Abstract
It is well appreciated that the evolutionary divergence of genes and genomes from a common ancestor ultimately leads to incompatibilities if those genomes are hybridized. Far less is known about the ability and nature of compensatory evolution to yield the recovery of function in hybrid genomes. Here the major capsid gene of the bacteriophage T7 (40-kb dsDNA) was replaced with the homologous gene of either T3 or K11, each 22% different at the protein level from the T7 homolog. Initial fitness was moderately impaired for the T3 exchange, but the K11 exchange was not viable without a compensatory change in the T7 scaffolding protein. Subsequent adaptation of the transgenic phages led to nearly complete fitness recoveries. Compensatory changes were few, mostly in the transgene and its main interacting partner, the scaffolding protein gene. The large magnitude of fitness recovery with relatively few mutations suggests that the fitness costs of hybridizations and horizontal gene exchanges between moderately diverged genomes can potentially be short-lived through compensatory evolution.
Keywords: genome, experimental evolution, adaptation, compensatory evolution, incompatibility, engineering
Divergence among lineages sharing a common ancestor is a defining property of evolution. However, the integrity of diverging lineages also depends on their eventual genetic incompatibility—speciation—whereby the genome of one lineage is no longer able to recombine and function in the genome of the other (Coyne and Orr 2004). Despite the overwhelming complexity of interactions occurring within genomes and the inherent difficulties in identifying which interactions hinder fitness, there has been considerable progress in understanding the early stages of these incompatibilities, both as individual cases of speciation in eukaryotes (Presgraves 2010) and in hybrid swarms of related viruses within hosts (Martin et al. 2011).
The flip side of this coin is that gene exchange has been rampant in evolution (PubMed lists more than 5000 references to “horizontal gene exchange”). Furthermore, gene exchanges are not detectable unless there has been considerable divergence; therefore, the detected cases are biased toward the extremes. Thus, we have an apparent contradiction in that gene divergence leads to genomic incompatibility yet successful gene exchange is nonetheless common. One solution is that the successful exchanges represent the tiny minority that do not create incompatibilities.
Alternatively, initial genomic incompatibilities need not be insurmountable. Once formed and provided that they are not fully sterile, hybrids can evolve to ameliorate and reverse the effects of genomic incompatibilities. Not surprisingly, there has been much less progress in understanding the evolutionary reversal of hybrid genomic incompatibility than in understanding the bases of the incompatibilities themselves. Reversals of incompatibilities may indeed be rare in nature, but it is at least important to know the evolutionary possibilities of reversal.
The same questions are relevant to synthetic biology. The creation of new genomes often involves combing parts from different genomes. A big challenge to combining parts is compatibility: functions of a genomic element that depend on interactions with other elements may not be preserved in a divergent genomic background. Understanding how to avoid and reduce the deleterious interactions among heterologous elements should enhance the success of synthesized genomes.
In this article, we explore evolutionary recovery in a recombinant, engineered viral genome (phage T7). The capsid gene, an essential structural gene, was replaced by its homolog from either of two relatives, diverged 22% at the protein level. The transgene was inserted so that its mRNA expression remained the same as in the intact, native phage. Thus, any deleterious effect of the transgene likely stems from protein−protein interactions rather than regulation. Initial fitness was depressed, the transgenic phage evolved to recover fitness, and the genetic changes associated with the recovery identified. The results suggest that rapid evolutionary recovery is possible with few compensatory changes.
The Model System: T7 and Its Relatives
T7 is a member of the Podoviridae, a group of Gram-negative bacterial viruses characterized by short, stubby tails. One genus of the Podoviridae includes viruses with intermediate sized dsDNA genomes (~40 kb) that encode a phage-specific RNA polymerase along with the more typical DNA metabolism and structural genes. A member of this genus, bacteriophage T7, has nearly 60 genes with little sequence overlap (Molineux 2006). Its most highly expressed gene (gene 10) encodes the capsid protein that interacts directly with at least two other proteins in the T7 protein network (see below). Four hundred fifteen copies of capsid protein are present in each capsid shell, making it the main component of the protein coat protecting the viral genome.
The capsid gene of T7 is translated in two forms, the major capsid,10A, and the minor capsid, 10B (henceforth we refer to the protein products of genes as gp #, where # is the gene number). The major capsid contains 345 amino acids (AA) and is essential. The minor capsid is nonessential and consists of the first 342 AA of major capsid plus a 56 AA C-terminal extension caused by ribosomal frameshifting near the 3′end of gene 10A that allows occasional read through of a stop codon. Minor capsid is produced in lower abundance (~5%) than major capsid, and abolition of the sequences unique to gene 10B has no detectable phenotype (Condron et al. 1991).
Here the full gene 10A/B sequence in T7 was replaced with its counterpart homologs from a T7 relative, either T3 or K11. These three phages are all members of the same genus, and all encode both major and minor forms of the capsid protein. For gp10A, AA identities are 78.6% for T7-T3, 77.4% T7-K11, and 81.7% T3-K11; for the complete gp10B, identities are 65.3%, 66.4%, and 68.3%, respectively; there is little conservation of AA sequence in the C-terminal extensions. The similar level of divergence between the two donor genes and between them and T7 means that the T3 and K11 capsid genes are evolutionarily independent replicates in this transgenic experiment.
Perhaps equally important to incompatibility as the dissimilarity of the transgenes themselves is the dissimilarity of the proteins with which they interact. If the interacting partners are fully conserved, any divergence of the transgene could be of little consequence. Although a pseudo-atomic structure is available for the T7 capsid protein (Ionel et al. 2011), reconstructions of procapsid and mature particles have been made from cryo-electron microscopic studies of T7 (Agirrezabala et al. 2007) and also from a marine relative P-SSP7 (Liu et al. 2010). The capsid protein is known to interact directly with the scaffold and portal proteins and possibly also contacts three to five other proteins. Interaction between the scaffold and capsid proteins is essential to the formation of capsid like structures.
The AA similarity among the scaffolding proteins in T7, T3, and K11 could highlight the degree to which this interaction is conserved. AA identities between the ~310 AA scaffolding proteins of the three phages are lower than for the capsid protein: 61% (T7-T3), 56% (T7-K11), and 58% (T3-K11). Thus, the transgenic capsid protein may exhibit structural defects in interacting with the T7 scaffold protein. The portal protein provides the only other well-known and understood direct interaction with the capsid protein. In the prohead, the dodecameric portal protein is inserted into one vertex of the gp10 icosahedron (Molineux 2006). Although this interaction is of importance, the slightly greater similarity in AA sequence of the three phages portal proteins [85% (T7-T3), 80% (T7-K11), 80% (T3-K11)] suggests that this interaction may be well conserved. On the basis of a comparison of AA similarity among the proteins, compensatory mutations in the portal gene might be less likely to evolve relative to those in the capsid or scaffold.
Methods
Strains
The bacterial and phage strains are described in Table 1. All are from the collection of I.J.M. or J.J.B.
Table 1. Bacteria, phage, and plasmids.
| Notation | Species/Type | Genotype | Purpose |
|---|---|---|---|
| IJ1133 | Escherichia coli | K-12 ΔlacX74 thi Δ(mcrC-mrr)102::Tn10 | Creation, adaptation and fitness assays for transgenic T7 phage |
| IJ511 | Escherichia coli | K-12 ΔlacX74 supE44 galK2 galT22 mcrA rfbD1 mcrB1 hsdS3 | Host for expression plasmids |
| T761 | Phage | A wild-type T7 adapted to high fitness on IJ1133 (Heineman et al. 2005) | Parent of T7 Δ10 |
| T3 | Phage | Wild-type (Genbank no. AJ318471) | Source of T3 gene 10 |
| K11 | Phage | Wild-type (Genbank no. NC011043) | Source of K11 gene 10 |
| T7 Δ10 | Phage | T761 with deletion of bp 22967-24162 | Parent of T7 transgenic phages with T3 or K11 gene 10 |
| T7-10(T3) | Phage | T761 with its gene 10 replaced by the T3 homolog | Focus of this study; used for adaptation |
| T7-10(K11) | Phage | T761 with its gene 10 replaced by the K11 homolog | Focus of this study; used for adaptation |
| pRT3 | Plasmid | T7 bp 22717-22966, followed by T3 bp 20891-22191, then T7 bp 24163-24432 inserted into pCRBlunt | Creation of T7 transgenic phages with T3 gene 10 |
| pRK11 | Plasmid | T7 bp 22618-22966, followed by 39 bp,a then K11 bp 21475-22784 and T7 bp 24163-24432 inserted into pCRBlunt | Creation of T7 transgenic phages with K11 gene 10 |
| pRK11-2 | Plasmid | T7 bp 22618-22966, followed by K11 bp 21475-21613 | Correction of initial transgenic T7-K11 phage with additional sequence between T7 promoter and K11 gene10 |
| pSW5 | Plasmid | A pUC derivative with T3 gp10A expression under control of T7 promoter | Growth of T7 Δ10 and EOP assay |
| pSWK11 | Plasmid | pSW5 with HindIII-EcoRI fragment, replaced with K11 bp 21450-22784 (K11 gene 10) | EOP assay, testing effect of scaffold mutation |
39 bp of what appears to be primer sequence was unintentionally a part of fragment insert.
Expression plasmids
Transgenic T7 phages were created via an intermediate T7 genome deleted for its major capsid gene (T7 Δ10). Because gene 10 is essential, a complementing plasmid that does not recombine with T7 Δ10, pSW5, was routinely used for propagation of T7 Δ10 (Condreay et al. 1989). A similar complementing plasmid replaced the ribosome binding site and coding sequences of T3 gene 10 with those of K11. Expression plasmids were maintained in IJ511.
Media
All adaptations and fitness assays used LB broth (10 g of NaCl, 10 g of Bacto Tryptone, 5 g of Bacto yeast extract per liter). Plates used 15 g of Bacto agar per liter LB broth and, when growing phages, were overlaid with soft agar (7 g of Bacto agar per liter LB broth).
Passage conditions and fitness assays
Adaptations were conducted at 37° using Escherichia coli host IJ1133 in aerated LB broth as in many other studies (Bull and Molineux 2008). Selection was for rapid phage growth using serial transfer: cells from frozen aliquots were inoculated into 10-mL cultures in 125-mL flasks, which were grown for 1 hr at 37° to a density of approximately 108 cells/mL. Typically, 105 phage were added, grown 20-60 min (until the phage density had reached at least 107/mL, but often until lysis), and then transferred directly to the next flask. Most phage generations in each culture experienced low levels of coinfection, but the final round of growth was often at high multiplicity, which allows recombination between genomes. The last culture of the day was treated with chloroform to kill cells and then used to start the first transfer on the subsequent day.
For all phages, fitness was measured as the log2 change in phage density per hour during serial transfer between flasks (the same transfer method used for adaptations). Each assay of fitness was determined by titers taken from cultures separated by multiple, consecutive transfers. A single fitness estimate was determined by the titer at the end of 1 hr of consecutive phage growth and the titer from the end culture after an additional hour of growth; growth before the measurement of the initial titer reduces the effects of life-cycle synchronization that occur when phage are first added to a culture (Bull et al. 2011). In contrast to passage conditions, phage densities during fitness assays were kept at least 10-fold below cell densities, avoiding exhaustion of hosts and allowing exponential phage growth throughout the assay.
Creating specific recombinant phages
Transgenic T7 phages were created in two steps. Both steps involved recombination of a T7 phage genome over a plasmid insert carrying the desired change. Using overlapping polymerase chain reaction (PCR), a segment of DNA for introducing a deletion or insert was created with flanking segments of T7 DNA matching the desired insert site in the T7 genome. The PCR fragment was cloned into a plasmid, verified by sequencing, and transformed into the host E. coli IJ1133. T7 was grown on the plasmid-bearing host to allow recombination. Under optimal conditions, recombinants constitute ∼5% of the phage progeny, and screening by plating or PCR was used to find the desired recombinant. In the first step, T761 was grown in the presence of a gene 10 deletion plasmid to generate T7 Δ10 (del 21450-22789); the lysate was plated on a host carrying the complementing plasmid, pSW5. Plaques were then screened for the presence of Δ10. In the second step, T7 Δ10 was grown in the presence of a plasmid carrying the transgenic T3 gene 10 flanked by T7 sequences, and recombinants containing T3 gene 10 were selected directly on a plasmid-free host.
The K11 transgenic phage had to be created using two plasmids. High-copy number plasmids carrying an intact K11 gene 10 appear to be deleterious to E. coli, and the plasmid pRK11 carries a 39-bp insertion, derived from a PCR primer, upstream of the 5′ end of the K11 gene 10. The T7 Δ10 recombinants tested that grew on plasmid-free hosts contained the 39 bp insert plus the wild-type K11 gene 10. A second plasmid, pRK11-2, was therefore constructed to delete the additional DNA by recombination. The desired phage was sequenced and shown to contain a precise replacement of T7 gene10A/B coding sequences by their K11 counterparts (nucleotides 21475-22784) without additional sequences; however a mutation in the scaffold gene (base 22349) was found to be present (see below).
A recombinant phage was created between T7Δ10 and T7-10(K11) to create a transgenic lacking gene 10 but containing the scaffold mutation at base 22349. The two genomes were digested with RsrII (which cuts near the 3′ end of gene 9), complementary fragments were ligated, and the mix was transfected into cells with pSW5 (T3 gene 10) to obtain isolates that were then tested on a host with pRK11 (K11 gene 10). This phage also carried a deletion of gene 4.5 (bases 13346-13919); gene 4.5 has no known function, complete deletions have no known phenotype, so we attribute the pRK11 plating phenotype of this phage to its gene 9 mutation.
Creating recombinant populations for assessing compensatory evolution
A mutation in an evolved transgenic phage could be strictly compensatory—beneficial only in the presence of the transgene—or it could be generally beneficial for growth of any genome in the conditions used. The two types of mutations can be distinguished by allowing recombination between the evolved transgenic phage and T7, followed by a brief period of adaptation of the recombinant pool (Rokyta et al. 2002). Any mutations that are beneficial regardless of which capsid gene is present, will also increase in frequency in a T7 phage containing the T7 capsid gene. However, those mutations only beneficial because they compensate for the presence of the transgene will be disfavored in the presence of the wild-type gene. If the transgene is lost during outgrowth, sequence comparison of the evolved transgenic phage with the evolved recombinant pool allows determination of mutations that are specifically compensatory for the transgene.
To facilitate recombination, a zone of mixed coinfection was created by cross-streaking phages on agar plates seeded with bacteria. After incubation at 37°, regions of the soft agar that contained both phage types were suspended and used as recombinant populations. Recombinant populations were typically propagated by serial transfer for 5 hr, allowing lysis before each transfer. This procedure thus allowed for occasional recombination with continued selection for beneficial mutations.
Sequence analysis
Nucleotide changes were determined with ‘454’ pyrosequencing technology, Sanger technology, or both. Sanger sequences were obtained using an ABI 3730 with BigDye v. 3.0, and reactions were performed and run at the ICMB core facility at the University of Texas. For 454 sequencing, genomic DNA was processed and run at the Genome Sequencing and Analysis facility at University of Texas. All sequence profiles were compiled and analyzed with DNAStar software. 454 sequencing of viral populations provides frequency estimates of nucleotide changes but not genomic linkages beyond a few hundred nucleotides. Sanger sequencing provides linkage on sequenced isolates but only provides approximate frequencies on sequenced populations. Nucleotide changes whose frequencies were less than 25% in 454 data were ignored to avoid errors associated with this technology. The genome reference for T7 nucleotide positions is GenBank V01146. Sequences unique to the transgene used genome reference positions in GenBank AJ318471 (T3) or GenBank NC011043 (K11). Both evolved transgenic populations were sequenced by 454; Sanger sequencing was used with the evolved, transgenic T3 population and with one clonal isolate from the evolved transgenic K11 population. All other sequences used Sanger technology (e.g., initial phages and recombinants). Where both 454 and Sanger sequences were applied, each method was used to help resolve ambiguities in the other.
Sequence alignments
A BLAST nonredundant protein search was initiated with T7 major and minor capsid proteins and T7 scaffold and portal proteins. All protein sequences of other viral origin with significant similarity to the T7 protein sequence (a score greater than 1E-80) were first aligned using MUSCLE (Edgar 2004), then manually corrected. Percent identity was calculated by multiplying the number of matches within a pair of protein sequences by 100 and dividing by the length of the aligned region, including gaps.
Results
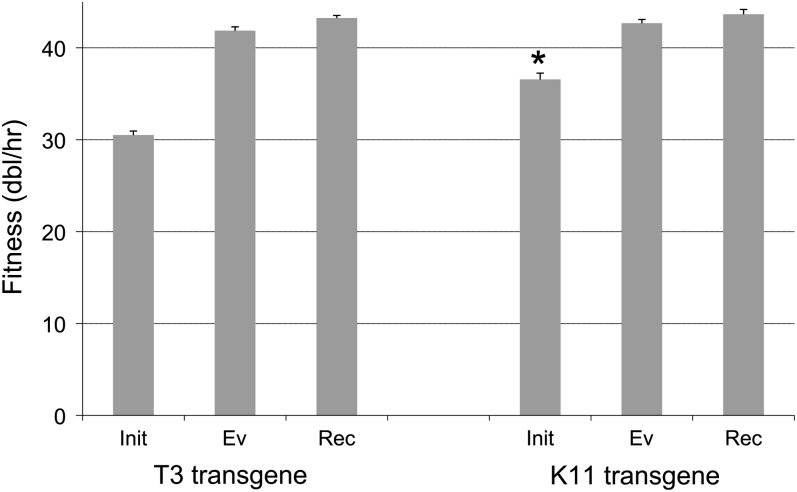
The T7 capsid gene was replaced with its homolog from two relative phages, T3 and K11, and each transgenic phage was then adapted for rapid growth. Fitnesses of evolved phages reached ~42 doublings per hour (Figure 1). The initial effect of incorporating T3 gene 10 did not appear to be profoundly deleterious: the fitness immediately after transgene insertion was 30.5. The initial fitness of the K11 transgenic phage was also high (36.6), although it was likely inviable before the acquisition of a change in the scaffold protein (see below, this section). After adaptation, the fitness of T7-10(T3) reached 41.9 ± 0.43, and the fitness of T7-10(K11) reached 42.7 ± 0.40. To compare these fitness limits with that attainable by intact T7, two approaches were used: (1) T7 was adapted to the conditions used here, and (2) a recombinant pool between intact T7 and each evolved transgenic phage was adapted for 5 hr. Final fitness of the pure T7 adaptation done for this study was 41.4, although it is known that T7 fitness can evolve to ~44 under these conditions (Bull et al. 2011); the fitness of the recombinant pools was more in line with the latter value, 43-44 (Figure 1). Both recombinant pools lost the transgene and retained the T7 capsid gene, thus revealing at least a slight fitness cost of the transgene in an otherwise purely T7 genome.
Figure 1.
Fitnesses of transgenic phages. Fitness is given as doublings/hr with 1 standard error. Two transgenic phages were created, both using a backbone of T7. One transgenic phage carried the T3 capsid gene (10A/B) in place of the T7 homolog, and the other carried the K11 capsid gene in place of the T7 homolog. For each transgenic phage line, fitness was measured at three points: immediately after creation (Init), after adaptation/evolution (Ev), and after the evolved phage was recombined with T7 and outgrown 5 hr (Rec). The greater fitness of Rec than of Ev is marginally significant for the T3 transgenic phage (P ≈ 0.048) but not for the K11 transgenic phage. However, because both Rec phages lost the transgene and regained the original T7 capsid protein, they must have greater fitnesses than their corresponding evolved transgenic phages. * Indicates that this initial phage carried a mutation (in the scaffold gene) that was essential for viability. Fitness of the initial transgenic phage lacking this change is thus much lower than shown, even negative, but such a phage could not be isolated. From left to right, mean fitness scores are 30.5, 41.9, 43.2, 36.6, 42.7, and 43.6. For reference, the fitness of a wild-type T7 adapted to these conditions was 41.4 ± 0.42. The T3 transgenic evolved phage was adapted for 45 hr, the K11 phage for 28 hr of serial transfer.
However, the full fitness cost of the transgenic replacement is not necessarily revealed by the fitness of plaque isolates immediately after the gene insertion. If the gene replacement is highly deleterious, plaques may contain compensatory mutations that rescue fitness because phages lacking such compensatory mutations are inviable or only weakly viable. It was previously known that the T3 major capsid gene complements a T7 defective in its major capsid gene (Condreay et al. 1989), and in this work we routinely propagated T7 Δ10 on a host supplying the T3 capsid protein. It was therefore not surprising that no compensatory mutations were observed in the initial T3 transgenic phage. However, phages in the first plaque found to contain the K11 transgene also carried a mutation in the scaffolding protein gene. To see whether this mutation was important for viability, a recombinant phage, T7 Δ10 *, was created by combining the left end of the K11 transgenic phage containing the mutated scaffold gene with the right end of the T7 Δ10 lacking a capsid gene. The titer of T7 Δ10 on an expression plasmid carrying K11 gene 10 (pSWK11), relative to T3 gene 10 (pSW5) was ~10−6, whereas T7 Δ10* was ~10−2 (data not shown). This test thus reveals a major benefit, essentiality, of the change in the scaffolding protein. It is also possible that the benefit is larger than the difference measured here because phages that appeared at 10−6 may well have carried their own compensatory mutations.
At the end of both adaptations, mutations accompanying the adaptations were tested for compensatory vs. noncompensatory status. Compensatory mutations were found in the transgenic capsid gene and in the scaffold gene of both adaptations (Table 2). For the most part, these substitutions caused the proteins to change away from consensus AAs. A few additional compensatory mutations were found in other parts of the genome with the T7-10(T3) adaptation, affecting genes with no known role in capsid assembly.
Table 2. Compensatory changes in evolved transgenic phages.
| Position | Change | Frequency, % | Gene/Promoter | AA Change |
|---|---|---|---|---|
| T3 transgenic phage | ||||
| 6553 | G->T | 100 | 1.3 (ligase) | G27W |
| 22506 | T->C | 59 | 9 (scaffold) | V186Aa |
| T3 20910 | G->C | 32 | T310 (capsid) | G7Aa |
| T3 21120 | A->G | 100 | T310 | E77Ga |
| 24204 | T del | 64 | Tϕ (terminator) | |
| K11 transgenic phage | ||||
| 22349b | G->A | 100 | 9 (scaffold) | A134Ta |
| K11 21610 | A->G | 85 | K11 10 (capsid) | T46Aa |
| K11 21716 | A->G | 81 | K11 10 | D81Ga |
| K11 22033 | A->G | 97 | K11 10 | T187Aa |
| K11 22408 | A->G | 73 | K11 10 | T312A |
Destroys consensus AA or changes to less common AA in homologs identified with BLAST (not corrected for phylogeny).
Present in initial isolate.
Discussion
T7 phage genomes were constructed with the native capsid gene replaced by the homolog of either of two relatives. At AA sequence differences of 22% between the native capsid protein and either replacement, only one of two transgenic genomes was viable; however, a single mutation rendered the second transgenic genome viable. Nearly full fitness recovery (relative to adapted wild-type) was achieved with five compensatory substitutions in each line.
Our primary interests here are in results that generalize beyond this system and, equally, the predictability of the evolution. Toward this goal, we can compare the T7 data to a study of Rokyta and Wichman (2009), in which the capsid gene was exchanged between microvirid phage genomes and the recombinant genomes adapted (microvirid have ~5 kb ssDNA genomes with 11 genes). In addition, various other types of modified T7 genomes have been adapted in vitro and analyzed (Bull and Molineux 2008). We consider four results of possible generality.
Magnitude of recovery
After relatively short adaptations, nearly full fitness recovery was attained by both transgenic T7 phages—from a substantial initial deficit, the fitness evolved to within 2 doubling/hr of the maximum attainable by the T7 parent. Three of the four adaptations of Rokyta and Wichman (2009) also resulted in a transgenic fitness equal to or above that of one of the adapted parent genomes. However, other types of genome modifications in T7—deletions of important genes and rearrangements of genome order—did not lead to near full recovery (Bull and Molineux 2008). Thus, incompatibilities caused by structural incompatibilities alone may be surmountable with minimal effort/adaptation. Lessons from evolution of novel bacterial plasmids with their hosts also support the ease of evolutionary fitness recovery (Bouma and Lenski 1988; Lenski et al. 1994).
Number of changes, magnitudes of effect
Despite profoundly low initial fitness for one of two transgenic phages, nearly full fitness recovery was achieved with five compensatory changes (and five compensatory changes also occurred in the line whose initial fitness was moderately high). We did not assess the individual effects of most substitutions but did establish that the first substitution of one adaptation had a profoundly large effect, converting an inviable genome (incapable of forming a plaque, thus a fitness less than 5 can be inferred from other T7 observations) into one with a fitness of 36.6 doublings/hr; the total contribution from four additional compensatory changes was merely 6.1 greater. Rokyta and Wichman (2009) provided a more thorough analysis of individual mutational fitness effects, observing effects of 5-12 doublings/hr for the first or second change, but much smaller effects for later ones. Large fitness effects of early changes and a modest number of compensatory changes is typical of other adaptations of modified T7 genomes (Bull and Molineux 2008). This point thus reinforces the first in suggesting that evolution to ameliorate incompatibilities can be rapid and is especially rapid early.
Interacting partners
The two known physical interactants with the T7 capsid protein are the scaffold protein and the portal protein. In one adaptation, all compensatory changes were in the capsid transgene and scaffold gene; in the other adaptation, three of five compensatory changes were in those two genes, whereas the other two compensatory changes were in elements with no known role in capsid assembly. In the four adaptations of Rokyta and Wichman (2009), 12 of 19 changes were in the transgenic capsid or a scaffold protein gene, and at least the first two changes in each adaptation were in those genes. Thus, the transgene itself and its structural partners seem to be the main locations of compensatory evolution. This result is not surprising and is backed by decades of genetics work on “second-site suppressors” to identify interacting partners; second-site suppressors are extremes of the type of compensatory changes studied here.
Limit of divergence tolerated
The two studies differ in the degree of divergence tolerated between the native gene and its heterologous replacement. The transgenes used here are 22% diverged from the T7 capsid at the protein level. Only one of the two constructs was viable at this level of divergence, suggesting that 22% may be near the limit for T7. In the microvirid study, the successful transgenes were diverged 8% from the native protein, and repeated efforts to introduce a transgene at ~25% divergence failed to create a viable hybrid (D. Rokyta and H. Wichman, personal communication). However, it is reported that the major capsid protein of microvirid phage G4 can complement an amber mutation in the ϕX174 major capsid gene but not vice versa (Borrias et al. 1979); the major capsid proteins of those two phages differ by 35% (using BLAST, see also Rokyta et al. 2006). The level of divergence tolerated may thus be similar between the two groups of phages, but there is a potentially large stochastic element in successful complementation, such that individual cases fail or succeed on the basis of specific details.
It is likely that many properties of the adaptations, such as the identities of the compensatory changes, will be predictable only by an understanding the molecular details of the system—knowing which residues interact for essential functions. Although the level of molecular detail for T7 (or any phage) is still superficial by this latter standard, progress toward this level of detail has been made on another front: a virtual model of the infection cycle exists, although it is one of regulation not of structure (Endy et al. 2000). Even lacking knowledge of such molecular details, however, additional studies at the level of Rokyta and Wichman (2009) and our study may lead to simple models for successfully predicting the broad requirements for transgenic viability and the pathways of compensatory evolution. Indeed, recent high-throughput sequencing efforts performed on pools of mixed viral infections have identified degrees of tolerated or favored recombinant genotypes on the basis merely of the abundances of different sequence combinations (Martin et al. 2011). Thus, useful inferences can come from bioinformatic patterns, rather than strictly from detailed biochemical studies.
This study and that of Rokyta and Wichman (2009) raise the possibility that compensatory evolution may operate fast enough to overcome some fitness valleys encountered by hybrids and horizontal gene exchanges, with some relevance to speciation (Coyne and Orr 2004). Clearly there is a limit to the amount of divergence tolerated, but a mere fitness decline upon incorporation of heterologous DNA may be short-lived. Indeed, the overall patterns observed here and in Rokyta and Wichman (2009) are broadly reminiscent of the genomic sorting observed when experimentally reconstructing polyploid hybrid ancestors of naturally occurring sunflowers (Rieseberg et al. 2003). In the very early stages of speciation, therefore, the re-uniting of mildly diverged populations may lead to evolution in two directions, one toward reinforcement of the differences but an opposing one of overcoming the incompatibilities. The latter is not likely to be effective except when very few strong incompatibilities exist, of course.
The results are also encouraging for synthetic biology. When combining multiple interacting systems into a single genome, it may often be that the combination of all systems at once is detrimental or even lethal. Modest amounts of compensatory evolution may overcome the incompatibilities, and furthermore, stepwise construction and recovery before introducing the next system may be a way of creating a viable, final product.
The construction of recombinant genomes by exchanging genes or even introducing new genes is now commonplace in virology, and, indeed, some of the prospects for such engineering are worrying (e.g., Jackson et al. 2001) or involve exchanging genes from highly virulent human pathogens (Twu et al. 2007). The results here suggest that there is considerable potential for evolution of fitness improvement in those recombinants, at least in what are highly impaired genomes at the outset.
Acknowledgments
We thank R. J. Bull for lab work that preceded this study. This work was supported by National Institutes of Health grant GM57756 (J.J.B.); J.J.B. also receives support as the Miescher Regents Professor at the University of Texas.
Footnotes
Communicating editor: D. J. Gresham
Literature Cited
- Agirrezabala X., Velázquez-Muriel J. A., Gómez-Puertas P., Scheres S. H., Carazo J. M., et al. , 2007. Quasi-atomic model of bacteriophage t7 procapsid shell: insights into the structure and evolution of a basic fold. Structure 15: 461–472 [DOI] [PubMed] [Google Scholar]
- Borrias W. E., Hagenaar M., Van Den Brekel R., Kühlemeijer C., Weisbeek P. J., 1979. Functional relationship between bacteriophages G4 and phi X174. J. Virol. 31: 288–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma J. E., Lenski R. E., 1988. Evolution of a bacteria/plasmid association. Nature 335: 351–352 [DOI] [PubMed] [Google Scholar]
- Bull J. J., Molineux I. J., 2008. Predicting evolution from genomics: experimental evolution of bacteriophage T7. Heredity 100: 453–463 [DOI] [PubMed] [Google Scholar]
- Bull J. J., Heineman R., Wilke C. O., 2011. The phenotype-fitness map in experimental evolution of phages. PLoS ONE 6: e27796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condreay J. P., Wright S. E., Molineux I. J., 1989. Nucleotide sequence and complementation studies of the gene 10 region of bacteriophage T3. J. Mol. Biol. 207: 555–561 [DOI] [PubMed] [Google Scholar]
- Condron B. G., Gesteland R. F., Atkins J. F., 1991. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 19: 5607–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endy D., You L., Yin J., Molineux I. J., 2000. Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc. Natl. Acad. Sci. USA 97: 5375–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman R. H., Molineux I. J., Bull J. J., 2005. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J. Mol. Evol. 61: 181–191 [DOI] [PubMed] [Google Scholar]
- Ionel A., Velázquez-Muriel J. A., Luque D., Cuervo A., Castón J. R., et al. , 2011. Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J. Biol. Chem. 286: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Ramsay A. J., Christensen C. D., Beaton S., Hall D. F., Ramshaw I. A. , 2001. Expression of mouse interleukin-4 by a recombinant ectromelia virus suppresses cytolytic lymphocyte responses and overcomes genetic resistance to mousepox. J. Virol. 75: 1205–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Simpson S. C., Nguyen T. T., 1994. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176: 3140–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang Q., Murata K., Baker M. L., Sullivan M. B., et al. , 2010. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 17: 830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. P, Lefeuvre P., Varsani A., Hoareau M., Semegni J. Y., et al. , 2011. Complex recombination patterns arising during geminivirus coinfections preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 7: e1002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux I. J., 2006. The T7 group, 277–301 The Bacteriophages, edited by Calendar R. Oxford University Press, Oxford, New York [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., Raymond O., Rosenthal D. M., Lai Z., Livingstone K., et al. , 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216 [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Wichman H. A., 2009. Genic incompatibilities in two hybrid bacteriophages. Mol. Biol. Evol. 26: 2831–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D. R., Badgett M. R., Molineux I. J., Bull J. J., 2002. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol. Biol. Evol. 19: 230–238 [DOI] [PubMed] [Google Scholar]
- Rokyta D. R., Burch C. L., Caudle S. B., Wichman H. A., 2006. Horizontal gene transfer and the evolution of microvirid coliphage genomes. J. Bacteriol. 188: 1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu K. Y., Kuo R.-L., Marklund J., Krug R. M., 2007. The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells. J. Virol. 81: 8112–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]