Abstract
Although various genetic tools have been developed and used as transgenes, the expression of the transgenes often is hampered by negative regulators. Disrupting such negative regulators of gene expression is potentially a way to overcome the common problem of low expression of transgenes. To find such regulators whose mutations enhance transgene expression in Caenorhabditis elegans, we took advantage of a newly developed reporter transgene, lin-11pAΔ::venus. This transgene induces expression of a fluorescent protein, Venus, in specific neurons including AIZ, where the expression was stochastic. The frequency of reporter expression in AIZ seemed to be correlated with the strength of transgene expression. By using this system, in which a moderate increase of expression was converted to all-or-none expression states, we describe here a forward genetic screen for mutations that enhance the expression of transgenes. Through the screen, we found that mutations in the pqe-1 gene, which encodes a Q/P-rich nuclear protein with an exonuclease domain, increase the chance of reporter expression in AIZ. The fluorescence intensity in RIC, in which all lin-11pAΔ::venus animals show reporter expression, was increased in pqe-1 mutants, suggesting that pqe-1 reduces the expression level of the transgene. Expression of transgenes with other promoters, 3′UTR, or reporter genes was also enhanced by the pqe-1 mutation, suggesting that the effect was not specific to a particular type of transgenes, whereas the effect did not seem to extend to endogenous genes. We propose that pqe-1 mutants can be used to increase the expression of various useful transgenes.
Keywords: Caenorhabditis elegans, transgene expression, pqe-1
Various genetic tools have been developed and used for probing and manipulating molecular and cellular functions in model organisms. For example, green fluorescent protein (GFP) and related fluorescent proteins are used to visualize gene expression or dynamics of proteins (Chalfie et al. 1994). Calcium indicators like cameleon (Miyawaki et al. 1997) and GCaMP (Nakai et al. 2001) were derived from fluorescent proteins and used to visualize intracellular calcium responses. Recently developed optogenetic tools, such as channelrhodopsin-2 (Boyden et al. 2005) and halorhodopsin (Zhang et al. 2007), are useful for manipulating the activity of neurons. These are powerful experimental tools, but it can be difficult to get them expressed in sufficient levels. Introducing transgenes in high copy numbers does not always lead to strong expression because repetitive sequences often cause gene silencing (Hsieh and Fire 2000). To increase the expression of transgenes without modifying the coded proteins, several methods could be applied. For example, optimization of codon usage within the coding sequence, by incorporating codons for the most abundant isoaccepting tRNA, is known to increase expression (Ikemura 1985). Recently, it was also reported that optimizing the codons to stabilize the mRNA around the ribosomal binding site effectively increases the gene expression in Escherichia coli (Kudla et al. 2009). In addition, refining the sequences of untranslated regions, 5′UTR and 3′UTR, or introducing synthetic introns also results in an increase of gene expression in eukaryotes by optimizing posttranscriptional processes (Le Hir et al. 2003; Mignone et al. 2002).
In the soil nematode Caenorhabditis elegans, transgenes injected into the germline form large heritable extrachromosomal arrays consisting of dozens to several hundreds of copies through recombination (Mello et al. 1991; Stinchcomb et al. 1985). However, these repetitive transgenes show lower expression levels than that expected from their copy numbers. This suppression is probably attributable to gene silencing of repetitive sequences because less-repetitive arrays formed by co-introduction of transgenes with excess C. elegans genomic DNA cause greater expression (Kelly et al. 1997). To characterize the mechanisms of gene silencing of repetitive arrays in somatic cells, screens for mutations that enhance the silencing of fluorescent reporter genes were previously performed. In the screens, mutations in a RING finger/B-Box factor TAM-1, a retinoblastoma-like protein LIN-35, and a bromodomain protein LEX-1 were all identified to enhance the silencing in somatic cells (Hsieh et al. 1999; Tseng et al. 2007). In contrast, mutations that enhance expression of transgenes have not been reported, to the best of our knowledge. Detecting subtle increases in reporter expression would be difficult because the expression level of transgenes in individual animals is variable even in a wild-type population. Although it is challenging, if we obtain mutations that enhances the expression of transgenes generally, we will be able to apply such mutations to increase the expression of useful genetic tools.
The nervous system of C. elegans comprises 302 neurons, each with specialized functions (White et al. 1982). Among those neurons, AIZ neurons receive inputs from several sensory neurons and act in the integration of thermosensory signals (Mori and Ohshima 1995). They are also indispensable for salt chemotaxis (Iino and Yoshida 2009). In the course of characterizing the function of AIZ, we produced a composite promoter lin-11pAΔ, which induced fluorescent reporter expression mainly in RIC and AIZ neurons. Surprisingly, the expression in AIZ was stochastic, whereas the expression in RIC was observed in all animals. The decision to express or not was affected apparently by the potential of gene expression that represent, for example, the copy number of the transgenes. A greater potential of gene expression results in a greater probability of expression in AIZ neurons. This expression system enabled the detection of a moderate increase, by approximately twofold, of fluorescent reporter expression by converting it to all-or-none expression states in AIZ neurons. We describe here a successful screen for mutations that generally enhance the expression of transgenes in somatic cells of C. elegans by taking advantage of this expression system.
Materials and Methods
Strains
C. elegans strains were cultured and maintained using standard procedure (Brenner 1974) except that NA22 E. coli strain was used as food source. The following strains were used: wild-type Bristol strain (N2), wild-type Hawaiian strain (CB4856), ttTi5605 II; unc-119(ed3) III, unc-119(ed3) III; cxTi10882 IV, pqe-1(pe334) III, pqe-1(pe335) III, pqe-1(pe336) III, pqe-1(ok1983) III, peIs303[lin-11pAΔ::venus, tdc-1p::mRFP, rol-6(d)] X, peIs304[lin-11pAΔ::venus, tdc-1p::mRFP, rol-6(d)] I, peIs1334[osm-10p::venus::unc-2(3′UTR), unc-122p::mCherry], peIs1336[osm-10p::venus::unc-54(3′UTR), unc-122p::mCherry], peIs1338[sra-6p::venus::unc-2(3′UTR), unc-122p::mCherry], peIs1339[sra-6p::venus::unc-54(3′UTR), unc-122p::mCherry], peIs1090[sra-6p::ChR2::venus, unc-122p::mCherry], mIs13[myo-2::GFP, pes-10::GFP, F22B7.9::GFP] I, peIs1323[lin-11pA::venus] II, and peIs1326[lin-11pA::venus] IV. pqe-1(pe334), pqe-1(pe335), and pqe-1(pe336) mutants were obtained in a forward genetic screen for mutants that increase the AIZ expression of peIs304. peIs303, peIs304, peIs1334, peIs1336, peIs1338, peIs1339, and peIs1090 were produced by the insertion of extra-chromosomal arrays into chromosomes by γ-ray irradiation or MMS (i.e., methyl methanesulfonate) treatment. Other strains were kindly provided from Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN). pqe-1(pe334), peIs303, peIs304, peIs1334, peIs1336, peIs1338, peIs1339, and peIs1090 were backcrossed to N2 (wild-type) more than three times before characterization.
Plasmid construction
Plasmids were generated by the GATEWAY system (Invitrogen) as described (Matsuki et al. 2006). The entry vectors that contain lin-11p (1338 bp) (Hobert et al. 1998), sra-6p (4005 bp) (Troemel et al. 1995), H20p (2479 bp), pqe-1p1 (2299 bp), and pqe-1p2 (2082 bp) were created by BP reactions (site-directed recombination) between a polymerase chain reaction (PCR)-amplified promoters and the pDONR201 plasmid. lin-11pA, lin-11pB, and lin-11pC were produced by cloning PCR-amplified fragments of 660 bp, 367 bp, and 361 bp, respectively, into DraI-digested pENTR-sra6p. lin-11pAΔ was produced by removing the 80 bp region of lin-11pA. The deletion was introduced by PCR amplification of the rest of the plasmid by primers 5′- TAGCGACTATGAATATCTAA-3′ and 5′-CTTGAATGTGTCCCCATCAT-3′, and then the product was circularized by ligating the ends.
The destination vectors that contain open reading frames (ORFs) of each gene were generated by cloning the cDNA fragment of the gene of interest into the pDEST vector. ORFs for venus, mRFP, and mCherry were amplified from pPD-venus [carrying a modified version of Venus (Nagai et al. 2002), which was a gift from T. Ishihara, Kyushu University, Fukuoka, Japan], pRSETb (a gift from R. Tsien, Stanford University School of Medicine), and pWD176 (carrying a optimized mCherry produced by K. Oegema, which was a gift from E. Jorgensen, and subcloned into pDEST by H. Ohno), respectively. The cDNAs of pqe-1 isoforms were amplified from reverse transcripts of total RNA. The pDEST plasmid of venus-fused pqe-1C was produced by inserting termination codon-deleted pqe-1C ORF in front of the venus gene of pDEST-venus. For the pDEST constructs with unc-2 3′UTR, a pDEST vector with 853 bp of unc-2 3′UTR, which was PCR-amplified by two primers, 5′-CAACCGTTTAACAAAGCCTCTC-3′ and 5′-TTGCGTTCTCCTCCAATTCT-3′, was used. The expression constructs were produced by LR reaction between the entry plasmids and the destination plasmids. Details of the use of the GATEWAY system are available at our web site: http://park.itc.u-tokyo.ac.jp/mgrl/IINO_lab/Gateway/Gateway_overview1.html.
Germline transformation
Transgenic strains were produced by injecting DNA into the gonad of adult animals as described (Mello et al. 1991). Expression constructs for lin-11p variants were injected at 30 ng/μL along with unc-122p::mCherry (10 ng/μL) as a transformation marker and pPD49.26 (60 ng/μL) as carrier DNA except that lin-11pAΔ::venus was injected at 30 ng/μL along with tdc-1p::mRFP (30 ng/μL) for marking RIC neurons and pRF4[rol-6(d)] (40 ng/μL) for inducing roller phenotype. For examination of the promoter activity of pqe-1, pqe-1p1::venus and pqe-1p2::mCherry were injected at 30 ng/μL each along with pPD49.26 (40 ng/μL). PQE-1C::Venus-expressing line was produced by injecting H20p::pqe-1C::venus (60 ng/μL) along with H20p::fib-1::mCherry (5 ng/μL), for marking the nucleoli, and pPD49.26 (35 ng/μL). For rescue experiments of pqe-1(pe334), a PCR-amplified 15-kb fragment (1 ng/μL) or promoter-fused pqe-1 cDNAs (10 ng/μL) was injected along with myo-3p::mRFP (10 ng/μL) as transformation marker and pPD49.26 (80 ng/μL). Germline transformation using the MosSCi method was performed as described (Frøkjaer-Jensen et al. 2008).
Assay of AIZ phenotypes
Unless otherwise noted, adult animals were prepared by placing six parent adults on each 6-cm Petri dish, which was seeded with NA22 E. coli, and letting the offspring grow at 20° for four days. The phenotypes of adults were assayed by counting randomly selected animals under a fluorescence stereomicroscope (Leica M165 FC). For rescue lines, the phenotypes were checked after we randomly selected animals that possessed the transformation marker. L1 larvae were prepared by incubating the eggs in M9 buffer for about 6 hours after lysing the 4-day-old adults with bleach mix (1N NaOH 0.5 mL, 0.2 mL household bleach [5% solution of NaClO]). Hatched L1 larvae were mounted on an agar pad, and the phenotypes were tested under a epifluorescence microscope (Axioplan2) with 20× or 40× objective.
Forward genetic screen
Mutagenesis was performed as described (Brenner 1974). A total of 18,000 F1 progeny of mutagenized peIs304 animals were divided into 12 groups; 3000 F2 embryos were collected from each group and raised to adults. The phenotypes of individual F2 adults were observed under a fluorescence stereomicroscope (Leica M165 FC) and AIZ-2ON animals were independently isolated from each group. The F3 progeny of each isolated animal were tested for whether the population shows high rate of AIZ-2ON phenotype. The lines with the most obvious phenotype were selected from each group. Mapping of the responsible mutations in the obtained strains was performed using the SNPs between the N2 Bristol and CB4856 Hawaiian wild-type (Wicks et al. 2001). After identifying the chromosomes in which the mutations resided, mapping was focused on pe334, pe335, and pe336, which showed linkage to chromosome III.
Quantification of fluorescence intensity
The total intensity of Venus in a cell was quantified in L1 larva as previously described (Yamada et al. 2010). Nonconfocal fluorescence images of target cells were taken by 100× objective for RIC, AIZ, and ASH neurons and 40× objective for posterior bulb of the pharynx with Zeiss Axioplan2 epifluorescence microscope equipped with a Hamamatsu ORCA-ER camera. The length of exposure was 10 ms for peIs1334, peIs1336, and peIs304[tdc-1p::mRFP]; 100 ms for peIs303, peIs304[lin-11pAΔ::venus], peIs1090, peIs1339, and mIs13; 200 ms for low copy−inserted transgenes, peIs1323 and peIs1326; and 500 ms for peIs1338. Photos were processed with Metamorph software (Molecular Devices), and the total intensity for the neuronal cell body of each animal was calculated by the equation below.
Total intensity was normalized by the average of the control conditions.
Quantitative real-time PCR
The total RNA of the worms was extracted from L1 larvae. The L1 larvae were obtained by incubating eggs for 12 hr in M9 buffer after collecting eggs from bleached 4-day-old adult animals. Proteins were removed and nucleic acid was recovered with Trizol (Invitrogen) followed by isopropanol precipitation. The nucleic acid extracts were processed with DNase I (QIAGEN) and RNeasy MinElute cleanup kit (QIAGEN) to remove the contaminating DNA and reverse-transcribed by PrimeScript RT reagent Kit (Takara). Quantification of the reverse transcripts was performed by using SYBR RT-PCR kit Perfect real time (Takara) and ABI PRISM7000 (Applied Biosystems) according to the manufactures’ protocol. A total of 200 ng of total RNA was reverse-transcribed to cDNA, and an equal amount of cDNA was subjected to a gene-specific PCR in a total volume of 25 μL. Serial dilutions of cDNA prepared from total RNA of wild-type worms were used to generate a standard curve. The relative quantity of venus transcripts was calculated using the amount of lmn-1 transcript as a reference, and the results of duplicated experiments for a same sample were averaged.
The primers used for the amplification of each gene were as follows: lmn1-F: 5′-CGTTCACCACCCACCAGAA-3′, lmn1-R: 5′-CAAGACGAGCTGATGGGTTATCT-3′ for lmn-1; venusRT-F: 5′- TGGAGAGGGTGAAGGTGATG-3′, venusRT-R: 5′- GACAAGTGTTGGCCATGGAA-3′ for venus; lin11-F: 5′-CGCAACACCCAAACCAACTC-3′, lin11-R: 5′-GAAACCACACCTGAATGACTCTC-3′ for lin-11; osm10-F: 5′-CAACCACAGAATGGCAATCG-3′, osm10-R: 5′-CGAGCCAAGTCTCTGCAATG-3′ for osm-10; myo2-F: 5′-TGAAGTTCAAGCAACGTCCA-3′, myo2-R: 5′-GTGGACGAGTCAAAGCCTTC-3′ for myo-2; cdc42-F: 5′-CTGCTGGACAGGAAGATTACG-3′, cdc42-R: 5′-CTCGGACATTCTCGAATGAAG-3′ for cdc-42; and pmp3-F: 5′-GTTCCCGTGTTCATCACTCAT-3′, pmp3-R: 5′-ACACCGTCGAGAAGCTGTAGA-3′ for pmp-3. The primers for cdc-42 and pmp-3 were adopted from the previous report (Hoogewijs et al. 2008). All primers were designed to include an intron in the PCR product amplified from the genomic DNA for each gene to distinguish them from the product amplified from the cDNA.
The copy numbers of transgenes were quantified by almost the same method. 10 ng of purified genomes were extracted from adult animals and used as templates. The primers used for the amplification of venus were: venus-copy-N-count-F: 5′-CCAGACAACCATTACCTGTCC-3′ and venus-copy-N-count-R: 5′-CATCCATGCCAAGTGTAATCC-3′. The relative copy number was calculated by normalizing by the PCR products that were amplified from a specific region of the genome by using two primers: copy-N-control-F: 5′-GTCGCGTTGAAGGTTCTCTC-3′ and copy-N-control-R: 5′-CTCCATTGAATCGTGACCAA-3′.
Statistical analyses
Error bars in the figures indicate SEM. The statistical analyses for AIZ phenotypes were performed with χ2 tests. In cases in which multiple times of tests were required, the analyses were corrected by the use of Bonferroni’s method. For the statistical analyses for quantified fluorescence intensity and transcript numbers, Student’s t-test or Dunnett’s test for multiple comparisons was performed.
Results
Truncated promoter of lin-11p, lin-11pAΔ, drives reporter expression in RIC and AIZ
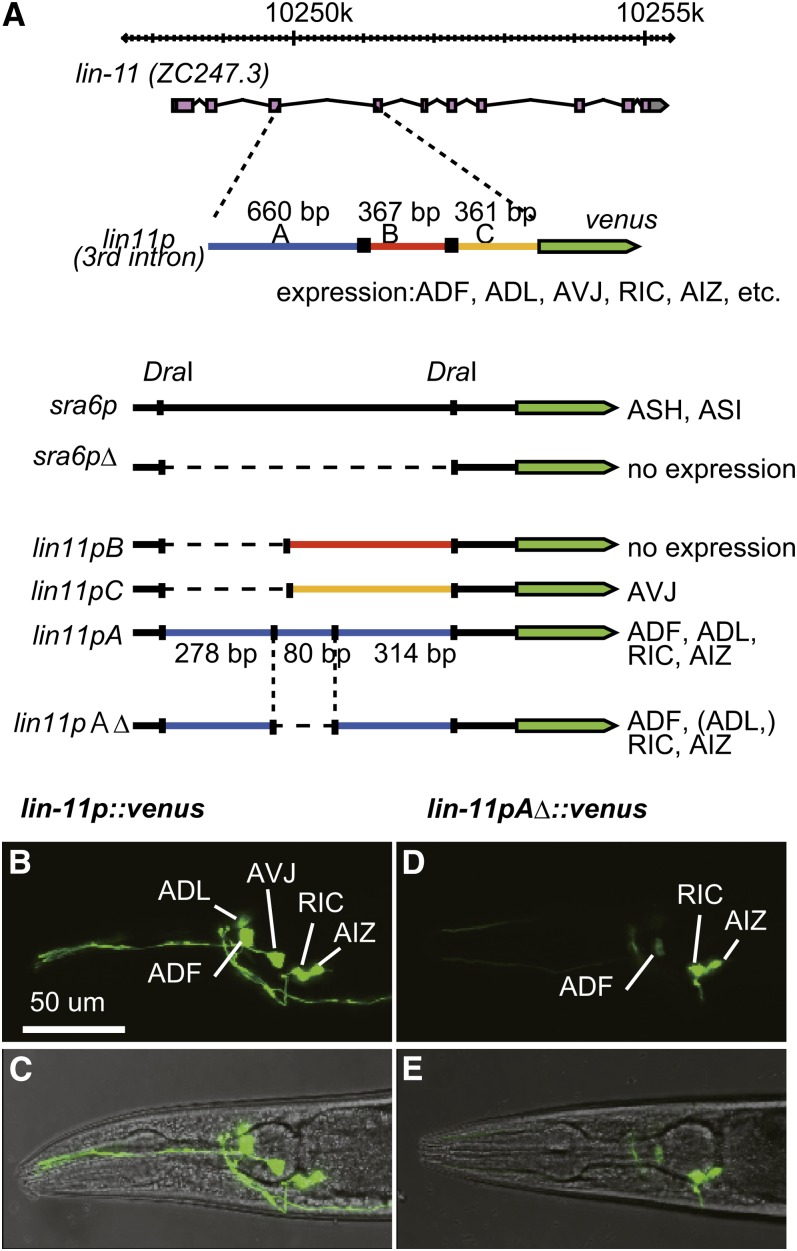
The third intron of the lin-11 gene, which encodes a LIM homeodomain transcription factor (Freyd et al. 1990), is one of the promoter/enhancers that drive gene expression in AIZ neurons and is denoted here as lin-11p. Consistent with the previous report, lin-11p::venus, in which a fluorescent reporter venus is fused to lin-11p, drove strong expression in ADL, ADF, AVJ, RIC and weak expression in RIF and AVG (Hobert et al. 1998) (Figure 1, A−C, and supporting information, Figure S1A and Figure S1B). However, only a small fraction of transgenic animals demonstrated reporter expression in AIZ. To finely characterize this unstable reporter expression in AIZ, we first tried to develop truncated promoter/enhancers to specify the expression in AIZ. The third intron of lin-11 had no obvious TATA box for transcription initiation, making it difficult to design truncated promoter/enhancers. Therefore, subregions of lin-11p, segments A, B, and C, were subcloned and connected to the TATA box of the promoter of sra-6 (Troemel et al. 1995) (Figure 1A). The segment C of the lin-11p drove expression in AVJ (Figure S1I, Figure S1J, Figure S1K, and Figure S1L), whereas the segment A drove expression in ADL, ADF, RIC, and AIZ (Figure S1E, Figure S1F, Figure S1G, and Figure S1H). In contrast, segment B drove no expression, suggesting that this segment includes no obvious enhancer function. We further truncated segment A and found that the truncated promoter, lin-11pAΔ, was sufficient for the expression in RIC and AIZ, whereas the expression in ADL and ADF was weakened in this promoter (Figure 1, D and E, Figure S1C and Figure S1D).
Figure 1.
Production of a promoter to restrict the reporter expression in RIC and AIZ. (A) Illustration of the construction of composite promoters. The 4-kb promoter of sra-6 was digested by a restriction enzyme, DraI, and ligated to exclude a 3.6-kb region. The resulting promoter sra-6pΔ did not induce expression of the fused reporter gene. The segments of lin-11p, A, B, and C, were introduced to the sra-6pΔ to produce lin-11pA, lin-11pB, and lin-11pC, respectively. lin-11pA was further truncated to produce lin-11pAΔ. (B−E) Expression pattern of lin-11p::venus and lin-11pAΔ::venus. The third intron of lin-11 (B and C) or the synthetic promoter lin11pAΔ (D and E) was fused to venus cDNA and introduced to wild-type animals to examine the promoter activity. The fluorescence image of the head and the image merged with bright field image of an adult animal are shown in (B and D) and (C and E), respectively.
lin-11pAΔ drives stochastic reporter expression in AIZ
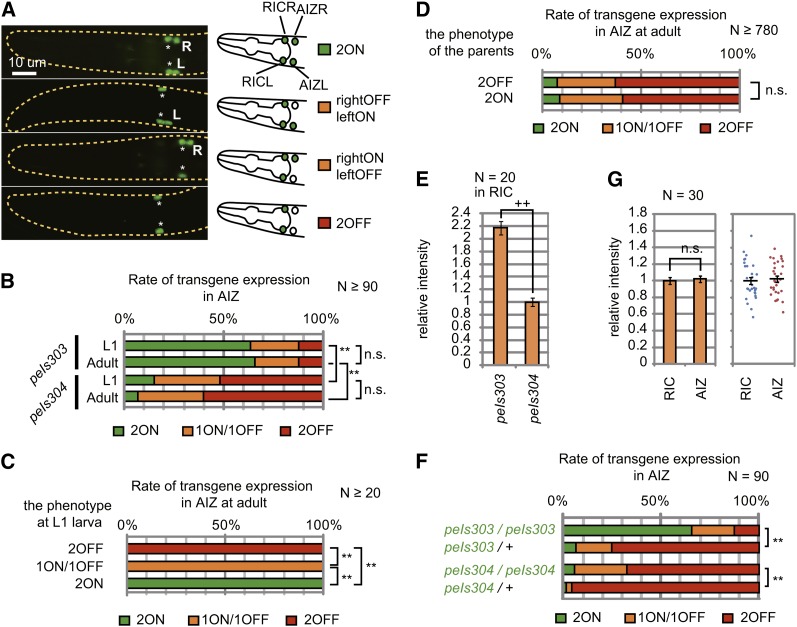
In C. elegans, transgenes introduced by microinjection are usually held as an extra-chromosomal array, which is randomly lost during cell divisions (Stinchcomb et al. 1985). To exclude the effect of mosaicisms, we integrated the extra-chromosomal transgenes of lin-11pAΔ::venus into the chromosomes by γ-ray irradiation. This treatment yielded two integrants, peIs303 and peIs304, in which the transgene lin-11pAΔ::venus was inserted into chromosome X and I, respectively. Both peIs303 and peIs304 animals showed stochastic expression in AIZ. C. elegans has two bilaterally symmetric AIZ neurons, AIZL and AIZR, and we found all four types of combinations of expression patterns, 2ON, rightOFF/leftON, rightON/leftOFF, and 2OFF in both integrants (Figure 2A; ON and OFF indicate that the expression is observed and not observed, respectively, in each cell). Because the numbers of rightOFF/leftON animals and rightON/leftOFF animals were almost the same, we did not distinguish them further and combined them as 1ON/1OFF. The distribution of AIZ phenotypes was different in the two integrant lines with a greater rate of expression in AIZ in peIs303 animals. On the other hand, there was no significant difference between L1 larvae, the first larval stage just after hatching, and adult animals in each strain (Figure 2B). When individual peIs304 animals were isolated at the L1 stage, sorted by their AIZ expression phenotypes and cultured further, they showed the same phenotypes 3 days later when they grew up to adults (Figure 2C). These results indicate that the phenotypes of AIZ expression are decided at the early stage of development and maintained throughout the individuals’ life. Furthermore, the progeny of isolated peIs304 individuals showed the distribution of phenotypes identical to their mother’s generation irrespective of their mother’s phenotype (Figure 2D and Figure S2). Therefore, the AIZ expression phenotypes are not inherited but decided by chance during embryogenesis.
Figure 2.
Genome-integrated lin-11pAΔ::venus drives stochastic expression in AIZ neurons. (A) The individual expression pattern of peIs304[lin-11pAΔ::venus] in an isogenic population of L1 larvae. All the animals in a population showed reporter expression in RIC neurons, whereas the expression in AIZ neurons were stochastic. The illustrations at the right side summarize the phenotype in the left picture. *, L, and R indicate RIC neurons, AIZL, and AIZR, respectively. (B) AIZ phenotype of peIs303 and peIs304 animals at L1 larval and adult stages. The L1 animals were observed within 6 hr from hatch and the adults were tested on the fourth day after putting the parents on the plates (n ≥ 90). (C) AIZ phenotype of adult peIs304 animals that were isolated at L1 larva and classified according to their AIZ phenotypes (n ≥ 20). (D) AIZ phenotypes of the offspring of the peIs304 animals with indicated phenotypes. The AIZ phenotypes of the offspring were tested at adult (n ≥ 780). (E) Normalized fluorescence intensity from peIs303 and peIs304 in RIC neurons of L1 larvae (n = 20, error bars represent SEM). (F) AIZ phenotypes of heterozygous integrant animals at adult. A dominant genetic marker, mIs13[myo-2p::gfp], was used to verify successful cross. (G) Normalized fluorescence intensity from peIs304 in RIC and AIZ neurons of L1 larva. The fluorescence intensity was quantified in the animals that showed reporter expression in AIZ neurons. Scatter plot in the right graph indicates the data of individual neurons (n = 30, Error bars represent SEM). **Significant differences at P < 0.001 by χ2 test; ++significant difference at P < 0.001 by Student’s t-test. n.s. indicates that they are not significantly different at P < 0.01 by χ2 test or Student’s t-test. χ2 tests in (B, C, and F) were corrected by Bonferroni’s method)
Probability of lin-11pAΔ::venus expression in AIZ is correlated with the strength of its expression in RIC
Through a series of observations of the reporter expression from peIs303 and peIs304, we noticed that peIs303 shows stronger reporter expression than peIs304. To clarify this, we compared the reporter expression in RIC because all animals harboring lin-11pAΔ::venus show reporter expression in RIC. The quantified fluorescence intensity in RIC of peIs303 was more than twice that of peIs304 (Figure 2E). Because the probability of reporter expression in AIZ was greater in peIs303 than in peIs304 (Figure 2B), these results suggest that the greater rate of “ON” expression of lin-11pAΔ::venus in AIZ in peIs303 animals may be caused by stronger expression induction potential of the transgene. To demonstrate the causal relationship between the potential of expression induction and probability of AIZ expression, we tested the AIZ phenotypes of heterozygotes of lin-11pAΔ::venus integrants. We crossed peIs303 or peIs304 homozygous animals to animals that carry a dominant genetic marker with wild-type background. As expected, resulting heterozygous animals, peIs303/+ and peIs304/+, showed lower rates of AIZ-ON phenotype than homozygous animals, peIs303/peIs303 and peIs304/peIs304, supporting the idea that the stronger expression induction potential of lin-11pAΔ::venus leads to a greater rate of AIZ-ON phenotype (Figure 2F).
It was possible that the reporter expression in AIZ was generally weaker than that in RIC, and therefore the reporter expression in some animals was below threshold of detection only in AIZ but not in RIC. However, this is not the case because the average fluorescence intensity of the reporter expression in AIZ among the animals in which we could observe the expression in AIZ was almost the same as the average intensity in RIC (Figure 2G). These results suggest that AIZ has two discrete expression states corresponding to ON and OFF and somehow the expression induction potential determines the probability of the ON state through a thresholding effect in AIZ (see Discussion). In contrast, the expression in RIC does not seem to show such transformation.
The difference of the expression level between peIs303 and peIs304 could be attributable to the difference in copy number of transgenes integrated into each genomic locus, the structures of the multicopy transgenes, or location of genomic insertion. To determine the copy number of transgenes, we compared the copy number of peIs303 and peIs304 to low copy genomic insertions produced by MosSCI. MosSCI, Mos1-mediated Single Copy transgene Insertion, is a technique to insert transgenes into Mos1 transposon−inserted genomic loci through repair of Mos1 excision events, resulting in low copy insertion of the transgenes (Frøkjaer-Jensen et al. 2008). By using this technique, we inserted lin-11pA::venus into two loci, ttTi5605 II and cxTi10882 IV, in the genome and obtained two alleles, peIs1323 and peIs1326, respectively. We extracted the genome from transgenic animals and compared the copy number of the venus gene by real-time PCR. Because MosSCI generally produces single copy insertion with less than 10% of duplicated insertion and the relative copy number of venus in peIs1326 was exactly twice that of peIs1323, we could speculate that peIs1323 was a single-copy insertion and peIs1326 was a duplicated-copy insertion (Figure S3). Compared with these low copy transgenes, the venus copy numbers in peIs303 and peIs304 were estimated to be 34(+/−8) and 35(+/−13), respectively. The estimated copy numbers of transgenes in peIs303 and peIs304 were not significantly different, and therefore the difference in AIZ phenotype may arise from other factors, such as the difference in the structure of the transgenes or different locations in the genome.
Mutations in the pqe-1 gene enhance the expression of lin-11pAΔ::venus
To screen for mutations that enhance the expression of transgenes in C. elegans, we took advantage of the expression system of peIs304, in which a moderate increase in expression induction potential is converted to a drastic change from AIZ-OFF to AIZ-ON through the thresholding effect observed in AIZ. We mutagenized peIs304 animals and screened for mutations that cause a greater rate of reporter expression in AIZ. Such a phenotype could be caused by an upward shift of the distribution of expression inducing potential. Of 12 independent mutations isolated by the screen, pe334, pe335, and pe336, were mapped to the center of chromosome III, whereas the others were mapped to the center of chromosome I. Because the mutations that were mapped to chromosome I showed strong linkage to the inserted transgene, peIs304, it was likely that these mutations occurred in the peIs304 allele itself.
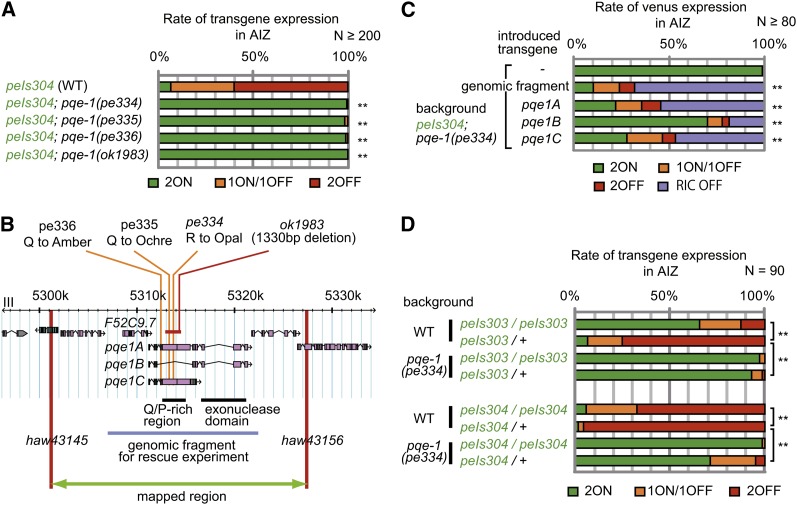
Therefore, we focused on the mutations on chromosome III for further analyses. Mutants of all three alleles showed almost 100% of AIZ-2ON phenotype (Figure 3A). Among those mutations, pe334 was mapped to a region of 26 kb between two SNPs, haw43145 and haw43156 (Figure 3B). There were only four genes in this interval, and among them we found a nonsense mutation in the pqe-1 gene. We concluded pqe-1 was responsible for the high frequency of AIZ-2ON phenotype for the following three reasons. First, the pe335 and pe336 mutants also possessed nonsense mutations in the pqe-1 gene (Figure 3B). Second, a 1330-bp deletion in the pqe-1 gene, ok1983, also caused a high frequency of AIZ-2ON phenotype when combined with peIs304 (Figure 3, A and B). Finally, the introduction of a PCR fragment spanning the pqe-1 gene and the adjacent gene F52C9.7, within which our mutants do not have any mutations, rescued the AIZ-2ON phenotype of the pe334 mutant (Figure 3, B and C). For the aforementioned reasons, we re-define the nonsense mutation in pqe-1 gene in each mutant as pqe-1(pe334), pqe-1(pe335), and pqe-1(pe336) (Figure 3B).
Figure 3.
Mutations in the pqe-1 gene enhance the reporter expression in AIZ from peIs304. (A) AIZ phenotypes of mutants, pe334, pe335, and pe336, isolated through mutant screen and an independently isolated deletion mutant of pqe-1, ok1983, at adult (n ≥ 200). (B) Locations of identified mutations. pe334, pe335, and pe336 were identified as Q to Opal, Q to Ochre, and Q to Amber, respectively. The mapped region and the genomic fragments used for rescue experiments in (C) are indicated. The pqe-1 gene has three main isoforms, which include either or both of the Q/P rich region and the exonuclease domain. (C) Rescue experiments for pqe-1(pe334) mutants. The cDNA of each isoform was expressed by a pan-neuronal promoter, H20p. The transgenes were introduced to mutants as extrachromosomal array by using myo-3p::mRFP as a transformation marker. Only the adult animals that carry the extrachromosomal array were used for the analysis. In this graph, the animals that lacked the reporter expression in both RIC and AIZ in either side were counted as RIC OFF. In addition, if a single cell showed expression in either side, that cell was assumed to be RIC (n ≥ 80). (D) AIZ phenotypes of heterozygous integrant animals in pqe-1(pe334) mutant background at adult. mIs13[myo-2p::gfp], was used to verify successful cross (n = 90). **Significant differences at P < 0.001 by χ2 test with Bonferroni’s correction.)
Along with the rescue of the AIZ phenotype, introduction of genomic pqe-1 fragments into the pqe-1(pe334) mutant also caused silencing of reporter in RIC, which was consistently expressed in the original peIs304; pqe-1(+) animals (Figure 3C, Figure S4A, Figure S4B, Figure S4C, Figure S4D, Figure S4E, Figure S4F, Figure S4G, and Figure S4H). The repression of the reporter in RIC may be caused by overexpression of the pqe-1 gene in the rescue constructs because the extra-chromosomal array also repressed the expression in RIC neurons, as well as AIZ neurons, when transferred into the pqe-1(+) background (Figure S4I). These results suggest that, when overexpressed, pqe-1 acts not only in AIZ but also in RIC and probably in other neurons to suppress the transgene expression regardless of whether they have threshold for expression. To further characterize the effect of pqe-1 on the AIZ phenotypes, we examined the effect of halved copy number of lin-11pAΔ::venus in pqe-1(pe334) mutant background. As described previously, the heterozygotes of lin-11pAΔ::venus, peIs303/+ and peIs304/+, show decreased expression in AIZ (Figure 2E). In the pqe-1(pe334) mutant background, the rate of reporter expressions in AIZ from both peIs303/+ and peIs304/+ were increased again, indicating that the mutation in pqe-1 reversed the effect of decreased transgene copy number (Figure 3D).
pqe-1 is a nematode-specific gene. Loss of function of this gene by itself shows no obvious phenotype but was reported to enhance the toxicity of overexpression of poly-glutamine(polyQ) (Faber et al. 1999, 2002). In these studies, the neurotoxic effect of mutant huntingtin, which causes neurodegenerative Huntington disease in humans, was examined in C. elegans. The polyQ region of mutant huntingtin, Htn-Q150, was ectopically expressed in ASH sensory neurons and mutations in pqe-1 was identified through forward genetic screen as mutations that enhance the neural degeneration by Htn-Q150. pqe-1 has at least three isoforms, PQE-1B with exonuclease domain, PQE-1C with Q/P rich region and a repeat of nuclear localization signals, and PQE-1A with all these domains [Figure 3B and Figure S5A; note that the naming of the variants are based on the previous report (Faber et al. 2002) and different from the isoform names in WormBase (http://www.wormbase.org)]. PQE-1C was sufficient for the polyQ toxicity-enhancing phenotype (Faber et al. 2002). To examine which isoforms are important for the high rate of AIZ-2ON phenotype, we conducted rescue experiments by expressing each isoform pan-neuronally in peIs304; pqe-1(pe334) mutants. As a result, we found that the expression of PQE-1A or PQE-1C isoform could rescue the AIZ-ON defect efficiently, as in the case for polyQ toxicity enhancing phenotype. In these rescue experiments, we observed reduction of reporter repression also in RIC, again probably by the effect of overexpression (Figure 3C). The rescue of the mutant phenotype by PQE-1A and PQE-1C isoforms was the same as the previous study, suggesting that these two phenomena may have common causes.
pqe-1 is expressed in various tissues and shows dot-like localization in the nuclei
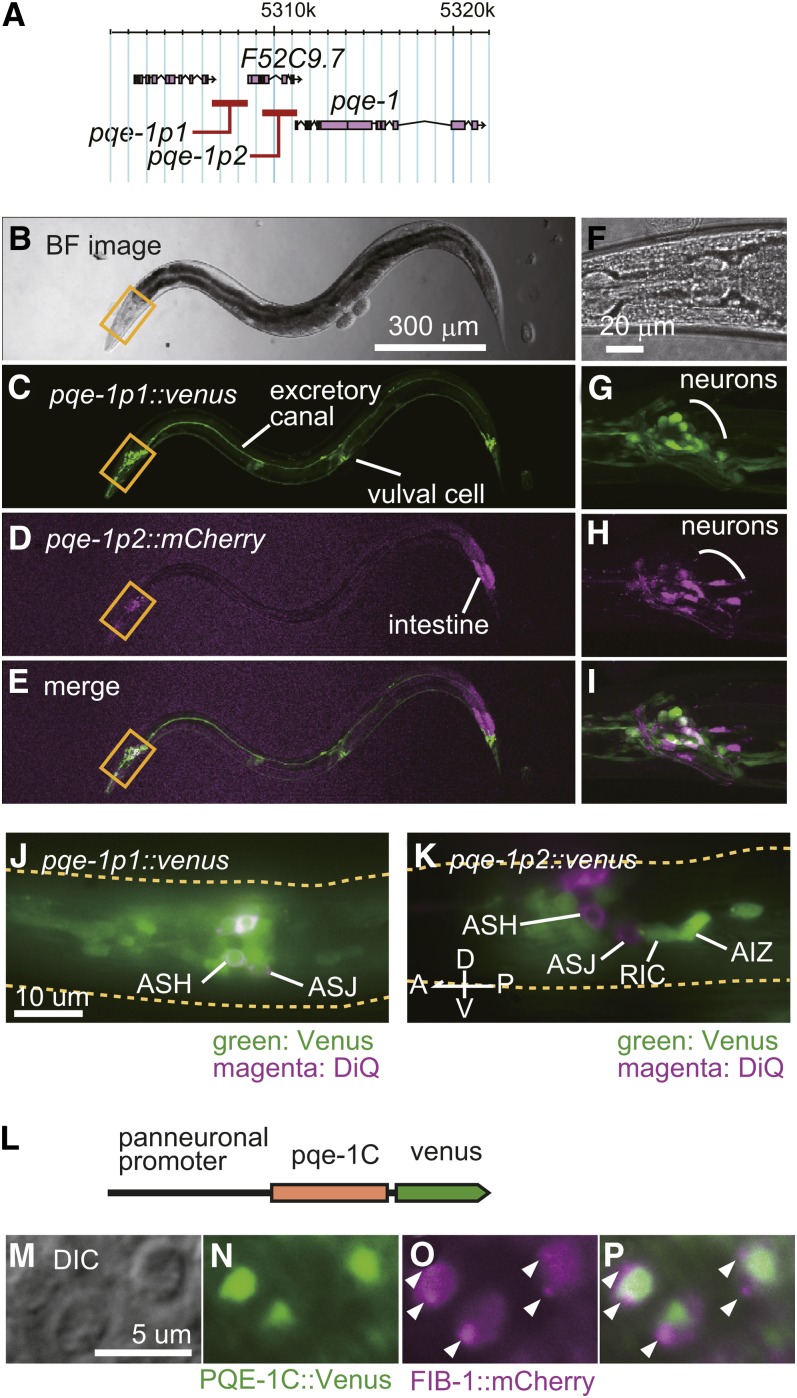
pqe-1 is predicted to form an operon with several other nearby genes (C. elegans Sequencing Consortium 1998). To examine the expression pattern of pqe-1, we tested two regions, pqe-1p1, which is located at upstream of the whole operon, and pqe-1p2, which is immediately upstream of pqe-1, for promoter activity (Figure 4A). We cloned a 2-kb region of each promoter and tested their expression induction by fusing to venus reporter. pqe-1p1 drove expression in excretory canal, vulval cells, and neurons at the head and the tail, whereas pqe-1p2 drove expression in intestine and neurons (Figure 4, B−I). The expressing cells were almost different between these two promoters with a small overlap (Figure 4, E and I). We confirmed that the pqe-1p1 drives reporter expression in sensory neurons including ASH, in which the polyQ toxicity enhancing phenotype of pqe-1 mutants was previously observed, whereas pqe-1p2 drives reporter expression in several other neurons, including RIC and AIZ (Figure 4, J and K). Previously, it was reported that PQE-1C protein shows a dot-like localization in the nucleus and functions cell-autonomously (Faber et al. 2002). Because we confirmed pqe-1 promoters drive reporter expression in AIZ, the effect of pqe-1 on AIZ phenotype may be also cell-autonomous. The most remarkable structure in the nuclei is nucleoli. To test whether PQE-1C localizes to the nucleoli, we expressed PQE-1C::Venus along with the nucleolus marker FIB-1::mCherry (Goldsmith et al. 2010; Lee et al. 2010) (Figure 4L−P). The intranuclear localization of PQE-1C::Venus (Figure 4N) was different from that of FIB-1::mCherry (Figure 4O), indicating that PQE-1 does not localize to the nucleoli. We speculate that the dots represent the aggregation of PQE-1C proteins because PQE-1C includes high content of glutamine, 112 of 572 amino acids in the Q/P rich region (Figure S5B), and polyQ proteins are known to have trends of aggregation (Williams and Paulson 2008).
Figure 4.
Expression pattern of pqe-1 gene and intracellular localization of PQE-1. (A) Promoters used for the expression of fluorescent reporters. pqe-1p1 and pqe-1p2 are the 2 kb upstream region of F52C9.7 and pqe-1, respectively. (B−I) Expression patterns of pqe-1p1::venus (C and G) and pqe-1p2::mCherry (D and H) in an adult animal. (E and I) are merged images of (C and D) and (G and H), respectively. (B and F) are bright-field images of (C−E) and (G−I), respectively. (F−I) are the magnified images of the head region shown by yellow squares in (B−E). (J and K) Expression patterns of pqe-1p1::venus (J) and pqe-1p2::venus (K) merged with the DiQ-stained cells (magenta: ASJ and ASH) in the head. (L-P) Nuclear localization of PQE-1C::Venus. A venus-fused pqe-1C cDNA is expressed by a pan-neuronal promoter, H20p (L). The dot-like localization of PQE-1C::Venus (N) and FIB-1::mCherry in the nucleus (O). (P) is the merged image of (N) and (O). (M) is the DIC image of the region in (N−P). Arrowheads in O and P indicate the nucleolar localization of FIB-1::mCherry.
pqe-1 affects the expression of various transgenes
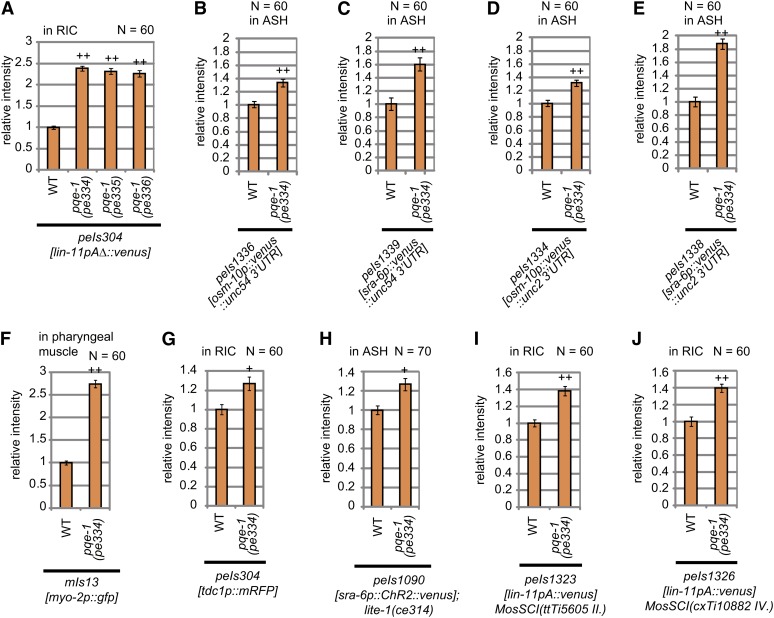
The suppression of peIs303 and peIs304 by pqe-1 overexpression in RIC suggested that pqe-1 diminishes the potential of expression induction in both RIC and AIZ (Figure 3C and Figure S4I). To confirm this, we quantified the intensity of the reporter fluorescence in RIC in the pqe-1 mutant background. As expected, we could detect a more than twofold increase in transgene expression in RIC of pqe-1 mutants (Figure 5A). The polyQ toxicity phenotype in the previous report was observed in ASH by expressing a polyQ region of huntingtin, Htn-Q150, by the promoter of osm-10 or sra-6 (Faber et al. 2002). To examine whether pqe-1 mutation also affects the reporter expression from these promoters, we produced transgenic strains with chromosomally integrated osm-10p::venus and sra-6p::venus, peIs1336 and peIs1339, respectively. Both of the transgenes showed stronger expression in ASH in the pqe-1(pe334) background (Figure 5, B and C). In this study and probably also in the previous report, the unc-54 3′UTR was used as the 3′UTR in the transgenes. To exclude the possibility that pqe-1 regulates the function of unc-54 3′UTR, we produced chromosomally integrated transgenic strains with another 3′UTR, unc-2 3′UTR. The transgenes with the 3′UTR of unc-2, peIs1334 and peIs1338, also showed increased expression in the pqe-1(pe334) background (Figure 5, D and E). We further examined the expression of a chromosomally integrated transgene, mIs13[myo-2p::gfp], which drives expression of another fluorescent reporter gene, gfp, in a non-neuronal tissue, namely the muscles in the posterior bulb of pharynx. The reporter expression by myo-2p::gfp also increased in the pqe-1(pe334) mutant background (Figure 5F, Figure S6A, Figure S6B, and Figure S6C). In addition, we examined the effect of pqe-1 mutation on the expression of mRFP, which was cloned from corals and has low sequence homology with GFP and its variants (Matz et al. 1999). The expression of mRFP from tdc1p::mRFP, which drives mRFP expression in RIM and RIC neurons and was included in peIs304 to help identification of RIC neurons, was also increased in the pqe-1(pe334) mutant background (Figure 5G). Therefore, pqe-1 affects many types of transgenes probably with no specificity of promoters, 3′UTRs, or coded genes. In addition to these integrants, we could observe increase in the expression of Channelrhodopsin-2 fused to Venus (Boyden et al. 2005), peIs1090[sra-6p::ChR2::venus], which can be used for optogenetic manipulation of ASH neurons (Figure 5H). These results suggest that pqe-1 mutations could be used as a tool for increasing the expression of useful transgenes in C. elegans.
Figure 5.
pqe-1 mutations enhance the expression of various transgenes. (A) Expression enhancement of peIs304[lin-11pAΔ::venus] in RIC neurons at L1 larva. The average fluorescence intensity in RIC neurons of each mutant is normalized by that of wild-type animals. (n = 60, Error bars represent the SEM; ++ indicates significant difference at P < 0.001 by Dunnett’s test) (B−J) Expression enhancement of transgenes in pqe-1(pe334) at L1 larva. The cells in which transgenes were expressed and fluorescence intensity was quantified are indicated at the top of the graphs. The normalized fluorescence intensity from transgenes, peIs1336[osm-10p::venus::unc-54 3′UTR], peIs1339[sra-6p::venus:unc-54 3′UTR], peIs1334[osm-10p::venus::unc-2 3′UTR], peIs1338[sra-6p::venus::unc-2 3′UTR], mIs13[myo-2p::gfp], peIs304[tdc-1p::mRFP], peIs1090[sra-6p::ChR2::venus], peIs1323, and peIs1326 are shown in (B), (C), (D), (E), (F), (G), (H), (I), and (J), respectively. The peIs304 transgene includes tdc-1p::mRFP, which induces mRFP expression in RIM and RIC, in addition to lin-11pAΔ::venus. In (G), the fluorescence intensity of mRFP was quantified. peIs1323 and peIs1326 are low copy insertions of lin-11pA::venus into the genome. (n ≥ 60, Error bars represent SEM; + and ++ show significant difference at P < 0.01, and P < 0.001, respectively, by Student’s t-test with Bonferroni’s correction.)
The chromosomally integrated transgenes used previously were formed by the insertion of multicopy tandem arrays into chromosomes. Multicopy transgenes are known to be regulated by multicopy recognition proteins. In C. elegans, several molecules, including a RING finger/B-Box factor TAM-1, a retinoblastoma-like protein LIN-35, and a bromodomain protein LEX-1, were reported to enhance the expression of multicopy transgenes (Hsieh et al. 1999; Tseng et al. 2007), although little is known about detailed mechanism of antisilencing by these regulators. Because recent progress of MosSCI techniques in C. elegans has enabled the production of low copy transgene insertions in the genome (Frøkjaer-Jensen et al. 2008), we examined whether the pqe-1 mutation also affects such low copy transgenes. We tested the effect of the pqe-1 mutation on peIs1323 and peIs1326, which were produced by the MosSCI technique (Figure S3). Even the low copy insertions showed stronger expression in the pqe-1(pe334) mutant background, indicating that the effect of PQE-1 also covers low copy transgenes produced by MosSCI (Figure 5, I and J). Altogether, 10 transgenes were tested and in all of them (10/10) the transgene expression was enhanced by the pqe-1 mutation (Figure 5).
To examine whether the enhancement of the reporter expression in pqe-1 mutants was attributable to the increase of transcripts, we performed RT-PCR assay against venus transcripts from peIs304[lin-11pAΔ::venus], peIs1336[osm-10p::venus::unc-54 3′UTR], peIs1334[osm-10p::venus::unc-2 3′UTR], peIs1338[sra-6p::venus::unc-2 3′UTR], mIs13[myo-2p::venus], and peIs1323[lin-11pA::venus](MosSCI) transgenes. As a result, we could detect a subtle increase of transcripts from peIs1338, mIs13, and peIs1323 in the pqe-1(pe334) background whereas no increase of transcripts was detected for peIs304, peIs1336, and peIs1334, even though the Venus fluorescence was increased (Figure S7A, Figure S7B, Figure S7C, Figure S7D, Figure S7E, and Figure S7F). PQE-1A and B isoforms possess an exonuclease domain with homology to REX1, RNA exonuclease homolog 1. Rex1 is known to function in the processing of 3′ ends of 5S rRNA and several tRNAs in yeast (Copela et al. 2008; Ozanick et al. 2009), suggesting that PQE-1 may also function in the regulation of components of the translation machinery. This idea is consistent with our observation that PQE-1 affects transgene expression irrespective of the promoter or the 3′UTR sequences. In this context, the increase of reporter transcripts from the peIs1338, mIs13, and peIs1323 transgenes in pqe-1 mutants may be a side effect of the increase of general translation. Alternatively, subtle increases in the transcripts, sometimes below threshold of detection, may be the primary cause of the pqe-1 defect. Notably, no increase in transcription was observed for any of the endogenous genes that we tested, suggesting that transgenes are specifically recognized by PQE-1 (Figure S7G, Figure S7H, Figure S7I, Figure S7J, and Figure S7K).
Discussion
The mechanisms of stochastic expression in AIZ
In this study, we observed two discreet states of reporter expression in AIZ neurons, denoted as AIZ-ON and AIZ-OFF. These states are apparently selected in a stochastic manner early in development and remain stable until adulthood. To explain the cause of this stochastic selection, we consider three possible mechanisms. First, this type of bistable behavior is often manifested by regulatory networks having either positive or double-negative feedback loops (Acar et al. 2005; Ferrell 2002; Mettetal et al. 2006; Ozbudak et al. 2004). The caveat is that in our case, the products of the transgenes are fluorescent proteins, which would not be expected to participate in a feedback loop. However, it is still possible that regulatory regions in the transgenes, other than the open reading frame, act in the feedback regulation. For example, if an unidentified miRNA is encoded in the composite promoter, lin-11pAΔ, it could form a feedback loop by down-regulating a putative regulator that, in turn, could negatively regulate the expression from the lin-11pAΔ promoter (Figure S8A). Second, binary transition of the gene expression unit is also possible. For example, cooperative switching in the chromatin compaction similar to the transition between heterochromatin and euchromatin can easily establish all-or-none expression states (Figure S8B). In this case, the greater potential for transcription probably results in a greater probability of the euchromatin form. Because the homologous chromosomes would be regulated independently, some fraction of cells should show intermediate reporter expression. However, we did not observe such a stepwise distribution of reporter expression (Figure 2G), making this chromatin regulation model less likely. Finally, we propose another model by epigenetic chromatin remodeling which does not accomplish all-or-none expression decision but fixes the rate of transcription at some level. Once the promoter activity is fixed, it can be translated to a quasi-binary output if a threshold effect is imposed. This threshold could be caused by a strong degradation system that degrades mRNA or protein product efficiently, but is limiting in its capacity. As a consequence, mRNA or protein product accumulates only when the rate of synthesis exceeds the degrading capacity, and above this threshold, the accumulation of the product steeply increases until it reaches the maximal capacity of the gene expression system (Figure S8C). In any case, some of the components of the regulatory system that cause binary expression of the fluorescent reporter must be operative in AIZ but absent in RIC. The regulation by pqe-1 appears to be part of the general mechanism shared by both neuronal types. Although the precise mechanisms of the peculiar characteristic of transgene expression in AIZ are still obscure, AIZ neurons are involved in multiple behaviors, and therefore the individuality of gene expression may be expressed as behavioral differences.
The function of PQE-1
In the previous report, pqe-1 was identified in a screen for mutations that enhance the toxicity of ectopic expression of the polyQ protein (Faber et al. 2002). As is the case in patients with Huntington’s disease, the expression of Htn-Q150, which is the N terminal part of mutant human huntingtin carrying an expanded polyQ repeat, in the ASH neurons caused neural death in C. elegans (Faber et al. 1999). On the basis of our study, the polyQ toxicity-enhancing phenotype in pqe-1 mutants could be caused by the enhancement of expression of Htn-Q150 in ASH neurons. The authors of the previous report observed no change in the fluorescence of the GFP reporter in pqe-1 mutants. However, a careful quantification of fluorescence intensity would have been required to reveal the effect of pqe-1 on the enhancement of transgene expression because pqe-1 mutations increase the transgene expression only by 2.5-fold at most, and the effect of pqe-1 mutation differs among integrants (Figure 5).
Our study suggested that pqe-1 regulates the expression of a wide range of transgenes, possibly through controlling the translation machinery or recognizing epigenetic markers on transgenes to suppress transcription. The rescue experiments with PQE-1 isoforms indicated that the Q/P-rich region was more important than the exonuclease domain (Figure 3, B and C). However, this Q/P-rich region is not really conserved even among the nematode species except that the domain includes high content of glutamine and proline, about 20% of glutamine and about 15% of proline (Figure S5B). PQE-1C isoform shows a dot-like localization in the nucleus (Figure 4, L−P) (Faber et al. 2002), and therefore Q/P-rich region probably induces aggregation and the aggregation may be important for controlling the function of the long isoform. Hypothesizing that the Q/P-rich region is required for the aggregation of PQE-1 for its function, it would be reasonable that rather than the amino acid sequences but the content of gluatmine and proline in Q/P-rich region are conserved among PQE-1 in related species.
The application of pqe-1 mutation to enhance the expression of transgenes in C. elegans
Because the PQE-1C isoform is strongly localized in the nucleus, it probably acts cell-autonomously as reported previously (Faber et al. 2002). We examined two candidate promoter sequences of pqe-1, pqe-1p1, and pqe-1p2, and observed the reporter expression in excretory canal, vulval cells, intestine, and many neurons. Although we did not find expression by these promoters in the pharyngeal muscle in which mIs13[myo-2p::gfp] is expressed, the pqe-1(pe334) mutation enhanced the transgene expression also in this tissue (Figure 5F and Figure S6). This discrepancy is probably caused by the insufficiency of the pqe-1 promoter regions that we used to examine the expression pattern of pqe-1; pqe-1 may be actually expressed in many other tissues, and in that case the pqe-1 mutations may have effects in a broader range of tissues than we have assumed.
The pqe-1 mutation enhanced expression of transgenes with no specificity to promoters, 3′UTRs, nor coded genes. In addition to this nonspecificity, the pqe-1 mutation can also affect low copy transgenes produced by MosSCI. Low expression level of transgenes sometimes hampers the use of transgenes in genetic manipulations. Specifically, many recently developed techniques in C. elegans such as genetic ablation (Chelur and Chalfie 2007), cell-specific knock-down (Esposito et al. 2007), and optogenetic manipulation (Nagel et al. 2005; Zhang et al. 2007) require high expression level of transgenes. We found no obvious defect in pqe-1 mutants other than enhancing the expression of transgenes, and therefore they would be a useful tool for maximizing the effect of transgenes. This strategy cannot be readily applied to other model organisms because pqe-1 is a nematode-specific gene. However, further exploration in other organisms may lead to the discovery of other genes similar to pqe-1 that also suppress general transgene expression.
Supplementary Material
Acknowledgments
We thank the Caenorhabditis Genetic Center, which is funded by National Institutes of Health National Center for Research Resources, for the C. elegans strains used in this study. We also thank members of our laboratory for useful advice and discussion. This work was supported by the Grant-in-Aid for Scientific Research (B) and Grant-in-Aid for Scientific Research on Innovative Areas “Systems Molecular Ethology” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Footnotes
Communicating editor: Brenda J. Andrews
Literature Cited
- Acar M., Becskei A., van Oudenaarden A., 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 [DOI] [PubMed] [Google Scholar]
- Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K., 2005. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018 [DOI] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C., 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805 [DOI] [PubMed] [Google Scholar]
- Chelur D. S., Chalfie M., 2007. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA 104: 2283–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela L. A., Fernandez C. F., Sherrer R. L., Wolin S. L., 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Di Schiavi E., Bergamasco C., Bazzicalupo P., 2007. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene 395: 170–176 [DOI] [PubMed] [Google Scholar]
- Faber P. W., Alter J. R., MacDonald M. E., Hart A. C., 1999. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. USA 96: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P. W., Voisine C., King D. C., Bates E. A., Hart A. C., 2002. Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc. Natl. Acad. Sci. USA 99: 17131–17136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell J. E., 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14: 140–148 [DOI] [PubMed] [Google Scholar]
- Freyd G., Kim S. K., Horvitz H. R., 1990. Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-11. Nature 344: 876–879 [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith A. D., Sarin S., Lockery S., Hobert O., 2010. Developmental control of lateralized neuron size in the nematode Caenorhabditis elegans. Neural Dev. 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., D’Alberti T., Liu Y., Ruvkun G., 1998. Control of neural development and function in a thermoregulatory network by the LIM homeobox gene lin-11. J. Neurosci. 18: 2084–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewijs D., Houthoofd K., Matthijssens F., Vandesompele J., Vanfleteren J. R., 2008. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J., Fire A., 2000. Recognition and silencing of repeated DNA. Annu. Rev. Genet. 34: 187–204 [DOI] [PubMed] [Google Scholar]
- Hsieh J., Liu J., Kostas S. A., Chang C., Sternberg P. W., et al. , 1999. The RING finger/B-box factor TAM-1 and a retinoblastoma-like protein LIN-35 modulate context-dependent gene silencing in Caenorhabditis elegans. Genes Dev. 13: 2958–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y., Yoshida K., 2009. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J. Neurosci. 29: 5370–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13–34 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics 146: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Murray A. W., Tollervey D., Plotkin J. B., 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Nott A., Moore M. J., 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28: 215–220 [DOI] [PubMed] [Google Scholar]
- Lee L. W., Lo H. W., Lo S. J., 2010. Vectors for co-expression of two genes in Caenorhabditis elegans. Gene 455: 16–21 [DOI] [PubMed] [Google Scholar]
- Matsuki M., Kunitomo H., Iino Y., 2006. Goalpha regulates olfactory adaptation by antagonizing Gqalpha-DAG signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103: 1112–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M. V., Fradkov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., et al. , 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17: 969–973 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettetal J. T., Muzzey D., Pedraza J. M., Ozbudak E. M., van Oudenaarden A., 2006. Predicting stochastic gene expression dynamics in single cells. Proc. Natl. Acad. Sci. USA 103: 7304–7309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F., Gissi C., Liuni S., Pesole G., 2002. Untranslated regions of mRNAs. Genome Biol 3: REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., et al. , 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388: 882–887 [DOI] [PubMed] [Google Scholar]
- Mori I., Ohshima Y., 1995. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature 376: 344–348 [DOI] [PubMed] [Google Scholar]
- Nagel G., Brauner M., Liewald J. F., Adeishvili N., Bamberg E., et al. , 2005. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15: 2279–2284 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., et al. , 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20: 87–90 [DOI] [PubMed] [Google Scholar]
- Nakai J., Ohkura M., Imoto K., 2001. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat. Biotechnol. 19: 137–141 [DOI] [PubMed] [Google Scholar]
- Ozanick S. G., Wang X., Costanzo M., Brost R. L., Boone C., et al. , 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak E. M., Thattai M., Lim H. N., Shraiman B. I., Van Oudenaarden A., 2004. Multistability in the lactose utilization network of Escherichia coli. Nature 427: 737–740 [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Shaw J. E., Carr S. H., Hirsh D., 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5: 3484–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Chou J. H., Dwyer N. D., Colbert H. A., Bargmann C. I., 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83: 207–218 [DOI] [PubMed] [Google Scholar]
- Tseng R. J., Armstrong K. R., Wang X., Chamberlin H. M., 2007. The bromodomain protein LEX-1 acts with TAM-1 to modulate gene expression in C. elegans. Mol. Genet. Genomics 278: 507–518 [DOI] [PubMed] [Google Scholar]
- White J. G., Horvitz H. R., Sulston J. E., 1982. Neuron differentiation in cell lineage mutants of Caenorhabditis elegans. Nature 297: 584–587 [DOI] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164 [DOI] [PubMed] [Google Scholar]
- Williams A. J., Paulson H. L., 2008. Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci. 31: 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Hirotsu T., Matsuki M., Butcher R., Tomioka M., et al. , 2010. Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science 329: 1647–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L. P., Brauner M., Liewald J. F., Kay K., et al. , 2007. Multimodal fast optical interrogation of neural circuitry. Nature 446: 633–639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.