Abstract
The kinetochore is a large structure composed of multiple protein subcomplexes that connect chromosomes to spindle microtubules to enable accurate chromosome segregation. Significant advances have been made in the identification of kinetochore proteins and elucidation of kinetochore structure; however, comparatively little is known about how cellular signals integrate with kinetochore function. In the budding yeast Saccharomyces cerevisiae, the cyclic AMP protein kinase A signaling pathway promotes cellular growth in response to glucose. In this study, we find that decreasing protein kinase A activity, either by overexpressing negative regulators of the pathway or deleting the upstream effector Ras2, improves the viability of ipl1 and spc24 kinetochore mutants. Ipl1/Aurora B is a highly conserved kinase that corrects attachment of sister kinetochores that have attached to the same spindle pole, whereas Spc24 is a component of the conserved Ndc80 kinetochore complex that attaches directly to microtubules. Unexpectedly, we find that kinetochore mutants have increased phosphorylation levels of protein kinase A substrates, suggesting that the cyclic AMP protein kinase A signaling pathway is stimulated. The increase in protein kinase A activity in kinetochore mutants is not induced by activation of the spindle checkpoint or a metaphase delay because protein kinase A activity remains constant during an unperturbed cell cycle. Finally, we show that lowering protein kinase A activity can rescue the chromosome loss defect of the inner kinetochore ndc10 mutant. Overall, our data suggest that the increased protein kinase A activity in kinetochore mutants is detrimental to cellular growth and chromosome transmission fidelity.
Keywords: budding yeast, protein kinase A, kinetochore, chromosome segregation, spindle checkpoint
Although the budding yeast Saccharomyces cerevisiae (S. cerevisiae) is able to utilize various carbon sources, glucose is the preferred carbon source. Upon addition of glucose to the cell, the cyclic AMP protein kinase A (cAMP-PKA) pathway is activated, and adenylate cyclase, called Cyr1 in yeast, catalyzes the synthesis of cAMP from ATP. In S. cerevisiae, the activity of adenylate cyclase is stimulated by the small G proteins Ras1 and Ras2 and a G protein coupled receptor system [supporting information, Figure S1, (Thevelein and de Winde 1999)]. Cdc25 and Sdc25 are two Ras guanine nucleotide exchange factor proteins that switch GDP-bound Ras to GTP-bound Ras, whereas Ira1 and Ira2 are redundant GTPase activating proteins that inactivate Ras by hydrolysis of the bound GTP to GDP (Dechant and Peter 2008; Tamaki 2007). The cAMP produced by adenylate cyclase binds Bcy1, which is an inhibitory subunit of PKA. In budding yeast, PKA is a heterotetramer of two catalytic subunits (any two of Tpk1, 2 or 3) and two Bcy1 subunits. Once cAMP binds Bcy1, the two catalytic subunits are released from Bcy1, which allows these subunits to phosphorylate multiple target proteins, including phosphorylation and inhibition of the stress response transcription factors Msn2 and Msn4 (Gelade et al. 2003). cAMP accumulation in yeast is downregulated by both a low (Pde1) and high (Pde2) affinity phosphodiesterase, which hydrolyze cAMP to AMP (Nikawa et al. 1987; Sass et al. 1986).
In addition to mediating the cellular response to glucose, the PKA pathway also has cell cycle regulatory roles. For example, PKA activity mediates mitotic arrest in response to DNA damage by regulating phosphorylation of Cdc20 (Searle et al. 2004). Cdc20 is a specificity factor of the anaphase promoting complex (APC), and multiple studies suggest that PKA is an inhibitor of the APC (Anghileri et al. 1999; Bolte et al. 2003; Heo et al. 1999; Irniger et al. 2000). The PKA pathway has also been implicated in chromosome segregation. Three studies have demonstrated a potential interaction between the kinetochore, a large structure composed of multiple protein complexes that connects spindle microtubules to chromosomes, and the cAMP-PKA pathway. The inner kinetochore is comprised of the CBF3 centromere-binding complex and a modified nucleosome (Choy et al. 2012; Westermann et al. 2007). Sgt1, which is required for assembly of the CBF3 inner kinetochore complex, physically interacts with Cyr1 and upregulates the activity of the cAMP-PKA pathway (Dubacq et al. 2002). Overexpression of negative regulators of the cAMP-PKA pathway rescues the lethality of kinetochore mutants (Li et al. 2005; Magtanong et al. 2011). However, no systematic analysis has been performed to analyze the effect of increasing or decreasing PKA activity on multiple kinetochore mutants, including spindle checkpoint active and inactive alleles, or what impact a kinetochore defect might have on PKA activity.
In this study, we find that decreasing PKA activity rescues the viability of strains carrying mutations in the highly conserved Ipl1/Aurora B kinase and the Ndc80 kinetochore complex. We also show that reduction of PKA activity rescues the chromosome loss defect of the inner kinetochore ndc10-1 mutant. Unexpectedly, we find that ipl1, spc24, and ndc10-1 kinetochore mutants have a high level of PKA activity that is not due to spindle checkpoint activation or metaphase arrest. We propose that the high level of PKA activity in kinetochore mutants is partially responsible for the chromosome loss and growth defects in these strains.
Materials and Methods
Yeast strains, plasmids, and media
The yeast strains used in this study are described in Table S1. The pRS326-PDE2 plasmid was obtained from a 2 μ yeast genomic DNA library (Connelly and Hieter 1996) as a high copy suppressor of spc24-9 lethality on 0.1M HU plates at 33° (Ma et al. 2007). The genomic DNA coordinates for the PDE2 insert are Chr XV 1011626–1019437. A subclone was constructed that contained only the PDE2 ORF (Chr XV 1013176–1015806), which also rescued spc24-9 lethality (data not shown). The BCY1 plasmid (p416-GPD-BCY1) and vector control (p416-GPD) were kind gifts from Kevin Morano (Trott et al. 2005). The RAS2val19 plasmid was a kind gift from Paul Herman (Ramachandran and Herman 2011). The liquid media were rich medium (YPD) or supplemental minimal medium (SC) (Kaiser et al. 1994). The plates for the carbon source spot assays were glucose (2%), glycerol (2%), acetate (1%), ethanol (3%), and YEP (no added carbon source except for the yeast extract and peptone used in YPD media).
Western blots
For Figure 3 and Figure 4, wild-type, spc24-10, spc24-9, spc24-8, spc24-8 mad2Δ, mad2Δ, ndc10-1, and ipl1-321 cells were grown to mid-logarithmic phase at 25° (Figure 3A) or the semipermissive temperature of 31.5° (Figure 3B) in YPD medium. Wild-type cells harboring the RAS2val19 plasmid were grown in SC-URA medium. 15 mL of culture was harvested, and the pellet was washed once with cold dH2O and lysed immediately. Forty micrograms of protein lysate was loaded on a 8% SDS-PAGE gel. For Figure 5A, wild-type cells were grown in YPD to log phase at 25°, synchronized in G1 with mating pheromone (α-factor, 5 μg/mL) for 2 hr, and released at 25°. Time points were taken at 0 min, 30 min, and every 15 min thereafter for 120 min. The pellet from each 15 ml of culture was washed once with cold dH2O, frozen at −80°, and lysed the next day. For Figure 5B, wild-type cells were grown to mid-logarithmic phase in YPD medium, then treated with 20 μg/mL nocodazole or without nocodazole (Log) for 2 hr at 25° before harvesting. Msn2 phosphorylation was detected with α-P-CREB (S133) antibody (1:1000, Cell Signaling), Msn2 protein was detected with an α-Msn2 antibody (1:10,000, kind gift from Francisco Estruch), Pgk1 protein levels were detected with an α-Pgk1 antibody (1:10,000, Invitrogen), phosphorylation of PKA substrates was detected with a phospho-PKA substrate antibody (α-sub,1:5,000, Cell Signaling #9624), and Clb2 was detected with an α-Clb2 antibody (1:5,000, Santa Cruz). For Figure 3, the blot was cut into two parts. The bottom part was probed with α-Pgk1; the top part was first probed with α-P-CREB, stripped for 30 min at 60° with Tris-SDS buffer [62.5 mM Tris HCl (pH 6.8), 2% SDS, 100 mM β-Mercaptoethanol], and then probed with α-Msn2. For Figure 4, the blot was probed with α-sub and then probed with α-Pgk1 after stripping. Images were captured with a ChemiDoc MP imaging system (Bio-Rad), and quantification was performed using Image Lab software (Bio-Rad). At least three independent experiments were performed for each Western blot. For Figures 3 and 4, a one-way ANOVA test was used to determine which mutants had significantly different phosphorylation levels from wild-type. For Figure 5, an ANOVA with Tukey’s post-hoc analysis was done to compare values from all time points, except for time point zero (which has lower PKA activity due to pheromone treatment), with each other.
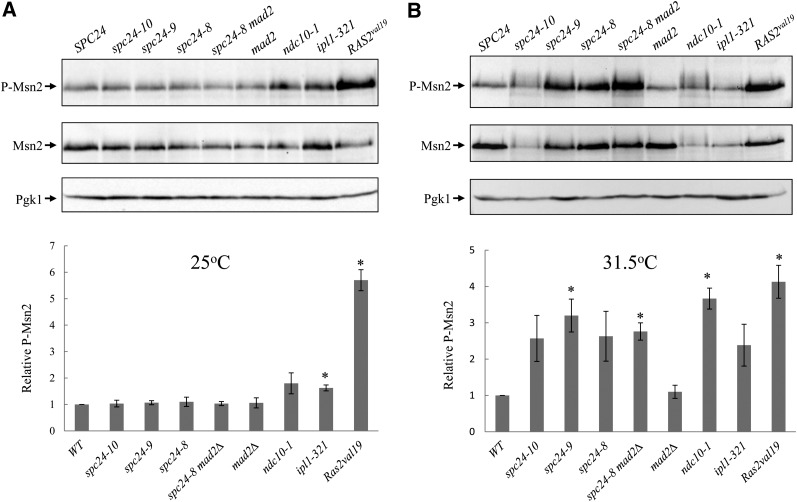
Figure 3.
Kinetochore mutants have higher levels of Msn2 PKA-dependent phosphorylation. Cells were grown to mid-logarithmic phase at 25°C (A) or the semipermissive temperature of 31.5°C (B) and lysed for immunoblot analysis. The blots were probed with α-P-CREB antibody to detect PKA-dependent Msn2 phosphorylation (P-Msn2) and α-Msn2 and α-Pgk1 as loading controls. One representative blot is shown out of three independent experiments. The bar graphs are the relative ratio of P-Msn2/total Msn2 analyzed from the three independent experiments. The value of 1 was assigned to the ratio of P-Msn2/total Msn2 in the wild-type cells. The asterisk represents values that are significantly different from the wild-type value (P < 0.05).
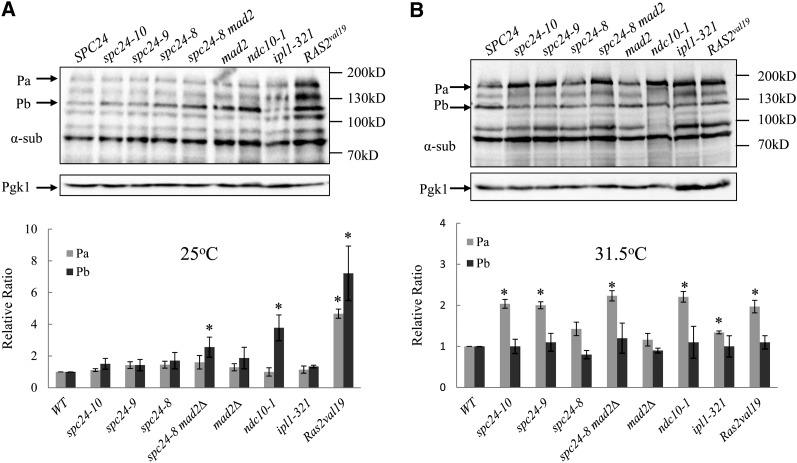
Figure 4.
PKA substrates have higher levels of phosphorylation in kinetochore mutants. (A) The lysates described in Figure 3 were probed with the anti-PKA substrate antibody (α-sub) and α-Pgk1 as a loading control. Two protein bands from the α-sub blot, Pa and Pb, were selected for quantification. One representative blot from three independent experiments is shown. Bar graphs are the relative ratio of Pa/Pgk1 or Pb/Pgk1 averaged from three independent experiments. The value of 1 was assigned to the ratio of Pa/Pgk1 or Pb/Pgk1 in the wild-type cells. The asterisk represents values that are significantly different from the wild-type value (P < 0.05).
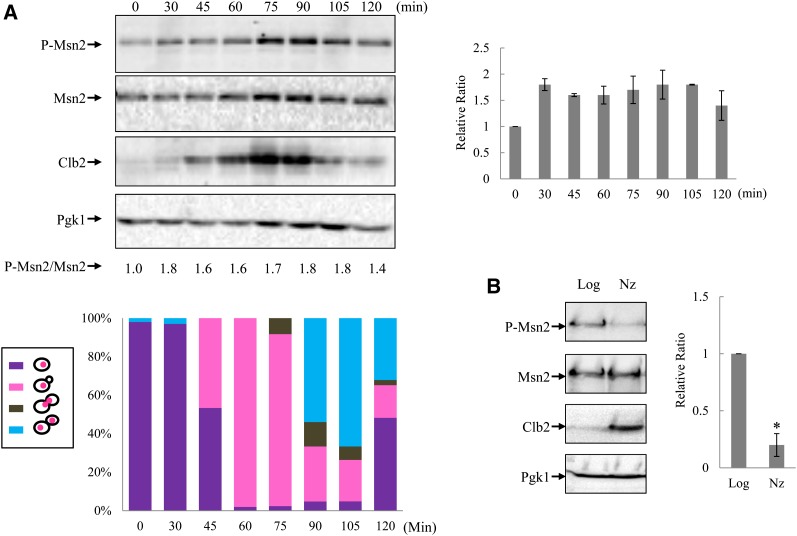
Figure 5.
Msn2 PKA-dependent phosphorylation is not cell cycle regulated. (A) Wild-type cells were synchronized in G1 with mating pheromone (α-factor, 5 μg/mL) for 2 hr and released at 25°C. Samples were taken at the indicated time points and analyzed for Msn2 PKA-dependent phosphorylation by immunoblotting (top panel) and for nuclear division by microscopy (bottom panel). The Western blot was probed with P-Msn2 (α-CREB), α-Msn2, α-Clb2, or α-Pgk1. The bar graph (top right) is the relative ratio of P-Msn2/total Msn2 analyzed from two independent experiments. A value of 1 was assigned to the ratio of P-Msn2/total Msn2 in the wild-type cells. (B) Wild-type cells were grown to mid-logarithmic phase at 25°C in YPD medium and either kept in log phase (Log) or treated with 20 μg/mL nocodazole (Nz) for 2 hr at 25°C. Western blots were probed with P-Msn2 (α-CREB), α-Msn2, and α-Clb2 as a marker for cell-cycle arrest. The asterisk represents the significant difference between the P-Msn2 levels upon Nz treatment vs. log phase cells (P < 0.05).
DAPI staining and budding index
For each time point in Figure 5A, 1 mL of cells was spun down at 2000 rpm, fixed with 70% ethanol for 1 hr at room temperature, resuspended in PBS, and stored at 4°. For microscopy imaging, cells were lightly sonicated and mixed well with DAPI (final concentration 50 ng/mL). Cells were categorized into four groups: (1) G1 cells with no bud; (2) G2/S cells with a small bud; (3) metaphase cells with the nucleus at the bud neck; and (4) anaphase/telophase cells with a divided nucleus. Two hundred cells were counted for each time point.
Chromosome fragment loss assay
ndc10-1 and ndc10-1 ras2Δ mutants carrying a chromosome fragment (CF) (CFIII TRP) were grown over night in SC-TRP media to select for the CF, and then ∼12,000 colonies were spread onto SC plates with limiting adenine as described (Koshland and Hieter 1987). ndc10-1 pRS416 CFIII TRP and ndc10-1 pRS416-BCY1 CFIII TRP strains were grown overnight in SC-TRP-URA media to select for the plasmid and CF, and then ∼6,000 colonies were spread onto SC-URA plates (to select for the plasmid) with limiting adenine. Plates were incubated at 25° for 3 days and then incubated for 2 days at 4° to develop the red pigment. Colonies that were all red (CF lost while plating), half red/half white (CF lost during first cell division), and total colonies were counted.
Results
Reduction of PKA activity alters the viability of kinetochore mutants:
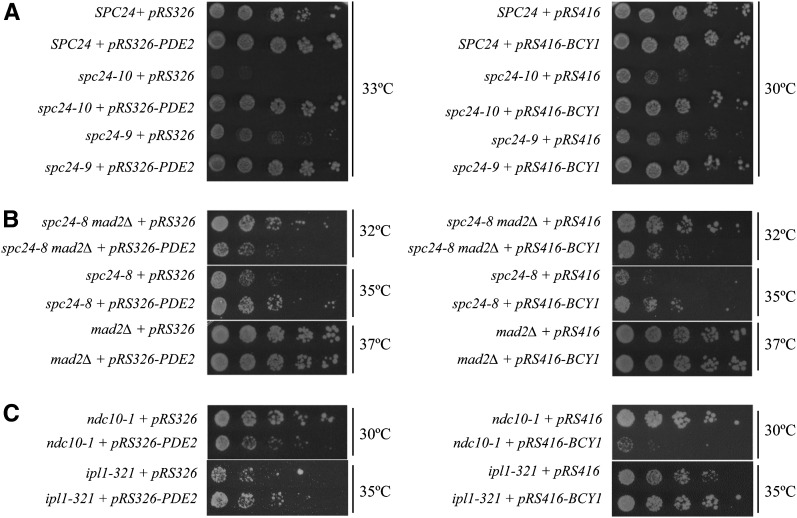
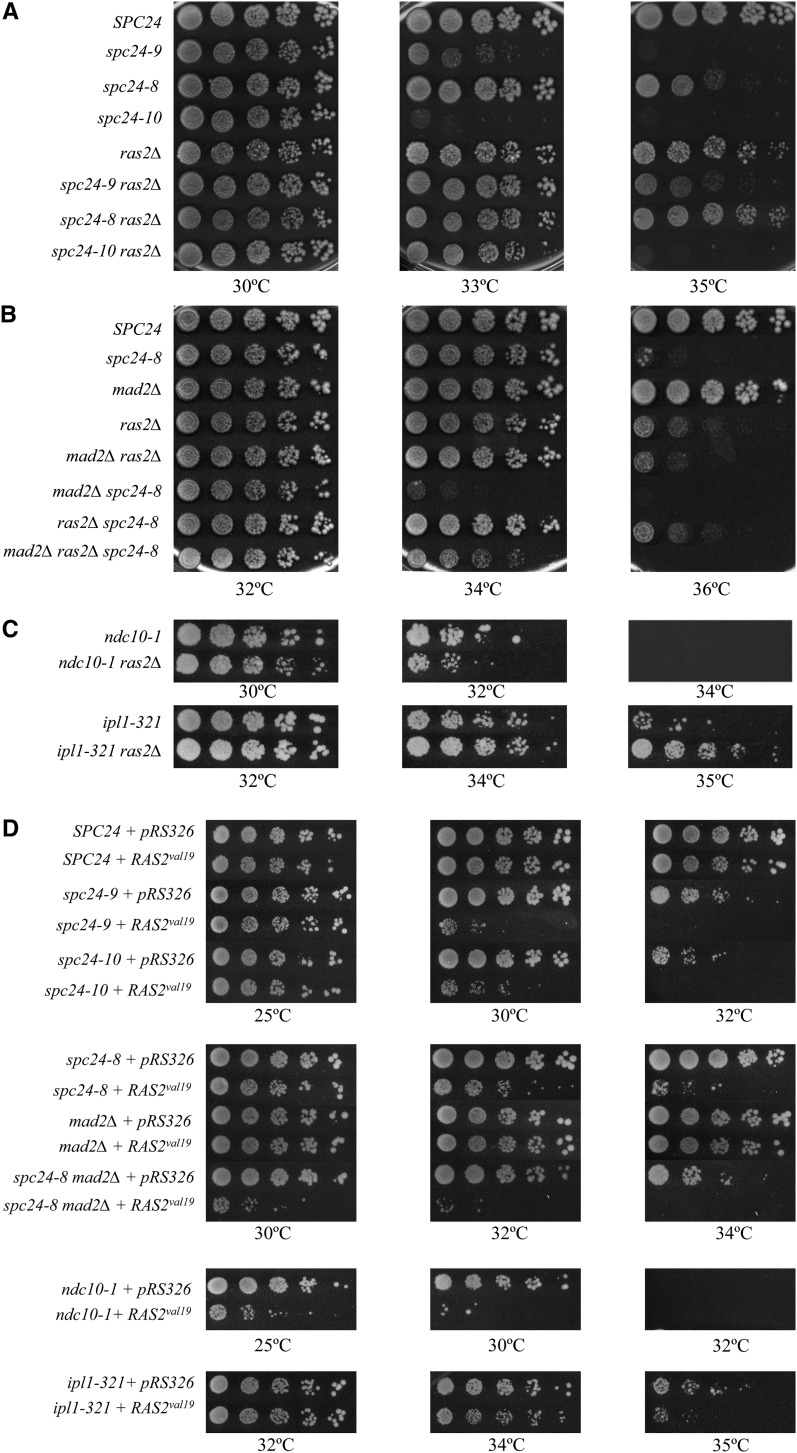
The Ndc80 complex is a highly conserved essential kinetochore complex composed of Ndc80, Nuf2, Spc24, and Spc25 in which the Ndc80/Nuf2 subcomplex binds directly to microtubules and the Spc24/Spc25 subcomplex interacts with the kinetochore (Tooley and Stukenberg 2011). We previously performed a genome-wide synthetic lethal (SL) screen with three phenotypically distinct alleles of the SPC24 gene and identified SL or synthetic sick interactions between multiple spc24 alleles and two negative regulators of the cAMP-PKA pathway, ira2Δ and pde2Δ (Montpetit et al. 2005). Consistent with our previous SL data, we found that overexpression of PDE2, which encodes the high affinity cAMP phosphodiesterase that converts cAMP to AMP (Sass et al. 1986), rescued the temperature-sensitive (Ts) lethality of spc24-8, spc24-9, and spc24-10 mutants (Figure 1, A and B). Likewise, overexpression of BCY1, which negatively regulates PKA, also rescued the Ts lethality of all three spc24 mutants (Figure 1, A and B) (Toda et al. 1987). The genetic interactions between negative regulators of the PKA pathway and spc24 mutants suggest that high PKA activity is particularly detrimental to strains with defective kinetochore-microtubule attachments.
Figure 1.
Inhibition of the cAMP-PKA pathway is beneficial to ipl1-321 and spc24 kinetochore mutants but detrimental to the ndc10-1 kinetochore mutant. (A–C) Cell dilution assay of wild-type cells and indicated mutants carrying the plasmid vectors pRS416-GPD (pRS416), pRS416-GPD-BCY1 (pRS416-BCY1), pRS326, or pRS326-PDE2 grown on SC-URA plates at the indicated temperatures for 2–4 days.
To determine whether other kinetochore mutants were affected by PKA activity, we overexpressed BCY1 and PDE2 in the ipl1-321 mutant, which suffers from inappropriate attachment of sister kinetochores to the same spindle pole (Biggins and Murray 2001; Cheeseman et al. 2002; Tanaka et al. 2002). We found that the growth of the ipl1-321 mutant was modestly rescued by BCY1 or PDE2 overexpression (Figure 1C). However, overexpression of BCY1 or PDE2 was detrimental to a ndc10-1 mutant that lacks an assembled kinetochore at a restrictive temperature [Figure 1C, (Goh and Kilmartin 1993)].
The small GTP-binding protein Ras2 interacts with adenylate cyclase and stimulates production of cAMP (Field et al. 1990; Suzuki et al. 1990). Consistent with the BCY1 and PDE2 overexpression data, we found that the double spc24ras2Δ and ipl1-321 ras2Δ mutants grew better than spc24 or ipl1-321 single mutants, whereas the ndc10-1 ras2Δ double mutant grew more poorly (Figure 2, A and C). Therefore, lowering PKA activity is beneficial to mutants that have kinetochore-microtubule attachment defects but is detrimental to strains that have no kinetochore assembled. To determine how kinetochore mutants respond to an increase in PKA activity, we introduced a dominant allele of Ras2, RAS2val19, which activates the PKA pathway, into ipl1-321, ndc10-1, and spc24 mutant strains. As expected, the temperature sensitivity of ipl1-321 and spc24 strains was exacerbated by the dominant RAS2val19 allele and, surprisingly, so was the Ts of the ndc10-1 strain (Figure 2D). Therefore increasing PKA activity is detrimental to mutants with either no kinetochore or kinetochore-microtubule attachment defects.
Figure 2.
Expression of the RAS2Val19 dominant allele is detrimental to all kinetochore mutants, whereas deletion of RAS2 rescues a subset of kinetochore mutants. (A–C) Cell dilution assay of the indicated strains grown on YPD plates at the indicated temperatures for 3 days. (D) Cell dilution assay of wild-type cells and the indicated mutants carrying the plasmid vectors pRS326 or pRAS2val19 grown on SC-URA plates at the indicated temperatures for 3 days.
The kinetochore monitors its state of microtubule attachment and signals to the spindle checkpoint to prevent anaphase onset in the presence of incorrectly attached or unattached kinetochores. Depending on the nature of the mutation, a kinetochore mutant may or may not activate the spindle checkpoint and arrest the cell cycle in metaphase. For example, of our three spc24 alleles, only spc24-8 cells are able to activate the spindle checkpoint at a restrictive temperature (Montpetit et al. 2005). To determine whether decreasing PKA activity can bypass the requirement for the spindle checkpoint, we impaired the spindle checkpoint in spc24-8 by deletion of the Mad2 checkpoint protein. The spc24-8 mad2Δ strain was SL at 34°, but the SL was rescued by deletion of RAS2, suggesting that lowering PKA levels may allow for partial bypass of the spindle checkpoint (Figure 2B). However, overexpression of PDE2 and BCY1 did not rescue the viability of the spc24-8 mad2Δ double mutant but instead exacerbated the growth defect (Figure 1B). In addition, increasing PKA activity by expressing RAS2va119 decreased the restrictive temperature of spc24-8 mad2Δ (Figure 2D). These data, combined with the fact that decreasing PKA activity rescues both checkpoint active (spc24-8) and checkpoint defective (spc24-9, spc24-10) kinetochore mutants, suggest that lowering PKA activity does not improve the growth of kinetochore mutants due to restoration of the spindle checkpoint.
Chromosome loss defects in ndc10-1 mutants are suppressed by deletion of RAS2
spc24, ndc10-1, and ipl1-321 mutants have defects in chromosome stability (Biggins et al. 2001; Goh and Kilmartin 1993; Montpetit et al. 2005). The chromosome fragment (CF) loss assay is a colony color-based sector assay to determine whether a strain is able to maintain a nonessential CF in the cell (Koshland and Hieter 1987). Colonies that contain the CF are white, whereas colonies that lose the CF are red. If a haploid cell loses the CF in the first cell division, the colony is half-red and half-white (red/white half-sector). We asked whether reduction of PKA activity was able to rescue the chromosome segregation defects of kinetochore mutants using the CF loss assay. We overexpressed BCY1 or deleted RAS2 in spc24-9, ndc10-1, and ipl1-321 strains carrying a CF and screened qualitatively for rescue of CF loss at a variety of temperatures compared with kinetochore mutant alone. We determined that ndc10-1 CF loss, at a permissive temperature, was rescued by both overexpression of BCY1 or deletion of RAS2, whereas ipl1-321 and spc24-9 CF loss was rescued by neither at a permissive or semirestrictive temperature. We next performed quantitative analysis of CF loss in ndc10-1 strains by growing cells in media to select for the CF, then plating on nonselective minimal media that contains limiting adenine, which enriches for the red color that arises when cells lose the CF. We scored for colonies that lost the CF in the first cell division (half-white, half-red) and for complete CF loss (red colonies, Table 1). The CF loss events in ndc10-1 were dramatically suppressed by deletion of RAS2 (∼20-fold) with a 1.3 × 10−2 CF loss rate in ndc10-1 compared with a 6.4 × 10−4 CF loss rate in ndc10-1 ras2Δ mutants (Table 1). Overexpression of BCY1 also suppressed CF loss rates in ndc10-1 cells by ∼4-fold (Table 1, 8.3 × 10−3 CF loss events in ndc10-1 cells compared with 1.9 × 10−3 CF loss events in ndc10-1 cells carrying BCY1). In addition to reduction of half sector colonies, inhibition of PKA activity reduced the frequency of red colonies (total CF loss) in ndc10-1 cells by ∼17-fold in the absence of RAS2 and ∼2-fold when BCY1 was overexpressed (Table 1). In summary, we have found that decreasing PKA activity rescues chromosome loss in ndc10-1 mutants at a permissive temperature.
Table 1. Reduction of PKA activity rescues CF loss in the ndc10-1 kinetochore mutant.
| Genotype (MATa) | Total Colonies | Chromosome Loss(Red) | Chromosome Loss in First Division (Red/White Half Sectors) |
|---|---|---|---|
| ndc10ΔHIS3 ndc10-1::kanMX6 CFIII | 11,632 | 6.9 × 10−2 (801)a | 1.3 × 10−2 (143) |
| ndc10ΔHIS3 ndc10-1::kanMX6 ras2ΔLEU2 CFIII | 11,118 | 4.1 × 10−3 (46) | 6.3 × 10−4 (7) |
| ndc10ΔHIS3 ndc10-1::kanMX6 CFIII pRS416 | 5,926 | 1.8 × 10−2 (104) | 8.3 × 10−3 (49) |
| ndc10ΔHIS3 ndc10-1::kanMX6 CFIII pRS416-BCY1 | 6,409 | 9.7 × 10−3 (62) | 1.9 × 10−3 (12) |
Numbers in brackets represent the total number of red or red/white colonies.
Kinetochore mutants have increased levels of PKA activity
One rational for the rescue of kinetochore defects when PKA activity is reduced is that PKA activity is elevated in kinetochore mutants. We employed two markers to determine if PKA levels are perturbed in spc24 mutants – the PKA dependent phosphorylation of the Msn2 transcription factor, and the phosphorylation profile of PKA substrates. We analyzed Msn2 phosphorylation in wild-type vs. kinetochore mutants using a phospho-CREB antibody that specifically recognizes PKA-dependent phosphorylation (Gorner et al. 2002). As a control, we analyzed Msn2 phosphorylation in wild-type strain expressing the dominant RAS2val19 allele. We quantified Msn2 PKA-specific phosphorylation compared with total Msn2 protein levels for all the samples. At a permissive temperature (25°) all spc24 mutants had similar Msn2 PKA-phosphorylation levels to wild-type cells (Figure 3A), whereas at a semipermissive temperature (31.5°), spc24-9 and spc24-8 mad2Δ cells had significantly higher (2.5- to 3-fold) Msn2 PKA-dependent phosphorylation levels compared with wild-type cells (Figure 3B). As expected, the phosphorylation of Msn2 in cells carrying RAS2val19 was dramatically increased by 5.7-fold at 25° and 4.1-fold at 31.5° compared with wild-type cells due to hyperactivation of the PKA pathway. Notably, ipl1-321 mutants had a 1.6-fold increase in Msn2 PKA-phosphorylation compared with the wild-type cells at 25°, and ndc10-1 had a 3.7-fold increase at 31.5°. Therefore, cells with defective kinetochore assembly or kinetochore-microtubule attachments have higher PKA activity. There was a reduction in total Msn2 protein levels in spc24-10, ndc10-1, and ipl1-321 cells, possibly due to a reduction in overall protein synthesis in these mutants at a semirestrictive temperature or because defects at the kinetochore affect Msn2 stability. Because Pgk1 levels are similar in all kinetochore mutants tested and spc24-10 mutants are not sensitive to cycloheximide (data not shown), we do not think that protein synthesis is reduced in spc24-10, ndc10-1, and ipl1-321 mutants.
Next, we examined the phosphorylation profile of PKA substrates in the kinetochore mutants using a phospho-PKA substrate antibody (α-sub). Strains were grown to log phase at permissive (25οC) and semipermissive (31.5°) temperatures, lysates were generated, and Western blot analysis was performed. A wild-type strain expressing RAS2val19 was used as control to detect PKA substrates. We used a chemiluminescence detection system and exposed the blot for a very short time (15 sec) before any protein band was saturated. Our image software detected six prominent PKA-phosphorylated substrates at 25° and five substrates at 31.5° that migrated between 70 kD and 200 kD (Figure 4, A and B). All six bands have a stronger signal in Ras2val19 compared with the wild-type cells at 25°, suggesting that these substrates are PKA phosphorylated. We quantified two proteins, Pa and Pb, as representative PKA substrates for all the strains. At 25°, all mutants have Pa phosphorylation levels similar to wild-type cells. However, when compared with wild-type cells, the phosphorylation level of Pb is increased by 2.6-fold in spc24-8 mad2Δ, 3.8-fold in ndc10-1 cells, and 7.2-fold in Ras2val19cells. At 31.5°, all the mutants, except for spc24-8 and mad2Δ mutants, have significantly increased phosphorylation of Pa by ∼2-fold compared with wild-type cells, whereas the phosphorylation of Pb does not change. We noticed that the 100 kD PKA substrate was absent from the ndc10-1 strain and the ∼160 kD band had reduced mobility (Figure 4B, ndc10-1 lane). Because Ndc10 is a 112 kD protein, we wondered whether Ndc10 was the 100 kD PKA substrate that was absent in ndc10-1 strains. However, we analyzed the PKA profile of strains carrying tagged versions of Ndc10 (Ndc10-13Myc and Ndc10-3HA), which should decrease mobility of the 100 kD band, and we did not detect any change in migration, suggesting that Ndc10 is not the 100 kD PKA substrate (data not shown). Therefore the 100 kD substrate could be an Ndc10-interacting protein that is destabilized at a semirestrictive temperature in the ndc10-1 strain. Overall, these data demonstrate that PKA substrates have increased levels of phosphorylation in multiple kinetochore mutant strains, suggesting that PKA activity is elevated in these strains.
PKA activity does not fluctuate during an unperturbed cell cycle
Strains defective in kinetochore function tend to accumulate in either metaphase or anaphase with a 2N population of DNA. Therefore, it is possible that we detect an increase in PKA activity in kinetochore mutants due to an overall increase in PKA activity in metaphase or anaphase. To address this possibility, we investigated Msn2 phosphorylation levels during the cell cycle. Wild-type cells were arrested in G1 phase at 25° with the mating pheromone α-factor, released into the cell cycle for 30 min and time points taken every 15 min until the cells had divided. We monitored Msn2 PKA-dependent phosphorylation, Msn2 protein levels, protein levels of the Clb2 mitotic cyclin, and nuclear division. At 0 min, Msn2 phosphorylation levels were low because mating pheromone inhibits adenylate cyclase (Liao and Thorner 1980). The metaphase-to-anaphase transition occurred between 75 and 90 min as Clb2 levels peaked and the nuclei divided (Figure 5A). Although Msn2 PKA-dependent phosphorylation appeared to increase at 75 to 90 min, so did Msn2 protein levels; therefore, when quantitated, no increase in Msn2 phosphorylation was detected at the metaphase-to-anaphase transition (Figure 5A). In fact, no significant difference was detected in Msn2 PKA-dependent phosphorylation between 30 and 120 min, suggesting that PKA activity remains constant during the cell cycle.
To specifically analyze Msn2 PKA-dependent phosphorylation in a population of metaphase-arrested cells, we treated wild-type cells with the microtubule depolymerizing agent nocodazole. Interestingly, the Msn2 PKA-dependent phosphorylation was significantly reduced upon nocodazole treatment (Figure 5B). In summary, our cell-cycle analysis suggests that Msn2 PKA-dependent phosphorylation does not specifically increase at the metaphase-to-anaphase transition. On the contrary, under conditions of cell-cycle arrest in G1 phase (mating pheromone treatment) or metaphase arrest (nocodazole treatment), Msn2 PKA-dependent phosphorylation is decreased. Therefore, our data suggest that the increase in PKA activity in kinetochore mutants is not simply due to an increase in PKA activity at the metaphase-to-anaphase transition.
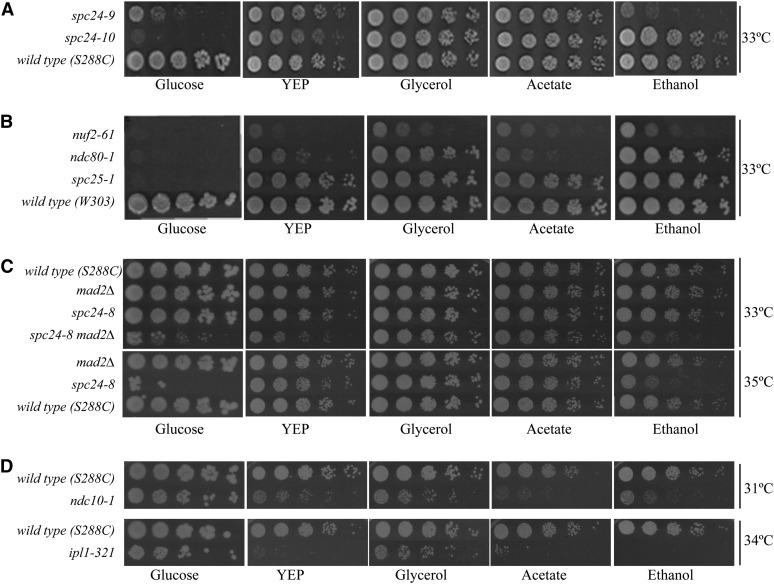
Nonfermentable carbon sources permit growth at a higher restrictive temperature for mutants of the Ndc80 complex
Cells grown in glucose media are presumed to have high PKA activity, whereas cells growing in nonfermentable carbon sources are presumed to have low PKA activity (Thevelein and de Winde 1999). Because reducing PKA activity increases the restrictive temperature of spc24 mutants, we tested the growth of spc24 mutants by plate assay on glucose vs. nonfermentable carbon sources (glycerol, acetate, and ethanol) or no added carbon source except for the yeast extract and peptone in the media (YEP) (Figure 6). spc24 mutants grew at a higher restrictive temperature on almost every media tested compared with glucose media (Figure 6, A and C). Notably, spc24-8 is rescued on nonfermentable carbon sources when the spindle checkpoint is active (spc24-8) or inactive (spc24-8 mad2Δ, Figure 6C). Therefore, exposing spc24 mutants to glucose, which triggers high PKA activity, is inhibitory to growth. We asked whether the glucose growth inhibition phenotype was specific to spc24 mutants or shared with other mutants in the Ndc80 complex. Ts mutants of other proteins of the Ndc80 complex, spc25-1, ndc80-1, and nuf2-61, were also rescued by growth on nonfermentable carbon sources (Figure 6B). To determine whether the glucose sensitivity was specific to the Ndc80 complex, we spotted ndc10-1 and ipl1-321 cells onto nonfermentable carbon sources (Figure 6D). Contrary to mutants in the Ndc80 complex, the fitness of the ndc10-1 mutant was optimal on glucose media, which is consistent with decreased fitness of ndc10-1 cells when PKA activity is reduced (Figures 1C, 2C, and 6D). Surprisingly, glucose was also the preferred carbon source of the ipl1-321 mutant, despite fact that lowering PKA activity improves the fitness of ipl1-321 cells (Figures 1C, 2C, and 6D). A previous report also found that the ipl1-2 mutant cannot be rescued by plating on media that slows growth, such as nonfermentable carbon sources (Tatchell et al. 2011). Because the growth rate of yeast is slower on nonfermentable carbon sources, it was possible that the rescue of spc24 mutants on nonglucose plates was due to a reduction in growth rate. Therefore, we tested growth of a subset of spc24 mutants on limiting nitrogen, cycloheximide, and rapamycin plates, all of which cause a reduction in growth rate. We did not detect any rescue of the spc24 Ts phenotype in any condition (data not shown). Therefore, spc24 mutants are rescued specifically by growth on nonglucose carbon sources and not generally by slowing cell growth rate.
Figure 6.
Strains carrying mutations in the Ndc80 kinetochore complex are rescued by growth on nonglucose carbon sources. Cell dilution assays were performed on the following media: 2% glucose, no added carbon source (YEP), 2% glycerol, 1% acetate, and 3% ethanol. (A) spc24-9, spc24-10 and isogenic wild-type strain (S288C background); (B) Ndc80 complex mutants nuf2-61, ndc80-1, spc25-1 and isogenic wild-type strain (W303 background); (C) spc24-8, mad2Δ, spc24-8 mad2Δ and isogenic wild-type strain (S288C); and (D) ndc10-1, ipl1-321 and isogenic wild-type strain (S288C). Plates were incubated at the indicated temperatures for 3 days. Different temperatures are shown depending on the restrictive temperature of each Ts allele.
Discussion
This work stemmed from the initial observation that decreasing cAMP-PKA activity, either by overproducing BCY1 or PDE2 or by deleting RAS2, rescued the lethality of spc24 kinetochore mutants (Figures 1 and 2). In addition, we had previously identified genetic interactions between pde2 and ira2, which are mutants of negative regulators of the cAMP-PKA pathway, and spc24 mutants (Montpetit et al. 2005). A simple model of these observations is that the cAMP-PKA pathway inhibits kinetochore function. However, our data suggest that the interaction between the cAMP-PKA pathway and the kinetochore is more complex. Inhibition of PKA activity is beneficial to the growth of spc24 and ipl1-321 mutants but detrimental to the ndc10-1 mutant, in which no kinetochore is assembled, and the spc24-8 mad2Δ double mutant, in which the spindle checkpoint has been abolished (Figure 1). In addition, all kinetochore mutants, except for the spindle checkpoint proficient spc24-8 mutant, have higher levels of PKA activity (Figures 3 and 4). The increased PKA activity is not due to accumulation of cells in metaphase or anaphase because during an unperturbed cell cycle, PKA-dependent Msn2 phosphorylation levels remain constant (Figure 5A). As well, metaphase-arrested cells have low levels of PKA activity (Figure 5B). Therefore, defects in the kinetochore, and not cell cycle arrest, cause an increase in PKA activity. Finally, we show that reduction of PKA activity rescues chromosome loss defects of the ndc10-1 mutant at a permissive temperature (Table 1). Therefore, the kinetochore is highly sensitive to fluctuations in PKA activity.
We demonstrate that, in addition to suppression of lethality by lowering PKA activity, mutants of the Ndc80 kinetochore complex are sensitive to glucose, and their growth defects are suppressed on nonfermentable carbon sources (Figure 6). It was shown previously that mutants of the APC are also suppressed by reducing Ras signaling and growth on nonglucose carbon sources (Irniger et al. 2000). Therefore, we expected that all kinetochore mutants that were rescued by reducing PKA signaling would preferentially grow on nonglucose carbon sources. However, we tested a variety of kinetochore mutants in addition to the mutants presented here and found no strict correlation between growth rescue by inhibition of PKA signaling and growth rescue on nonglucose carbon sources. For example, a strain carrying a mutation in CTF13, which codes for a protein in the CBF3 inner kinetochore complex, displayed no growth changes upon overexpression of BCY1 but was rescued by growth on glycerol or galactose media (data not shown). The growth of the ipl1-321 mutant is improved upon deletion of RAS2 or overexpression of BCY1 but not when plated on nonfermentable carbon sources (Figures 1, 2, and 6). Although the viability of all mutants of the Ndc80 kinetochore complex was rescued by reducing PKA signaling or plating on a nonglucose carbon source, mutants of other kinetochore complexes, such as the CBF3, COMA, and Dam1 complexes, displayed variable phenotypes (data not shown). Therefore, the sensitivity of kinetochore mutants to fluctuations in PKA activity is not strictly correlated to the presence of glucose in the media.
The chromosome loss defect of ndc10-1 cells is reduced by 20-fold upon deletion of RAS2 and 4-fold overexpression of BCY1 at 25° (Table 1). However, when grown at 30°, we find that overexpression of BCY1 causes lethality to ndc10-1 strains and that ndc10-1 ras2Δ strains grow more poorly than do ndc10-1 strains at 32° (Figures 1C and 2C). At a permissive temperature, kinetochore complexes still associate with centromere DNA in the ndc10-1 mutant, whereas at a restrictive temperature, no kinetochore complexes are able to assemble on the centromere (He et al. 2001; Janke et al. 2001; Nekrasov et al. 2003; Ortiz et al. 1999). Therefore, lowering PKA activity only rescues CF loss of the ndc10-1 mutant when there is an assembled kinetochore that is able to attach to microtubules. Perhaps this is why we did not detect rescue of CF loss in ipl1-321 or spc24-9 mutants in which kinetochore-microtubule attachment is impaired.
We find that the ipl1-321 growth defect is suppressed by overexpression of PDE2 and BCY1 or by deletion of RAS2, all of which lower PKA activity (Figures 1C and 2C). In addition, expression of the dominant RASval19 allele is detrimental to ipl1-321 growth (Figure 2D). The rescue of ipl1-321 growth defect by lowering PKA activity is intriguing in light of recent data demonstrating that reduction of target of rapamycin (TOR) complex 1 (TORC1) activity also suppresses the growth defect of an ipl1-2 mutant (Tatchell et al. 2011). TOR and PKA are the two major signaling pathways that activate cell growth in response to nutrients by regulating processing, such as translation, ribosome biogenesis, and glucose metabolism (Smets et al. 2010; Soulard et al. 2009). TORC1 and PKA have been shown to regulate common target proteins, and recent data demonstrate that TOR can activate PKA toward a subset of substrates (Soulard et al. 2010). Reduction of TORC1 activity suppressed the chromosome loss defect in ipl1-2 mutants, whereas we did not detect rescue of CF loss in ipl1-321 mutants upon reduction of PKA activity (Tatchell et al. 2011). Nonetheless, our data combined with Tatchell et al. (2011) strongly support a link between nutritional status and kinetochore function.
Activation of the PKA pathway is known to be inhibitory to the APC, possibly via phosphorylation of Cdc20 (Anghileri et al. 1999; Bolte et al. 2003; Irniger et al. 2000; Searle et al. 2004). For example, the growth defect of apc10-22 mutants, which stabilizes Pds1, is suppressed by deletion of RAS2 (Irniger et al. 2000). Stabilization of Pds1 (securin) prevents chromosome separation due to inhibition of Esp1 (separase). The APC is also inhibited, and Pds1 is stabilized upon activation of the spindle checkpoint by the interaction of the Mad2 checkpoint protein with Cdc20 (Clarke and Bachant 2008). However, no study has addressed the interaction between the PKA pathway and the spindle checkpoint. In this work, we monitored Msn2 PKA-dependent phosphorylation in cells arrested in metaphase due to induction of the spindle checkpoint upon nocodazole treatment (Figure 5B). We found that Msn2 PKA-dependent phosphorylation is reduced under these conditions, suggesting that PKA activity may be reduced during the spindle checkpoint. Interestingly, the only kinetochore mutant that did not display high levels of PKA activity was spc24-8, which arrests in metaphase due to activation of the spindle checkpoint (Montpetit et al. 2005) (Figures 3 and 4). It is possible that reducing PKA levels suppresses spc24 and ipl1 mutants due to spindle checkpoint activation; however, this does not explain why the viability of the spc24-8 mutant, which is spindle checkpoint proficient, is improved by reducing PKA activity (Figures 1 and 2). Activation of the PKA pathway causes phosphorylation of the Msn2 nuclear localization sequence on serine residues, prevention of Msn2 nuclear import, and restoration of Msn2 to the cytoplasm (Gorner et al. 1998, 2002; Jacquet et al. 2003). Inhibition of PKA activity upon nocodazole treatment might evoke a general stress response that results in dephosphorylation of Msn2 and subsequent localization to the nucleus. In fact, microarray studies performed after treatment of yeast cells with benomyl, another microtubule poison, demonstrated that expression of Msn2 genes is induced (Lucau-Danila et al. 2005). Whether inhibition of PKA activity rescues spc24 mutants due to activation of a Msn2-dependent stress response remains to be tested.
Because it is difficult to separate the spindle checkpoint response from a general cell stress response upon addition of microtubule poisons, deletion of the Mad2 spindle checkpoint protein is an alternative method to assess the spindle checkpoint. We find that overexpression of BCY1 and PDE2 only rescues viability of strains that have a partially assembled kinetochore and Mad2 present in the cell (Figure 7). For example, overexpression of BCY1 and PDE2 rescues the growth defect of the spc24-8 (spindle checkpoint active) mutant but is detrimental to the growth of the spc24-8 mad2Δ (spindle checkpoint defective) mutant (Figures 1B and 7). The APC specificity factor Cdc20 is phosphorylated on PKA consensus sites after DNA damage, and inactivation of PKA accelerates Pds1 destruction (Searle et al. 2004). If phosphorylation of Cdc20 by PKA prevents an interaction between Cdc20 and Mad2, then decreasing PKA activity may enrich the Cdc20-Mad2 interaction and spindle checkpoint response. If Mad2 is not present, then decreasing PKA activity may accelerate Pds1 destruction, which would force spc24-8 mad2 cells into anaphase with defective chromosome attachments. One complication to this argument is that deletion of RAS2 rescues the growth defect of both spc24-8 and spc24-8 mad2Δ strains (Figures 2B and 7). Likely, there is more than one mechanism by which a reduction in PKA activity rescues viability of kinetochore mutants.
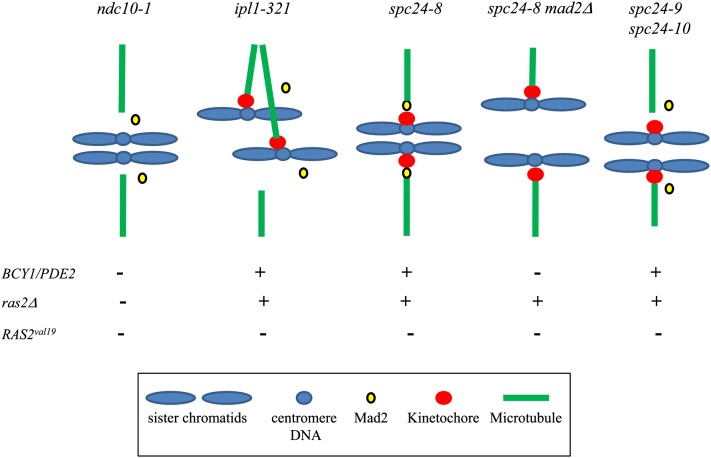
Figure 7.
Summary of kinetochore and PKA interactions. At a restrictive temperature, the ndc10-1 mutant does not assemble a kinetochore; therefore, microtubules cannot attach and Mad2 cannot be recruited. The ipl1-321 mutant has functional kinetochores that attach to the same pole, but Mad2 is not recruited (Gillett et al. 2004). The spc24-8 mutant is checkpoint active; therefore, Mad2 is presumed to be properly localized to the kinetochore. The spc24-8 mad2Δ mutant lacks Mad2 and is spindle checkpoint defective; however, kinetochores are still present. The state of the kinetochore-microtubule interaction in spc24-8 mad2Δ cells has not been investigated. The kinetochores in spc24-9 and spc24-10 mutants have defects in microtubule attachment and are presumed to mislocalize Mad2 as the spindle elongates in both mutants, despite attachment defects. In addition, similar mutants in the Ndc80 complex mislocalize Mad2 (Gillett et al. 2004). Genetic interactions upon overexpression of BCY1 or PDE2, deletion of RAS2 (ras2Δ), or expression of RAS2val19 in each kinetochore mutant is represented with a plus (+) sign if growth defects were improved and minus (−) sign if growth defects were exacerbated.
An unexpected finding from our study is that kinetochore mutants have increased levels of PKA activity when grown at a semipermissive temperature (Figures 3 and 4). These data suggest that defects in the kinetochore may trigger activation of the PKA pathway, possibly via components of the cAMP-PKA pathway, that are nuclear localized. Bcy1, the PKA regulatory subunit, is predominantly in the nucleus when cells are grown in glucose (Griffioen et al. 2000). It remains to be determined whether a nuclear pool of Bcy1 or a nuclear-localized PKA catalytic subunit, such as Tpk1, interacts with the kinetochore and whether disruption of kinetochore function releases active Tpk1. It has been demonstrated that loss of one copy of BCY1 in a diploid strain increases the rate of chromosome loss, suggesting that Bcy1 may be important for kinetochore function (Magtanong et al. 2011). How the kinetochore impinges upon the cAMP-PKA pathway and whether this function is conserved in higher eukaryotes will be an interesting subject of future research.
Supplementary Material
Acknowledgments
We would like to thank Merisa Mok for helping with colony counting for the CF loss assays. We would also like to thank Tony Bretscher for the gift of the ras2Δ strain, Francisco Estruch for the gift of the Msn2 antibody, Paul Herman for the gift of the Ras2val19 plasmid, Brie Lavoie for the gift of the ipl1-321 strain, Kevin Morano for the gift of the BCY1 plasmid, and Kristin Baetz for critical comments on the manuscript. K. Ho was supported by a Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best Canada Graduate Scholarship. This work was supported by CIHR operating grant MOP-84242 to V. Measday. V. Measday is a Canada Research Chair in enology and yeast genomics.
Footnotes
Communicating editor: M. Tyers
Literature Cited
- Anghileri P., Branduardi P., Sternieri F., Monti P., Visintin R., et al. , 1999. Chromosome separation and exit from mitosis in budding yeast: dependence on growth revealed by cAMP-mediated inhibition. Exp. Cell Res. 250: 510–523 [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A. W., 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D. L., Murray A. W., 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159: 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte M., Dieckhoff P., Krause C., Braus G. H., Irniger S., 2003. Synergistic inhibition of APC/C by glucose and activated Ras proteins can be mediated by each of the Tpk1–3 proteins in Saccharomyces cerevisiae. Microbiology 149: 1205–1216 [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., et al. , 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163–172 [DOI] [PubMed] [Google Scholar]
- Choy J. S., Mishra P. K., Au W. C., Basrai M. A., 2012. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim. Biophys. Acta. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D. J., Bachant J., 2008. Kinetochore structure and spindle assembly checkpoint signaling in the budding yeast, Saccharomyces cerevisiae. Front. Biosci. 13: 6787–6819 [DOI] [PubMed] [Google Scholar]
- Connelly C., Hieter P., 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R., Peter M., 2008. Nutrient signals driving cell growth. Curr. Opin. Cell Biol. 20: 678–687 [DOI] [PubMed] [Google Scholar]
- Dubacq C., Guerois R., Courbeyrette R., Kitagawa K., Mann C., 2002. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1: 568–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Xu H. P., Michaeli T., Ballester R., Sass P., et al. , 1990. Mutations of the adenylyl cyclase gene that block RAS function in Saccharomyces cerevisiae. Science 247: 464–467 [DOI] [PubMed] [Google Scholar]
- Gelade R., Van de Velde S., Van Dijck P., Thevelein J. M., 2003. Multi-level response of the yeast genome to glucose. Genome Biol. 4: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillett E. S., Espelin C. W., Sorger P. K., 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh P. Y., Kilmartin J. V., 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121: 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Martinez-Pastor M. T., Estruch F., Ammerer G., et al. , 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W., Durchschlag E., Wolf J., Brown E. L., Ammerer G., et al. , 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffioen G., Anghileri P., Imre E., Baroni M. D., Ruis H., 2000. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem. 275: 1449–1456 [DOI] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K., 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195–206 [DOI] [PubMed] [Google Scholar]
- Heo S. J., Tatebayashi K., Ikeda H., 1999. The budding yeast cohesin gene SCC1/MCD1/RHC21 genetically interacts with PKA, CDK and APC. Curr. Genet. 36: 329–338 [DOI] [PubMed] [Google Scholar]
- Irniger S., Baumer M., Braus G. H., 2000. Glucose and ras activity influence the ubiquitin ligases APC/C and SCF in Saccharomyces cerevisiae. Genetics 154: 1509–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Renault G., Lallet S., De Mey J., Goldbeter A., 2003. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 161: 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz J., Lechner J., Shevchenko A., Shevchenko A., et al. , 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20: 777–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis S., Mitchell A., 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Koshland D., Hieter P., 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155: 351–372 [DOI] [PubMed] [Google Scholar]
- Li J. M., Li Y., Elledge S. J., 2005. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with the ras/protein kinase A pathway and a novel subunit required for Ask1 association. Mol. Cell. Biol. 25: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H., Thorner J., 1980. Yeast mating pheromone alpha factor inhibits adenylate cyclase. Proc. Natl. Acad. Sci. USA 77: 1898–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucau-Danila A., Lelandais G., Kozovska Z., Tanty V., Delaveau T., et al. , 2005. Early expression of yeast genes affected by chemical stress. Mol. Cell. Biol. 25: 1860–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., McQueen J., Cuschieri L., Vogel J., Measday V., 2007. Spc24 and Stu2 promote spindle integrity when DNA replication is stalled. Mol. Biol. Cell 18: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L., Ho C. H., Barker S. L., Jiao W., Baryshnikova A., et al. , 2011. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat. Biotechnol. 29: 505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B., Thorne K., Barrett I., Andrews K., Jadusingh R., et al. , 2005. Genome-wide synthetic lethal screens identify an interaction between the nuclear envelope protein, Apq12p, and the kinetochore in Saccharomyces cerevisiae. Genetics 171: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V. S., Smith M. A., Peak-Chew S., Kilmartin J. V., 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 4931–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Sass P., Wigler M., 1987. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 3629–3636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J., 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13: 1140–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Herman P. K., 2011. Antagonistic interactions between the cAMP-dependent protein kinase and Tor signaling pathways modulate cell growth in Saccharomyces cerevisiae. Genetics 187: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P., Field J., Nikawa J., Toda T., Wigler M., 1986. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83: 9303–9307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle J. S., Schollaert K. L., Wilkins B. J., Sanchez Y., 2004. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 6: 138–145 [DOI] [PubMed] [Google Scholar]
- Smets B., Ghillebert R., De Snijder P., Binda M., Swinnen E., et al. , 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr. Genet. 56: 1–32 [DOI] [PubMed] [Google Scholar]
- Soulard A., Cohen A., Hall M. N., 2009. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Soulard A., Cremonesi A., Moes S., Schutz F., Jeno P., et al. , 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21: 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Choe H. R., Nishida Y., Yamawaki-Kataoka Y., Ohnishi S., et al. , 1990. Leucine-rich repeats and carboxyl terminus are required for interaction of yeast adenylate cyclase with RAS proteins. Proc. Natl. Acad. Sci. USA 87: 8711–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H., 2007. Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng. 104: 245–250 [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., et al. , 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108: 317–329 [DOI] [PubMed] [Google Scholar]
- Tatchell K., Makrantoni V., Stark M. J., Robinson L. C., 2011. Temperature-sensitive ipl1–2/Aurora B mutation is suppressed by mutations in TOR complex 1 via the Glc7/PP1 phosphatase. Proc. Natl. Acad. Sci. USA 108: 3994–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M., de Winde J. H., 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33: 904–918 [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Scott J. D., et al. , 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley J., Stukenberg P. T., 2011. The Ndc80 complex: integrating the kinetochore’s many movements. Chromosome Res. 19: 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott A., Shaner L., Morano K. A., 2005. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 170: 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G., 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76: 563–591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.