Abstract
Genotype by environment interaction is a phenomenon that a better genotype in one environment may perform poorly in another environment. When the genotype refers to a quantitative trait locus (QTL), this phenomenon is called QTL by environment interaction, denoted by Q×E. Using a recently developed new Bayesian method and genome-wide marker information, we estimated and tested QTL main effects and Q×E interactions for a well-known barley dataset produced by the North American Barley Genome Mapping Project. This dataset contained seven quantitative traits collected from 145 doubled-haploid (DH) lines evaluated in multiple environments, which derived from a cross between two Canadian two-row barley lines, Harrington and TR306. Numerous main effects and Q×E interaction effects have been detected for all seven quantitative traits. However, main effects seem to be more important than the Q×E interaction effects for all seven traits examined. The number of main effects detected varied from 26 for the maturity trait to 75 for the heading trait, with an average of 61.86. The heading trait has the most detected effects, with a total of 98 (75 main, 29 Q×E). Among the 98 effects, 6 loci had both the main and Q×E effects. Among the total number of detected loci, on average, 78.5% of the loci show the main effects whereas 34.9% of the loci show Q×E interactions. Overall, we detected many loci with either the main or the Q×E effects, and the main effects appear to be more important than the Q×E interaction effects for all the seven traits. This means that most detected loci have a constant effect across environments. Another discovery from this analysis is that Q×E interaction occurs independently, regardless whether the locus has main effects.
Keywords: barley, Markov chain Monte Carlo, mixed model, quantitative trait locus, Q×E interaction
Genotype by environment interaction is a phenomenon that genotype effects may significantly differ across environments. When the genotype refers to a quantitative trait locus (QTL), G×E interaction is renamed as QTL by environment interaction, denoted by Q×E. The magnitude of Q×E interaction for a particular locus is measured by the deviation of the estimated QTL effect in a specific environment from the average estimated QTL effect across environments. A QTL with a small Q×E interaction is more stable than a QTL with a larger Q×E interaction. A more stable QTL has a wider inference space and can be used in plant breeding across all environments. On the contrary, an unstable QTL can only be used in the environment from which it is detected.
Q×E interactions for quantitative traits in crops across multiple environments have been ubiquitous. Boer et al. (2007) reported that most of the detected QTL controlling grain yield and grain moisture of maize were influenced by temperature differences during critical stages of the growth. Li et al. (2010) found that several QTL governing growth trajectories of soybean cause significant genotype-environment interaction for plant height. Paterson et al. (2003) showed that genetic control of cotton fiber quality was markedly affected both by general difference between growing seasons and by specific differences in water management regimes. Yang et al. (2007) found that majority of the QTL detected for nine quantitative traits of wheat interact with drought stress and well-watered conditions. As for barley, G×E or Q×E interactions have been reported by several investigators (Cherif et al. 2010; Hayes et al. 1993; Piepho 2000; Tinker et al. 1996).
Three models have been proposed for detection of Q×E interaction. The simplest method is the standard two-way ANOVA (Pillen et al. 2003; Tinker and Mather 1995), in which Q×E interaction is tested one marker at a time. A slightly more advanced method is the split-plot ANOVA (Utz and Melchinger 1996; Utz et al. 2000), in which each genotype is considered to be a main plot, and observations from different environments play the role of split-plots. The common practice for Q×E analysis is to conduct interval mapping (Lander and Botstein 1989) or composite interval mapping (Zeng 1994) for each environment separately and then to compare the QTL mapping results for these environments (Li et al. 2007). If a QTL is detected in some environments but not in others, Q×E interaction is implied. This approach is subject to the same drawback as the ANOVA in terms of ignoring the covariance of the residuals because data are analyzed one environment at a time. Multivariate repeated measurement analysis is a more advanced method of Q×E analysis. Phenotypes of the same trait measured in different environments are treated as “different traits.” This multivariate approach to QTL mapping was first proposed by Jiang and Zeng (1995). The advantage of this method is that the variance-covariance matrix of the residuals can be incorporated into the model. So far, only unstructured covariance matrix (full positive definite matrix) has been considered under the multivariate QTL mapping. Recently, Piepho (2005) proposed a mixed-model approach to modeling highly structured covariance matrix and showed that the mixed-model approach outperforms all the methods described above.
Chen et al. (2010) recently developed a new Bayesian shrinkage method for estimating and testing Q×E interactions. This method estimates all main effects and Q×E interactions simultaneously in a single model. When the number of environments is large, Chen et al. (2010) used the variance of the estimated QTL effects across multiple environments as a measure of the Q×E interaction. This approach has tremendously simplified Q×E studies. In addition, they incorporated the permutation analysis into the Q×E study and provided a significance test for each effect. Chen et al. (2010) used the yield trait of barley as an example to demonstrate the application of the new method. Numerous main and Q×E study interaction effects were detected for that trait. This well-known barley data set was developed by the North American Barley Genome Mapping Project (Tinker et al. 1996). In the barley experiment, the investigators measured a total of seven quantitative traits from multiple environments and a total of 127 mapped markers that cover 1500 cM of the barley genome. Incorporating all 127 markers into a single model, Chen et al. (2010) only analyzed one of these traits. To capture the whole-genome marker information, we inserted a pseudo marker every 5 cM for a marker interval greater than 5 cM, and the total number of loci was 304. Then we analyzed all seven traits with 304 markers using the new method to draw general conclusions and reported the results in this study. As the genome coverage of the markers was quite high, no QTL would be missed. A general conclusion about the relative importance of Q×E in these quantitative traits is drawn, which can help us understand the genetic architecture of the complex quantitative traits to guide practical breeding programs of barley aimed at improving crop production in difficult environments.
MATERIALS AND METHODS
Experimental data
The data were retrieved from the North American Barley Genome Mapping Project (NABGMP) website (http://gnome.agrenv.mcgill.ca/). The experimental design and results were reported by Tinker et al. (1996). For the article to be self-contained, we briefly summarized the experiment here. This experiment consisted of a doubled haploid population with lines derived from a cross between two Canadian two-row barley lines, Harrington and TR306. Seven traits were investigated in the project: yield, heading, maturity, height, lodging, kernel weight, and test weight. The 145 lines were grown in a range of environments: yield was measured in 28 environments, heading in 29, maturity in 15, height in 27, lodging in 17, kernel weight in 25, and test weight in 28 environments. A total of 127 mapped markers covered 1500 cM of the barley genome (seven chromosomes). The genotype of each marker was coded as −1 for the TR306 allele and +1 for the Harrington allele. A missing genotype was coded as 0. The data contained about ∼4.9% missing marker genotypes and a few missing phenotypes.
We inserted a pseudo marker every 5 cM of the genome for a marker interval greater than 5 cM. Including the inserted pseudo markers, the total number of loci subject to analysis was 304. Missing marker and pseudo marker genotypes were inferred from flanking marker information using the multipoint method (Jiang and Zeng 1997). For example, the genotype indicator variable for line j at marker k is defined as
| (1) |
Let be the conditional probability of inferred from all markers of the current chromosome. The conditional expectation for a missing is
| (2) |
which will replace if is missing. It should be noted that each marker (true or pseudo) will be treated as a putative QTL in this research, i.e., only the effects of markers (true and pseudo) will be estimated. If a QTL is located between two markers, its effect would be picked up by the two flanking markers. Hereafter, we use markers (including true and pseudo markers) and putative QTL interchangeably.
The missing phenotype for line j in the ith environment was drawn from the normal distribution with mean and variance , where is the average of phenotypes of all lines in the ith environment and is the variance of phenotypes across lines in the ith environment. The seven traits were analyzed separately in seven analyses. The numbers of environments whose data contribute to the analysis for all traits are given in Table 2.
Table 2. Summary statistics for the seven agronomic traits of barley in the Harrington×TR306 double haploid population using Bayesian mapping.
| Trait | ||||||||
|---|---|---|---|---|---|---|---|---|
| Height | Heading | Kernel Weight | Lodging | Maturity | Test Weight | Yield | Average | |
| 27 | 29 | 25 | 17 | 15 | 28 | 28 | 24.12 | |
| 72 | 75 | 51 | 73 | 26 | 73 | 63 | 61.86 | |
| 4 | 29 | 23 | 43 | 22 | 35 | 31 | 26.71 | |
| 4 | 6 | 10 | 22 | 7 | 13 | 10 | 10.29 | |
| 72 | 98 | 64 | 94 | 41 | 95 | 84 | 78.28 | |
| 1.0000 | 0.7653 | 0.7969 | 0.7766 | 0.6341 | 0.7684 | 0.7500 | 0.7845 | |
| 0.0556 | 0.2959 | 0.3594 | 0.4574 | 0.5366 | 0.3684 | 0.3690 | 0.3489 | |
| 0.2368 | 0.2664 | 0.1678 | 0.2401 | 0.0855 | 0.2401 | 0.2072 | 0.2063 | |
| 0.0132 | 0.0954 | 0.0757 | 0.1414 | 0.0724 | 0.1151 | 0.1020 | 0.0879 | |
| 0.2368 | 0.3224 | 0.2105 | 0.3092 | 0.1349 | 0.3125 | 0.2763 | 0.2575 | |
| 2.23 | 0.74 | 2.02 | 2.77 | 0.4493 | 1.27 | 13.61 | 3.2985 | |
| 0.60 | 0.30 | 0.81 | 9.4343 | 0.06 | 0.90 | 171.88 | 26.28 | |
, number of environments; ,number of main effects; ,number of Q×E effects; , total number of effects; , number of both effects (main and Q×E effects); , proportion of the number of main effect over the total number of effects; , proportion of the number of Q×E effects over the total number of effects; , proportion of the number of main effects over the total number of loci (pseudo and true markers) (); , proportion of the number of Q×E effects over the total number of loci;, proportion of the number of effects over the total number of loci; , the absolutely value of the largest main effect; , the absolute value of the largest Q×E effect.
Statistical method
Let be the phenotypic values for line j collected in m environments. The multivariate model for is
| (3) |
where is a vector of intercepts, is a numerically coded genotypic indicator variable for line j at locus k, for k = 1,…, p, where is the number of markers, including true and pseudo markers, is an m×1 vector of QTL effects for the m environments and the residual error vector is assumed to be multivariate normal with density , where θ is an m×m variance –covariance matrix. The variance- covariance structures of residual errors considered are identity, diagonal and factor analytic structured matrices.
For locus k, the mean of the marker effects cross the environments represents the main effects and the variance of the marker effects represents the interaction. We used
| (4) |
to represent the uniformity of the QTL effects cross environments. When there is no interaction, we expect that and thus . Since the method is a Bayesian method, prior and posterior distributions of parameters are required in the analysis. These distributions have been described in detail by Chen et al. (2010) and thus are not provided here in this report.
MCMC sampling
The MCMC-implemented Bayesian analysis developed by Chen et al. (2010) was used for parameter estimation. The overall Markov chain contained 160,000 iterations. The first 40,000 iterations were deleted from the post MCMC analysis (burn-in deletion). Thereafter, the observations were saved in every 40 iterations to reduce the autocorrelation among different observations of the posterior samples. Therefore, the final posterior sample for analysis contained 3000 observations. Posterior means and other posterior statistics were obtained from the posterior sample of the 3000 observations.
Model selection
We used five different variance-covariance structures to analyze the seven traits, which were (1) the homogeneous variance , (2) the heterogeneous variance , where is a diagonal matrix with each diagonal element representing the residual error variance for the trait in that environment, (3) the first-order factor analytic structure , where B is factor loading, (4) the second-order factor analytic structure , and (5) the third-order factor analytic structure . In order to select the optimal covariance structures for fitting the each trait, we used the Bayesian information criteria (BIC) to evaluate the performance of the five models. The BIC score was calculated using
| (5) |
where L is the likelihood function, p is the number of parameters, and n is the sample size. The BIC scores for the seven traits are shown in Table 1. As seen in Table 1, the minimum BIC scores of the lodging and maturity traits are under the first-order factor scenario and those of the remaining traits with the heterogeneous residual variance structure model reach the minimum values. These indicate that the first-order factor analytic model outperforms the other models for lodging and maturity traits and the heterogeneous residual variance structure model for the remaining traits. Therefore, we will analyze the lodging and maturity traits using the first-order factor analytic model and the remaining traits with heterogeneous residual variance structure model in the subsequent studies.
Table 1. BIC scores of the five variance-covariance structures for the seven agronomic traits of barley in the Harrington×TR306 double haploid population.
| Covariance Structure | BIC | ||||||
|---|---|---|---|---|---|---|---|
| Height | Heading | Kernel Weight | Lodging | Maturity | Test weight | Yield | |
| Homogeneous | 20149.77 | 10284.11 | 15294.42 | 17855.92 | 7546.815 | 13379.04 | 38325.64 |
| Heterogeneous | 19298.47 | 9915.423 | 14940.67 | 16969.93 | 6613.732 | 12531.28 | 36940.23 |
| First-order factor | 19343.48 | 10081.89 | 14984.32 | 16890.66 | 6447.124 | 13059.46 | 37035.67 |
| Second-order factor | 19371.16 | 10289.86 | 15237.74 | 16905.9 | 6914.439 | 13630.45 | 37285.38 |
| Third-order factor | 19388.94 | 10289.82 | 15193.52 | 16925.5 | 6488.625 | 13144.99 | 37173.47 |
Permutation test
To test the significance of the QTL effects, we conducted a permutation test to generate the null distribution of each main effect and each Q×E interaction. In the permutation analysis, we repeated the MCMC sampling method but randomly shuffled the phenotypes to artificially destroy the association between the markers and the phenotypes. This permutation test for Bayesian shrinkage analysis was proposed by Che and Xu (2010). We adopted this permutation analysis to draw the critical values for significance tests. The number of iterations for the permutation analysis was the same as that described for the original data analysis. The posterior distribution for each and was inferred from the permuted posterior samples to draw the null distribution. An actual estimate of beyond the 0.5% and 99.5% interval indicates significant main effect while an actual estimate of larger than the 99% of the null distribution indicates significant interaction.
RESULTS
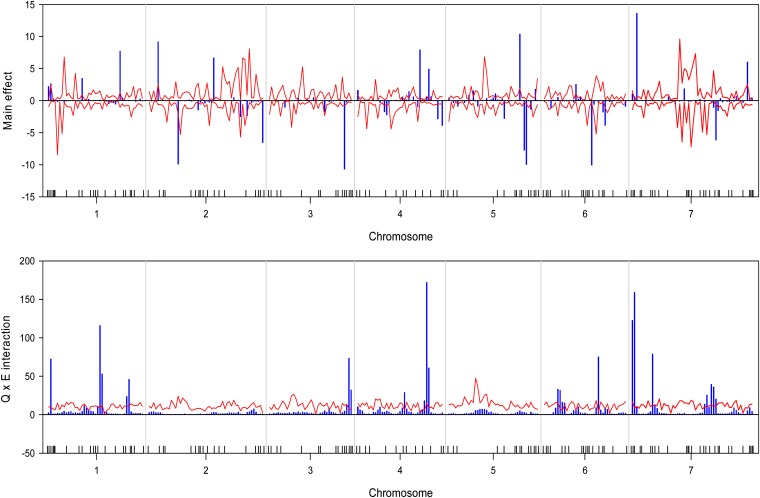
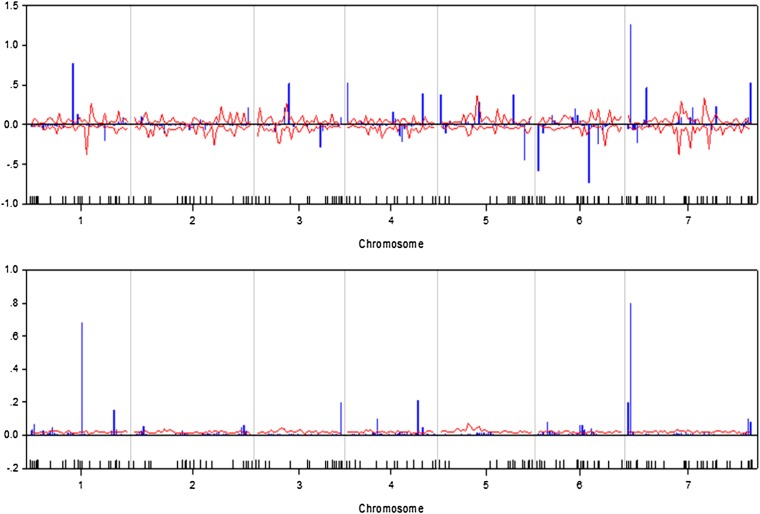
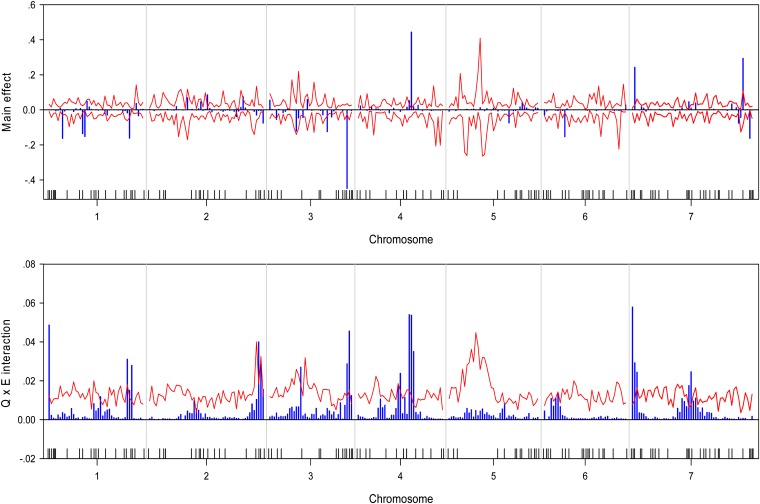
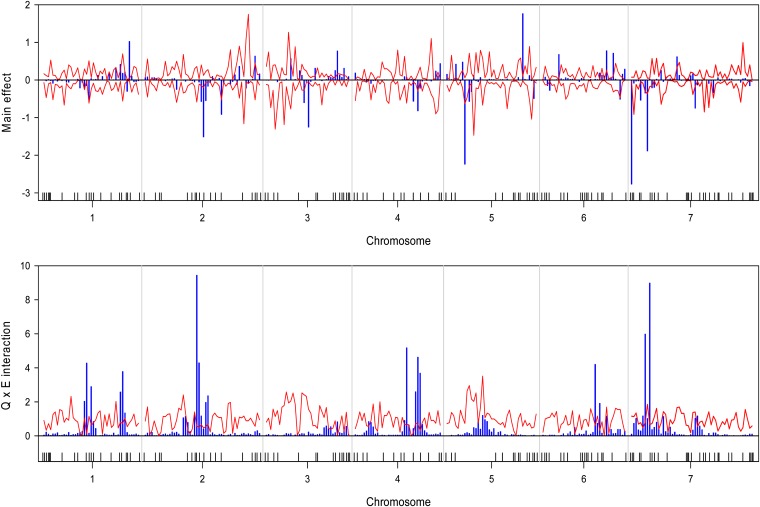
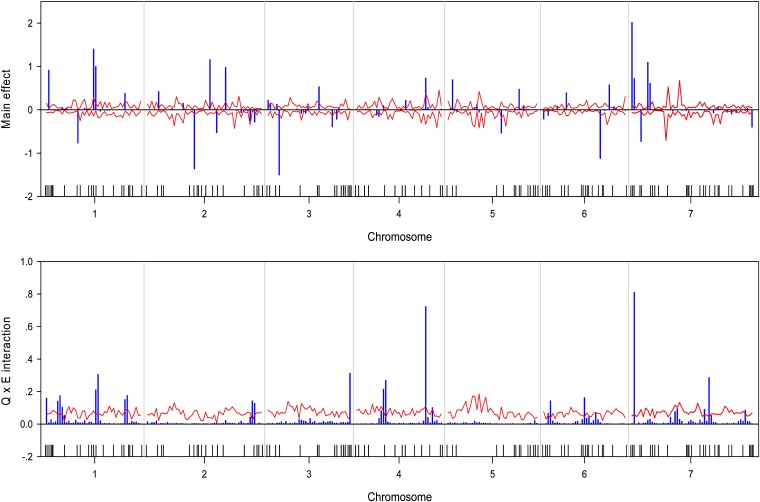
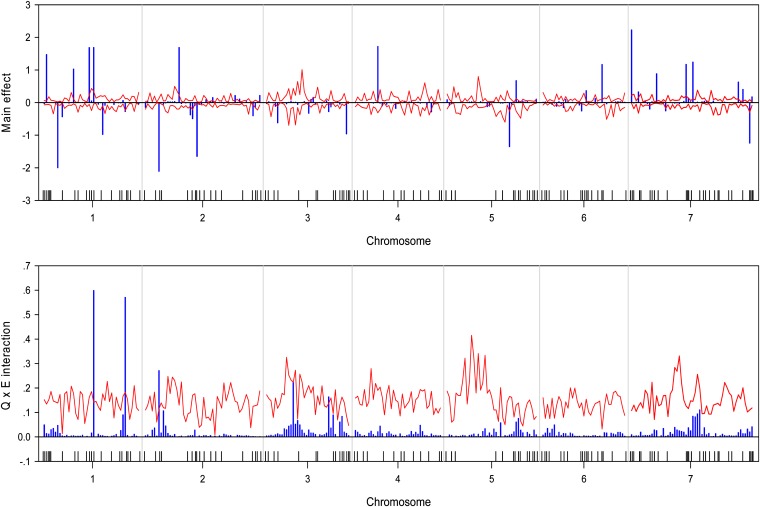
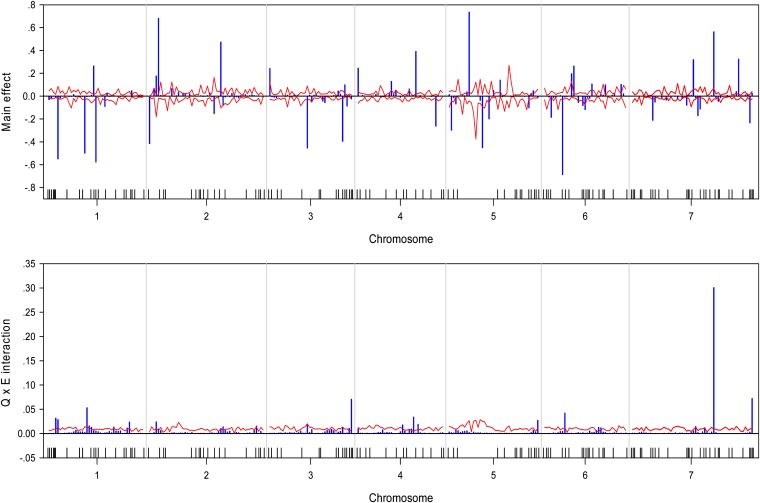
We wrote a SAS/IML program to implement the Bayesian analysis. The program took about 36∼54 CPU hours to complete the entire analysis for each trait on a Pentium PC with a 3.6-GHz processor and 3.00 GB RAM. The estimated QTL effect profiles, including the main effect and interaction, are depicted in Figure 1 for the trait yield. The figure also includes the 99% confidence interval profiles of the null distribution generated from the permutation analysis. The estimated QTL effect profiles for the remaining six traits are shown in Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, and Figure 7. QTL main and interaction effects have been detected for each of the seven traits. However, no regions of the genome show evidence of a locus affecting all traits. One region near the end of the plus arm of chromosome 7 affected six traits (excluding heading), but the magnitudes of effects varied across traits. The results of this analysis are summarized in Table 1. The number of main effects varied from 26 (maturity) to 75 (heading) with an average of 61.86. Apparently, heading has the most detected effects with a total of 98 effects (75 main and 29 Q×E effects). Among the 98 effects, 6 loci had both the main and Q×E effects. Among the total number of detected loci, on average, 78.5% of the loci show the main effects, and 34.9% of the loci show Q×E interaction. This means that main effects contribute more of the genetic variance than the Q×E interaction effects. Other summary statistics can be found in the last column of Table 2. Overall, we detected many loci with either the main or the Q×E effects and the main effects appear to be more important than the Q×E interaction for all the seven traits. This means that most detected loci have a constant effect across environments. Another discovery from this analysis is that Q×E interaction occurs independently, regardless whether the locus has main effect or not. Traditional analysis that evaluates Q×E only on those loci that show significant main effects should be abandoned because that approach will fail to detect a large number of Q×E effects that show no main effects. Table 2 and Figures 1–7 give the summary of the discovery of this study. Detailed information regarding the actual estimated effects and locations of the significance loci are provided in the tables in File S1.
Figure 1.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the yield trait. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 2.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the test weight trait. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 3.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the maturity trait. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 4.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the lodging trait. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 5.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the kernel weight. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 6.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for plant height. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Figure 7.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the date of heading. Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects and the curves (in red) represent the 99% confidence intervals generated from a permutation analysis. The ticks on the horizontal axis indicate the marker positions.
Responding to a reviewer’s comment, we also used the mixed-model approach of Piepho (2005) to analyze these data for comparison. This method is single-point analysis. The linear model is
| (6) |
where is the general mean at the kth locus, is the QTL main effect at the kth locus (treated as fixed effect), is the mth environmental main effect at the kth locus (treated as random), is the interaction effect in the mth environment at the kth locus (treated as random).
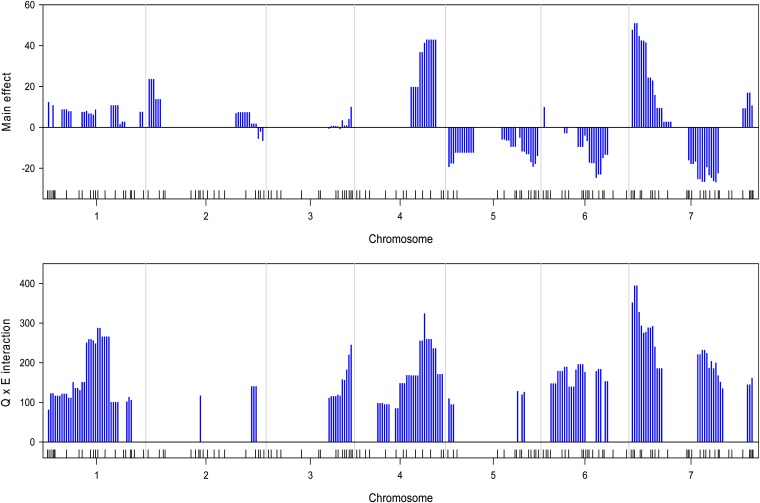
The mixed-model approach of Piepho (2005) was implemented using the MIXED procedure of the SAS system (SAS Institute, Cary, NC). As an example, we only presented in detail the result of the trait named yield. The estimated QTL main effects and Q×E interaction effects are depicted in Figure 8, where the top panel gives the estimated QTL main effects and the bottom panel shows the estimated Q×E interaction effects. This approach validated most of loci with QTL and Q×E interaction effects detected by the Bayesian analysis except a few places where the Bayesian analysis had extra peaks. For example, the two QTL with the effects in opposite directions in the middle of chromosome 2 detected by the Bayesian analysis disappeared in the mixed-model approach of Piepho (2005). Moreover, the magnitudes of the estimated QTL main effects and Q×E interaction effects are larger than those estimated by the Bayesian analysis. This comparison showed that the Bayesian analysis may be advantageous over the mixed-model approach. Note that this study is to report the Bayesian analysis of the barley data, not for the purpose of comparing different methods.
Figure 8.
Estimated main QTL effects (upper panel) and Q×E interaction effects (lower panel) across the barley genome for the yield trait using the mixed method approach of Piepho (2005). Chromosomes are separated by the vertical reference lines. The needles (in blue) represent the effects.
DISCUSSION
We used the new Bayesian method developed by Chen et al. (2010) for Q×E interaction. In the original study, the authors used one trait (yield) as an example to demonstrate the method and test the computer program. In this study, we emphasize the application of this new method to Q×E detection for all seven traits measured in the experiment. This is an application study using an advanced statistical method. The 8 main effects and 18 Q×E interaction effects detected by Chen et al. (2010) for yield were all validated by this study. Furthermore, we detected more markers with main and Q×E interaction effects. The difference between the original study of Chen et al. (2010) and this study deserves further discussion. Chen et al. (2010) used 127 markers (all true markers), and they only estimated 127 main effects and 127 Q×E interaction effects. In this study, we used 304 markers (127 true markers and 177 pseudo markers) and estimated 304 main effects and 304 Q×E interaction effects. The additional effects we detected were largely contributed by the inserted pseudo markers. For example, many additional peaks that appeared in this study coincided with pseudo markers in large marker intervals. It was impossible for Chen et al. (2010) to detect these additional effects because they only analyzed the 127 true markers, not the 177 pseudo markers. Comparison of the yield trait between this study and that of Chen et al. (2010) clearly demonstrated the advantage of the pseudo marker insertion approach.
In the original barley QTL mapping experiment, the authors (Tinker et al. 1996) provided some preliminary results about Q×E interactions using the interval mapping (Lander and Botstein 1989) and composite interval mapping approaches (Zeng 1994). The difference between the interval mapping and the Bayesian mapping in the context of Q×E is that the Bayesian method uses a multivariate (multiple environments in this case) and multiple QTL model. All parameters and significance tests are performed simultaneously within the same model. Advantages of the simultaneous analysis over the one-environment-and-one-marker analysis are 3-fold: (a) conclusions are drawn from the result of the same analysis, rather than from separate analyses; (b) significance tests are conducted simultaneously in one experiment, and thus no adjustment is required regarding the experiment-wise Type I error rate; and (c) more effects can be detected using the joint analysis. We examined the Q×E interaction effects detected from the ad hoc interval mapping studies by the original authors of the barley experiment (Tinker et al. 1996) and found that almost all effects detected by Tinker et al. (1996) were also detected in this study. The comparison is summarized in Table 3. Note that the numbers of QTL main effects and Q×E effects were counted directly from Figure 2 of Tinker et al. (1996). In addition, we detected more Q×E effects (see the tables in File S1 for detailed information about the results).
Table 3. The numbers of QTL and Q×E interactions for the seven agronomic traits of barley in the Harrington×TR306 double haploid population detected by the interval mapping approach and validated by the Bayesian approach.
| Trait | Main Effect | Q×E Interaction Effect | |||
|---|---|---|---|---|---|
| Height | 6 | 6 | 5 | 2 | |
| Heading | 10 | 10 | 20 | 20 | |
| Kernel weight | 6 | 6 | 18 | 16 | |
| Lodging | 6 | 6 | 10 | 9 | |
| Maturity | 4 | 4 | 5 | 4 | |
| Test weight | 6 | 6 | 16 | 16 | |
| Yield | 10 | 10 | 20 | 18 | |
, number of QTL main effects detected using interval mapping by Tinker et al. (1996); , number of main effects validated using Bayesian mapping; , number of Q×E interaction effects detected using interval mapping by Tinker et al. (1996); , number of Q×E interaction effects validated using Bayesian mapping.
In contrast to common Q×E studies (Boer et al. 2007; Cherif et al. 2010; Hayes et al. 1993; Jansen et al. 1995; Li et al. 2007, 2010; Paterson et al. 2003; Piepho 2000; Tinker and Mather 1995; Tinker et al. 1996; Yang et al. 2007) in which QTL effects from each environment are estimated and tested, here we used the variance (a single value) of the QTL effects across all environments as a measurement of Q×E interaction. The test is now based on a single value of variance for each locus. This automatically converts the traditional fixed-model analysis into a random model analysis because we are now estimating and testing a variance component. This random model treatment has eliminated many problems related to the fixed-model analysis. For example, there is little concern about model saturation (overfitting) under the random model framework. When the number of environments increases, the number of parameters to be tested remains the same because there is still only one variance per locus.
In this research, we used the permutation tests proposed by Chen et al. (2010) for drawing the empirical critical values of the test statistics to decide the significance of the detected QTL effects. In theory, significance test are not required in Bayesian analysis. However, the purpose of this study was to evaluate the genome-wide QTL main effects and Q×E interaction effects. We expected that some estimated QTL main effects and Q×E interaction effects might be detected by chance and thus wanted to exclude the noisy effects. In addition, the Bayesian mapping of Chen et al. (2010) was developed based on a hierarchical model and it is a hybrid method between the Bayesian and the frequentist approaches. The asymptotic theory derived based on the frequentist approach may not apply to the hierarchical model. Therefore, we employed the permutation test to separate the true QTL from the spurious QTL.
Understanding the relative importance of Q×E interaction is useful in plant breeding. The way we formulated the Q×E interaction is unique because it is measured as a variance component. The variance across environments reflects the stability of a QTL, in which a larger variance indicates a lower stability of the QTL. From the breeding point of view, understanding Q×E interaction is important in marker-assisted selection and breeding. A QTL with less or no Q×E interaction can be utilized in a broad range of conditions, whereas QTL with significant Q×E interaction can only be used in the specific environment in which it is detected.
Supplementary Material
Acknowledgments
We greatly thank two anonymous reviewers for their useful comments and suggestions on an earlier version of the manuscript. This project was supported by the Agriculture and Food Research Initiative of U.S.D.A. National Institute of Food and Agriculture 2007-02784 (to S.X.).
Footnotes
Communicating editor: B. S. Yandell
Literature Cited
- Boer M. P., Wright D., Feng L., Podlich D. W., Luo L., et al. , 2007. A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177: 1801–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X., Xu S., 2010. Significance test and genome selection in Bayesian shrinkage analysis. Int. J. Plant Genomics 2010: Article ID 893206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao F., Xu S., 2010. Mapping environment specific quantitative trait loci. Genetics 186: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherif M., Rezgui S., Devaux P., Harrabi M., 2010. Genotype × environment interactions and heritability of quantitative resistance to net blotch in Tunisian barley. Journal of Plant Breeding and Crop Science 2: 110–116 [Google Scholar]
- Hayes P., Liu B., Knapp S., Chen F., Jones B., et al. , 1993. Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor. Appl. Genet. 87: 392–401 [DOI] [PubMed] [Google Scholar]
- Jansen R., Ooijen J., Stam P., Lister C., Dean C., 1995. Genotype-by-environment interaction in genetic mapping of multiple quantitative trait loci. Theor. Appl. Genet. 91: 33–37 [DOI] [PubMed] [Google Scholar]
- Jiang C., Zeng Z. B., 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Zeng Z. B., 1997. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica 101: 47–58 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Huang Z., Xu M., Wang C., Gai J., et al. , 2010. Functional mapping of genotype-environment interactions for soybean growth by a semiparametric approach. Plant Methods 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu C., Jiang G., Wang L., He Y., 2007. Dynamic analyses of rice blast resistance for the assessment of genetic and environmental effects. Plant Breed. 126: 541–547 [Google Scholar]
- Paterson A., Saranga Y., Menz M., Jiang C. X., Wright R., 2003. QTL analysis of genotype× environment interactions affecting cotton fiber quality. Theor. Appl. Genet. 106: 384–396 [DOI] [PubMed] [Google Scholar]
- Piepho H. P., 2000. A mixed-model approach to mapping quantitative trait loci in barley on the basis of multiple environment data. Genetics 156: 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepho H. P., 2005. Statistical tests for QTL and QTL-by-environment effects in segregating populations derived from line crosses. Theor. Appl. Genet. 110: 561–566 [DOI] [PubMed] [Google Scholar]
- Pillen K., Zacharias A., Leon J., 2003. Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theor. Appl. Genet. 107: 340–352 [DOI] [PubMed] [Google Scholar]
- Tinker N., Mather D., 1995. Methods for QTL analysis with progeny replicated in multiple environments. Journal of Agricultural Genomics 1. Available at: http://wheat.pw.usda.gov/jag/papers95/paper195/jqtl15.html
- Tinker N., Mather D., Rossnagel B., Kasha K., Kleinhofs A., et al. , 1996. Regions of the genome that affect agronomic performance in two-row barley. Crop Sci. 36: 1053–1062 [Google Scholar]
- Utz H., Melchinger A., 1996. PLABQTL: a program for composite interval mapping of QTL. Journal of Agricultural Genomics. Available at: http://wheat.pw.usda.gov/jag/papers96/paper196/indexp196.html [Google Scholar]
- Utz H. F., Melchinger A. E., Schon C. C., 2000. Bias and sampling error of the estimated proportion of genotypic variance explained by quantitative trait loci determined from experimental data in maize using cross validation and validation with independent samples. Genetics 154: 1839–1849 [PMC free article] [PubMed] [Google Scholar]
- Yang D. L., Jing R. L., Chang X. P., Li W., 2007. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Triticum aestivum L.) stems. Genetics 176: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.