype; 308 nuclei from 4 embryos) or their pbl2/pbl+ or pbl+/pbl+ siblings at the same stage of development (pbl+; 382 nuclei from 6 embryos). Scale bar represents 5 µm for all panels. Note that both pbl+ and pbl- embryos show several nuclei with three and four signals for the dodeca repeat, which are presumed to result from separation of sister chromatids (Bateman and Wu 2008).
Discussion
Here we describe a genome-wide RNAi-based screen that examined changes in MSL localization in cultured cells. Although our prime motivation in carrying out this screen was to better understand genes involved in dosage compensation in Drosophila, the altered MSL localization that we observed upon knockdown of some genes was consistent with changes in somatic homolog pairing, shifting the focus of our investigation. Through visual scoring of our screen images, we identified 59 dsRNAs that cause an increase in the proportion of nuclei with more than one MSL staining body. Our follow-up screen using DNA FISH targeting the 359 bp repeat on the X chromosome confirmed that 17 of our SN hits increase the percentage of nuclei with multiple FISH signals above a high stringency threshold. We conclude that the activities of the 17 genes identified by our FISH screen are required for normal levels of somatic homolog pairing.
In a parallel investigation, Joyce and colleagues performed a genome-wide FISH-based screen for factors that affect somatic pairing in cultured Kc167 cells. They identified 40 “pairing promoting” genes whose disruption via RNAi led to a significant increase in the percentage of nuclei with multiple FISH signals (Joyce et al. 2012). Notably, 13 of the 17 genes identified in our screen were also tagged as significant hits in their study, thereby validating our methodology and further supporting the importance of these genes to somatic homolog pairing.
Previous analysis has shown that regions of heterochromatin and euchromatin pair at different levels in cultured cells, suggesting that these types of chromatin might be influenced by different pairing mechanisms (Williams et al. 2007). Although our FISH analysis focused on heterochromatic regions, our primary screen was based on binding of the MSL complex, which is enriched over transcribed genes in regions of euchromatin across the X chromosome (Alekseyenko et al. 2006). Thus, it is likely that the 17 genes highlighted by our screen affect pairing of both heterochromatic and euchromatic regions. However, we cannot exclude the possibility that the extra MSL staining bodies that we observed in our primary screen could reflect mislocalization of MSL complexes rather than unpairing of whole X chromosomes. Confirmation of the influence of our screen hits on euchromatin will await a detailed analysis of pairing using FISH probes targeting unique euchromatic sequences.
Of the 47 SN hits that we tested by FISH, 30 did not show a significant disruption of pairing of the 359 bp repeat. This may be due in part to the stringent cutoff that we established in order to consider a dsRNA as significant in this analysis. Of the SN hits that did not show significant disruption of somatic homolog pairing by FISH, 13/30 caused an overall decrease in pairing of the 359 bp repeat relative to the mean of water-reated controls, but did not meet our statistical cutoff. We tested one of these genes, Pr-set7, and found a significant effect on pairing of the 359 bp repeat in a focused analysis. Thus, it is possible that additional candidates from the SN hit list have a modest effect on somatic homolog pairing, but in depth analyses will be required to substantiate their significance. For other SN hits that did not decrease pairing of the 359 bp repeat, it is possible that changes in reagents, including S2 cell population and dsRNA sequences, or effects that are specific to euchromatin and/or the MSL complex could account for the lack of correlation between the two analyses.
Somatic homolog pairing and the cell cycle
How do the genes identified in our screen influence somatic homolog pairing? The majority of these genes are important for progression through the cell cycle, playing diverse roles that include cytokinesis, chromosome congression, spindle organization, and regulation of replication. Complicating the interpretation of our data, disruption of many of these genes alters the ploidy of dsRNA-treated cells (Bettencourt-Dias et al. 2004; Bjorklund et al. 2006; Somma et al. 2002). We did not see a significant relationship between pairing of the 359 bp repeat and average nuclear size among our FISH hits, nor did we see evidence of altered ploidy in staged pbl mutant embryos where dodeca pairing is decreased, suggesting that decreases in somatic homolog pairing were not directly caused by increased chromosome complements. Moreover, a prior comparison of somatic homolog pairing between diploid, tetraploid, and even partially hexaploid cell lines showed that the percentage of nuclei with a single FISH signal is strikingly similar in each type of cell (Williams et al. 2007), implying that pairing mechanisms can accommodate changes to chromosome number. Notably, in their parallel analysis, Joyce et al. (2012) found that the loss of somatic homolog pairing caused by some dsRNAs targeting pairing promoters was Cap-H2–dependent, as simultaneous loss of Cap-H2 activity could restore pairing to wild-type levels. We therefore believe that the changes in ploidy resulting from dsRNA treatment cannot fully explain our observations. Rather, it appears that perturbation of diverse cell-cycle processes negatively impacts somatic homolog pairing, in some cases via a mechanism that requires Cap-H2.
Several prior studies have analyzed the influence of the cell cycle on somatic homolog pairing, with varying conclusions depending on the system analyzed and methods used. For example, analysis of BrdU-labeled larval CNS cells showed that both euchromatic and heterochromatic pairing is disrupted during progression through S phase and remains at reduced levels of pairing through G2; higher levels of pairing are then reestablished in G1 of the next cell cycle (Csink and Henikoff 1998). In contrast, DNA FISH targeting the histone gene complex in rapidly cycling early embryos has shown that pairing of this locus remains high through G2 and the early stages of M phase and is reduced as sister chromatids separate during anaphase (Fung et al. 1998). Analysis of cultured Kc167 cells showed a similar pattern to that of early embryos, with no discernable difference in pairing of a euchromatic locus among G1, S, and G2 subpopulations, but a presumed disruption during later stages of M phase (Williams et al. 2007). Thus, the relationship between somatic homolog pairing and the cell cycle appears to be complex, with apparent differences among cell types in addition to possible locus-specific effects. Excitingly, the genes identified by our screen provide genetic tools for future analyses to dissect the mechanisms that coordinate somatic homolog pairing with events of the cell cycle.
Pr-set7 and somatic homolog pairing
Disruption of Pr-set7 via dsRNA knockdown caused an overall increase in signals per nucleus for both MSL localization and FISH targeting the 359 bp repeat, implying that Pr-set7 activity is necessary for wild-type levels of somatic homolog pairing. Excitingly, an independent analysis also found changes in higher-order chromatin organization that are consistent with a disruption of homolog pairing in response to loss of Pr-set7 function (E. F. Joyce and R. Steward, personal communication). As with our other screen hits, previous studies have implicated Pr-set7 in several cell-cycle processes, with demonstrated roles in replication during S phase and in chromatin condensation during M phase (reviewed by Brustel et al. 2011; Wu and Rice 2011). In Drosophila, loss of function mutations in Pr-set7 cause activation of a DNA damage checkpoint and subsequent G2 arrest (Sakaguchi and Steward 2007). Thus, loss of Pr-set7 could impact somatic homolog pairing via disruption of the cell cycle as we have postulated for our stronger screen hits. However, other mechanisms involving a more direct role for Pr-set7 in somatic homolog pairing are possible. For example, the proposed role of Pr-set7 in chromatin compaction (reviewed by Yang and Mizzen 2009) could more directly impact pairing at the level of chromatin structure. Similarly, histone modification by Pr-set7 could provide binding sites for other factors that influence pairing. Indeed, a human homolog of the tumor suppressor lethal (3) malignant brain tumor (l(3)mbt) drives chromatin compaction through binding of methylated H4K20 (Trojer et al. 2007), and loss of function mutations in l(3)mbt have been shown to disrupt pairing of polytene chromosomes in Drosophila salivary glands (Riede 1997). A more detailed analysis of Pr-set7 function will help to differentiate between its possible modes of influence on somatic homolog pairing.
Somatic homolog pairing and nuclear organization
Somatic homolog pairing is just one component of the multifaceted structure of the interphase genome. Additional influences on genome organization and dynamics include the formation of chromosome territories, the partitioning of active and inactive genomic regions to separate domains, and functional interactions of chromosomal regions with nuclear compartments, such as transcription factories, Cajal bodies, insulator bodies, and polycomb bodies (reviewed by Bantignies and Cavalli 2011; Branco and Pombo 2007; Schneider and Grosschedl 2007). Novel high-throughput techniques, such as Hi-C, have begun to provide genome-wide descriptions of many types of intra- and interchromosomal interactions in diverse species (Lieberman-Aiden et al. 2009; Sexton et al. 2012); however, the sequence identity between homologous chromosomal regions complicates the ability of Hi-C and similar 3C-based approaches to reliably uncover interhomolog interactions. A more complete understanding of interphase nuclear structure will require that we decipher the dynamic interplay between each of these influences on chromosome folding under changing environmental conditions.
Supplementary Material
Acknowledgments
We are grateful to the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School, the Bloomington Drosophila Stock Center at Indiana University, David Knecht for help designing the ImageJ script used in our automated analysis, and Christian Belisario for assistance. Special thanks to Eric Joyce, Ting Wu, and Ruth Steward for sharing information prior to publication and for helpful comments, and to Giovanni Bosco for valuable comments on this manuscript. This project was supported by National Institutes of Health grants R37 GM045744 to M.I.K., 5P20 RR016463-12 and 8P20 GM103423-12 to J.R.B., and by National Science Foundation grant 1024973 to B.G.M.
Footnotes
Communicating editor: K. S. McKim
Literature Cited
- Alekseyenko A. A., Larschan E., Lai W. R., Park P. J., Kuroda M. I., 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20: 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher C. P., Guggiari M., Brors B., Augui S., Clerc P., et al. , 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299 [DOI] [PubMed] [Google Scholar]
- Bantignies F., Cavalli G., 2011. Polycomb group proteins: repression in 3D. Trends in genetics. TIG 27: 454–464 [DOI] [PubMed] [Google Scholar]
- Bateman J. R., Wu C.-T., 2008. A genomewide survey argues that every zygotic gene product is dispensable for the initiation of somatic homolog pairing in Drosophila. Genetics 180: 1329–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote J. M., Lucchesi J. C., 1980. Male-specific lethal mutations of Drosophila melanogaster. Genetics 96: 165–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., et al. , 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432: 980–987 [DOI] [PubMed] [Google Scholar]
- Bjorklund M., Taipale M., Varjosalo M., Saharinen J., Lahdenpera J., et al. , 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Fu R., Theurkauf W. E., Hawley R. S., 2008. Components of the RNAi machinery that mediate long-distance chromosomal associations are dispensable for meiotic and early somatic homolog pairing in Drosophila melanogaster. Genetics 180: 1355–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Kiger A. A., Armknecht S., Kerr K., Hild M., et al. , 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Branco M. R., Pombo A., 2007. Chromosome organization: new facts, new models. Trends Cell Biol. 17: 127–134 [DOI] [PubMed] [Google Scholar]
- Brustel J., Tardat M., Kirsh O., Grimaud C., Julien E., 2011. Coupling mitosis to DNA replication: the emerging role of the histone H4-lysine 20 methyltransferase PR-Set7. Trends Cell Biol. 21: 452–460 [DOI] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., et al. , 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J. C., Worby C. A., Simonson-Leff N., Muda M., Maehama T., et al. , 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97: 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink A. K., Henikoff S., 1998. Large-scale chromosomal movements during interphase progression in Drosophila. J. Cell Biol. 143: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I. W., 2002. Transvection effects in Drosophila. Annu. Rev. Genet. 36: 521–556 [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G. R., Foley E., O’Farrell P. H., 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14: 1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert U. S., Kiger A. A., Richter C., Perlman Z. E., Perrimon N., et al. , 2004. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2: e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., van Cappellen W. A., Theil A. F., van Drunen E., Jaspers N. G., et al. , 2005. Dynamics of relative chromosome position during the cell cycle. Mol. Biol. Cell 16: 769–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., Odell G. M., Edgar B. A., 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint, pp. 149–300 in The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Francastel C., Schubeler D., Martin D. I., Groudine M., 2000. Nuclear compartmentalization and gene activity. Nat. Rev. Mol. Cell Biol. 1: 137–143 [DOI] [PubMed] [Google Scholar]
- Fritsch C., Ploeger G., Arndt-Jovin D. J., 2006. Drosophila under the lens: imaging from chromosomes to whole embryos. Chromosome Res. 14: 451–464 [DOI] [PubMed] [Google Scholar]
- Fung J. C., Marshall W. F., Dernburg A. F., Agard D. A., Sedat J. W., 1998. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J. Cell Biol. 141: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Larschan E., Peng S., Park P. J., Kuroda M. I., 2009. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nat. Struct. Mol. Biol. 16: 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart W. M., 1982. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79: 2636–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemkow M. J., Verveer P. J., Arndt-Jovin D. J., 1998. Homologous association of the Bithorax-Complex during embryogenesis: consequences for transvection in Drosophila melanogaster. Development 125: 4541–4552 [DOI] [PubMed] [Google Scholar]
- Gerlich D., Beaudouin J., Kalbfuss B., Daigle N., Eils R., et al. , 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112: 751–764 [DOI] [PubMed] [Google Scholar]
- Hamada F. N., Park P. J., Gordadze P. R., Kuroda M. I., 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T. A., Smith H. F., Bosco G., 2008. Chromosome alignment and transvection are antagonized by condensin II. Science 322: 1384–1387 [DOI] [PubMed] [Google Scholar]
- Heard E., Bickmore W., 2007. The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 19: 311–316 [DOI] [PubMed] [Google Scholar]
- Hime G., Saint R., 1992. Zygotic expression of the pebble locus is required for cytokinesis during the postblastoderm mitoses of Drosophila. Development 114: 165–171 [DOI] [PubMed] [Google Scholar]
- Jordan P., 2006. Initiation of homologous chromosome pairing during meiosis. Biochem. Soc. Trans. 34: 545–549 [DOI] [PubMed] [Google Scholar]
- Joyce E., Williams B., Xie T., Wu C.-t., 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8: e1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., Southworth J. W., 2002. Transvection in Drosophila. Adv. Genet. 46: 399–420 [DOI] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A. A., Baum B., Jones S., Jones M. R., Coulson A., et al. , 2003. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeman J. M., Russell R. C., Tan M. H., Petillo D., Westphal M., et al. , 2008. Somatic pairing of chromosome 19 in renal oncocytoma is associated with deregulated EGLN2-mediated [corrected] oxygen-sensing response. PLoS Genet. 4: e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M. M., Booker M., Silver S. J., Friedman A., Hong P., et al. , 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 3: 833–838 [DOI] [PubMed] [Google Scholar]
- Lehner C. F., 1992. The pebble gene is required for cytokinesis in Drosophila. J. Cell Sci. 103: 1021–1030 [DOI] [PubMed] [Google Scholar]
- Lewis E. B., 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225–239 [Google Scholar]
- Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., et al. , 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Phillips C. M., Bhalla N., Weiser P., Villeneuve A. M., et al. , 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123: 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee B. D., 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim. Biophys. Acta 1677: 165–180 [DOI] [PubMed] [Google Scholar]
- Otsu N., 1979. A threshold selection method from gray-level histograms. IEEE Trans. Sys. Man. Cyber. 9: 62–66 [Google Scholar]
- Prokopenko S. N., Brumby A., O’Keefe L., Prior L., He Y., et al. , 1999. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 13: 2301–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko S. N., Saint R., Bellen H. J., 2000. Tissue distribution of PEBBLE RNA and pebble protein during Drosophila embryonic development. Mech. Dev. 90: 269–273 [DOI] [PubMed] [Google Scholar]
- Ramadan N., Flockhart I., Booker M., Perrimon N., Mathey-Prevot B., 2007. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat. Protoc. 2: 2245–2264 [DOI] [PubMed] [Google Scholar]
- Riede I., 1997. Proliferative genes induce somatic pairing defects in Drosophila melanogaster and allow replication. Cancer Genet. Cytogenet. 97: 143–154 [DOI] [PubMed] [Google Scholar]
- Sage B. T., Csink A. K., 2003. Heterochromatic self-association, a determinant of nuclear organization, does not require sequence homology in Drosophila. Genetics 165: 1183–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi A., Steward R., 2007. Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J. Cell Biol. 176: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I., 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27: 353–365 [PubMed] [Google Scholar]
- Schneider R., Grosschedl R., 2007. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 21: 3027–3043 [DOI] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., et al. , 2012. Three-dimensional folding and functional organization principles of the drosophila genome. Cell. 148: 458–472 [DOI] [PubMed] [Google Scholar]
- Silahtaroglu A. N., Tommerup N., Vissing H., 2003. FISHing with locked nucleic acids (LNA): evaluation of different LNA/DNA mixmers. Mol. Cell. Probes 17: 165–169 [DOI] [PubMed] [Google Scholar]
- Smith E. R., Eisen A., Gu W., Sattah M., Pannuti A., et al. , 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95: 3561–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma M. P., Fasulo B., Cenci G., Cundari E., Gatti M., 2002. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13: 2448–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M. A., Wisotzkey R. G., Newfeld S. J., 2001. A screen for modifiers of decapentaplegic mutant phenotypes identifies lilliputian, the only member of the Fragile-X/Burkitt’s Lymphoma family of transcription factors in Drosophila melanogaster. Genetics 157: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sybenga J., 1999. What makes homologous chromosomes find each other in meiosis? A review and an hypothesis. Chromosoma 108: 209–219 [DOI] [PubMed] [Google Scholar]
- Thomson I., Gilchrist S., Bickmore W. A., Chubb J. R., 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14: 166–172 [DOI] [PubMed] [Google Scholar]
- Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., et al. , 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928 [DOI] [PubMed] [Google Scholar]
- Walter J., Schermelleh L., Cremer M., Tashiro S., Cremer T., 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 160: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. R., Bateman J. R., Novikov N. D., Wu C. T., 2007. Disruption of topoisomerase II perturbs pairing in drosophila cell culture. Genetics 177: 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Rice J. C., 2011. A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle 10: 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wang W., Kong X., Congdon L. M., Yokomori K., et al. , 2010. Dynamic regulation of the PR-Set7 histone methyltransferase is required for normal cell cycle progression. Genes Dev. 24: 2531–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Tsai C. L., Lee J. T., 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Yang H., Mizzen C. A., 2009. The multiple facets of histone H4-lysine 20 methylation. Biochem. Cell Biol. 87: 151–161 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., et al. , 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.