Abstract
Porcine circovirus type 2 (PCV2) is associated with postweaning multisystemic syndrome in pigs, whereas the ubiquitous related porcine circovirus type 1 (PCV1) is nonpathogenic. Corroborating an earlier observation in PCV2, Rep and Rep’ proteins encoded by ORF1 are essential for the initiation of PCV2 replication. Cap protein encoded by ORF2 has a potential causative role in the initiation of PCV2 replication and contains a type-specific epitope. The putative ORF3 of PCV2 oriented in the opposite direction within ORF1 is unknown. In this study, ORF3-encoding protein of PCV2 was expressed in vitro as a fusion protein (GST-ORF3 protein), and monoclonal antibodies (MAbs) to the PCV2-ORF3-encoding protein were generated and biologically characterized. The mRNA transcript of ORF3 was characterized during a productive infection in PK-15 cells, and the PCV2 infectious DNA clone lacking ORF3 was constructed. GST-ORF3 protein, with an approximate molecular weight of 37.7 kDa, was obtained from the Escherichia coli transformed with the recombinant vector pGEX-4T-1-F3 after codon optimization of ORF3 DNA sequence. Four MAbs reacted strongly to the ORF3-encoding protein expressed in PK-15 cells in immunohistochemical staining. The mRNA transcript of ORF3 was confirmed in RT-PCR, Northern blot, and sequencing analyses. The progeny PCV2 virions were not revealed in the PK-15 cells transfected by the PCV2 infectious DNA clone without ORF3. These results demonstrate that the ORF3 of PCV2 can be transcribed and expressed and that ORF3-encoding protein plays a pivotal role in viral replication.
Introduction
The circoviruses are a family of small non-enveloped icosahedral viruses infecting parrots, geese, canaries, pigeons, and pigs.(1–6) Porcine circovirus (PCV) isolated as a persistent contaminant from a porcine kidney cell line is non-pathogenic for experimentally infected pigs and is designated PCV1.(3,7–9) In 1991 a new disease, postweaning multisystemic wasting syndrome (PMWS), was found in pigs.(10,11) A novel strain of PCV was isolated from pigs with PMWS and named PCV2.(12–15) Pathogenic and phylogenetic studies show that PCV1 and PCV2 belong to two different genotypes.(16)
PCV has an ambisense, single-stranded, closed-circular genome of 1759 bp for PCV1, 1767 bp, and 1768 bp for PCV2.(12,16–19) The overall DNA sequence homology within the PCV1 or PCV2 isolates is greater than 90%, while the homology between PCV1 and PCV2 is only about 76%.(12,17) The genomic DNA of both PCV1 and PCV2 has a similar genomic organization, containing 11 predicted open reading frames (ORFs).(17) ORF1 and ORF2, oriented in opposite directions, are the two major ORFs in PCV1 and PCV2. ORF1 encodes Rep protein in viral DNA replication, while ORF2 encodes an immunogenic capsid protein.(20,21) Cheung and associates further reported that 9 PCV2-specific RNAs and 12 PCV1-specific RNAs were detected during productive infection in PK-15 cells. The ORF3 gene of PCV2 identified in 2005 encodes an 11.9 kDa protein, which completely overlaps the ORF1 gene and is oriented in the opposite direction. The novel protein is not essential for PCV2 replication but can induce apoptosis in infected cells and plays an important role in viral pathogenesis by its apoptotic activity.(22,23)
Until now the role and expression of ORF3-encoding protein were unknown in PCV2 replication. In this study, we report for the first time the expression of the ORF3-encoding protein of PCV2 in prokaryotic and eukaryotic cells, and the creation of a monoclonal antibody (MAb) to the PCV2-ORF3-encoding protein. The role of ORF3 in PCV2 replication was characterized in the PK-15 cell line by the construction of a PCV2 infectious DNA clone without ORF3.
Materials and Methods
Virus, cell line, and antiserum
The PCV2 isolate HZ0201was originally isolated from a superficial inguinal lymph node sample of a pig with naturally occurring PMWS.(19) The PCV1 virus (ISUVDL PK-15 2000) was kindly provided by Dr. K.J. Yoon (Veterinary Diagnostic Laboratory, Iowa State University, Ames, IA). The PCV1-free PK-15 cell line was kept in our laboratory and maintained in minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal calf serum (FCS, Gibco BBL, New York, NY) at 37°C with 5% CO2. Swine PCV2-positive serum was described previously.(24)
Expression of ORF3-encoding protein (GST-ORF3 protein) in E. coli
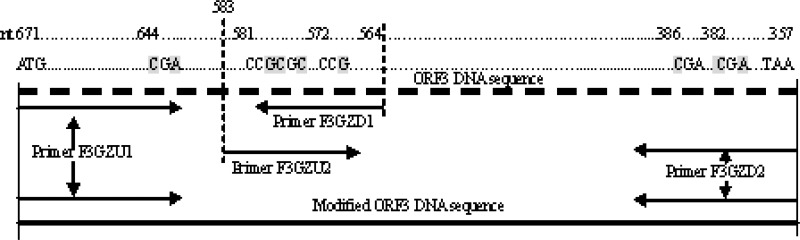
To generate the recombinant ORF3-encoding protein, PCV2 virions from the infected PK-15 cells were purified through sucrose gradient and diluted with the nuclease-free ddH2O. The recombinant vector pGEX-4T-1-F3 was constructed as follows. Briefly, a modified 108 bp fragment (nt 671 to 564) was amplified with the primers F3GZU1 containing the Eco RI site and F3GZD1 (Table 1) from the DNA of PCV2 strain HZ0201 (Genbank accession no. AY188355), and another modified 227 bp fragment (nt 583 to 357) was amplified with the primers F3GZU2 and F3GZD2 containing the Xho I site (Table 1). Then the full ORF3 fragment of 315 bp (nt 671 to 357) was amplified by PCR from the 108 bp and 227 bp fragments with the primers F3GZU1 and F3GZD2 (Fig. 1). The PCR product was digested with Eco RI and Xho I and cloned into the vector pGEX-4T-1 (Amersham, Pharmacia Biotech AB, Uppsala, Sweden). The resultant plasmid (pGEX-4T-1-F3) was transformed into E. coli BL21 (Invitrogen, Carlsbad, CA) and sequenced. The expression was induced by isopropylthio-β-D-galactoside (IPTG, Amersham) according to the manufacturer's protocol.
Table 1.
Oligonucleotide Primers in This Study
| Primer | Primer sequencea | Application |
|---|---|---|
| F3GZU1 | >5′-GCGAATTCATGGTAACCATCCCACCACTTGTTTCTCGAT-3′ | Amplification of ORF3 DNA sequence |
| F3GZD1 | <5′-TGTGCGGCCAGCGCGGTGTGGTAA-3′ | Rare codon modification of ORF3 DNA sequence |
| F3GZU2 | >5′-TACCACACCGCGCTGGCCGCACAATGA-3′ | Rare codon modification of ORF3 DNA sequence |
| F3GZD2 | <5′-GGCCTCGAGTTACTTATTGAATGTGGAGCTTCGCGATCGCAA-3′ | Amplification of ORF3 DNA sequence |
| F3TF3 | <5′-GCGGATCCTTACTTATTGAATG-3′ | pEGFP construction |
| F671 | >5′-ATGGTAACCATCCCACCACTTG-3′ | RT-PCR |
| R357 | <5′-TTACTGATGGAGTGTGGA-3′ | RT-PCR |
| F-sac2 | >5′-GAACCGCGGGCTGGCTGAACTTTTGAAAGT-3′ | Amplification of full-length PCV2 genome |
| R-658 | <5′-TTCTTCACCTTGGTAACCATC-3′ | Mutation of ORF3 initiation codon |
| F661 | >5′-ATGGTTACCAAGGTGAAGAAGT-3′ | Mutation of ORF3 initiation codon |
| R-sac2 | <5′-GCACCGCGGAAATTTCTGACAAACGTTACA-3′ | Amplification of full-length PCV2 genome |
Primer direction is designated at beginning of sequence.
FIG. 1.
Modified schematic map of PCV2 ORF3 DNA sequence expressed in E. coli. The mutated nucleotide is in gray highlight. A fragment of 108 bp in length was obtained with the primers F3GZU1 and F3GZD1, while a fragment of 227 bp in length was amplified with the primers F3GZU2 and F3GZD2. Modified DNA sequence of ORF3 was generated by primers F3GZU1 and F3GZD2 from fragments of 108 bp and 227 bp.
Purification of the recombinant GST-ORF3 protein
Purification of GST-ORF3 protein was performed by the following two methods. In the first method, the supernatant of the cell lysates containing GST-ORF3 protein was loaded to HiTrap affinity column (Amersham) according to the manufacturer's protocol, and GST-ORF3 protein was purified. In the other method, the purified GST-ORF3 protein was generated by cutting off the specific band after SDS-PAGE. Briefly, the lysates were electrophoresed through a 15% SDS-PAGE gel. After staining with Coomassie blue, the interest gel band was cut off. The recombinant GST-ORF3 protein was released from the specific gel within a dialyzer (Serva, Heidelberg, Germany) by electrophoresis in protein electrophoresis buffer (25 mM Tris base, 192 mM glycine, 3.5 mM SDS). After that, the purified protein was concentrated with PEG20000. Total protein concentration was determined by the Bradford assay with bovine serum albumin as a standard.
Preparation of MAb against ORF3-encoding protein and isotyping
The MAbs against ORF3-encoding protein were produced as described previously.(24) Briefly, the SPF BALB/c mice were immunized subcutaneously (S/C) with complete Freund's adjuvant (Sigma-Aldrich, St. Louis, MO)-GST-ORF3 protein emulsion (50 μg GST-ORF3 protein per mouse). Three weeks later, the mice were injected intraperitoneally (i.p.) with incomplete Freund's adjuvant-GST-ORF3 protein emulsion. After another 3 weeks, each mouse was given 0.1 mg GST-ORF3 protein i.p. The mice were subsequently euthanized 3 days after the third immunization, and their splenocytes were collected and fused with SP2/0 myeloma cells. Hybridoma supernatants were screened by an indirect ELISA for the presence of the specific antibodies using GST-ORF3 protein, recombinant Cap protein of PCV2,(24) the purified PCV2 and PCV1 virions, and cell lysates of E. coli BL21 strain containing the parental pGEX-4T-1 vector, PK-15 cells, and RPMI 1640 medium as antigens. Finally, analysis of MAb IgG subtype was performed with standard procedures illuminated by the protocols of SBA Clonotyping™ System/HRP (Southern Biotechnology Associates, Birmingham, AL).
SDS-PAGE and Western blot analyses
Bacterial lysates and the purified GST-ORF3 protein were separated by SDS-PAGE on 15% polyacrylamide gels and were transferred onto nitrocellulose membranes (Amersham) in a transfer buffer (20 mM Tris-HCl, 190 mM glycine, 20%[vol/vol] methanol [pH 8.3]) using a semi-dry transfer unit (Amersham) at 50 V for 1 h. The membranes were blocked with 5% skimmed milk in TTBS (Tris-buffered saline containing 0.05% Tween-20) overnight and incubated with rabbit anti-GST polyclonal antibody (1:1000, Zymed Laboratories, South San Francisco, CA), diluted hybridoma supernantants (1:10) for 2 h at 37°C, respectively. After three washes in TTBS, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG or goat anti-mouse IgG (KPL, Kirkegaard & Perry Laboratories, Gaithersburg, MD) for 1 h. Color development was performed using 3,3’-5,5’-tetramethylbenzidine (Promega, Madison, WI).
Virus neutralization assay
To access the virus neutralization (VN) activity of the MAbs, a 100 μL volume of serial ten-fold dilutions (10−1 to 10−6) of MAb in MEM was mixed respectively with an equal volume 10−4.75 TCID50/mL PCV2 virions and was placed in an incubator at 37°C for 3 h. Following this incubation, the mixture was mixed with PK-15 cells (2×104 cell/mL) dispersed from confluent monolayers at a ratio of 2:8, and this suspension was added to each well in a 96-well plate. After a further incubation for 72 h, the cell monolayers were stained by an immunohistochemical method (IHC) as described elsewhere.(25) Briefly, 72 h after infection, the infected PK-15 cells were rinsed three times in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH7.4]), and fixed in PBS containing 4% paraformaldehyde (pH 7.4) at room temperature for 30 min. After being washed twice in PBS, the PK-15 cells were incubated with PBSTW (0.05% Tween-20, 0.1% Triton X-100, 4% skimmed milk in PBS) for 1 h at 37°C. After three washes in PBST (0.25% Tween-20 in PBS), the cells were incubated with MAbs to PCV2 capsid protein for 90 min. After they were washed three times with PBST, the cells were incubated in HRP-labeled goat anti-mouse IgG for 1 h. Color development was carried out with 3-Amino-9-ethylcarbazole (Sigma).
Expression of ORF3-encoding protein in eukaryotic cells
To express ORF3-encoding protein in eukaryotic cells, ORF3 DNA sequence of PCV2 containing the Eco RI and the Bam HI sites was amplified by PCR from the genomic DNA of PCV2 isolate HZ0201 with the primers F3GZU1 and F3TF3 (Table 1). Afterwards, the PCR products of 315 bp in length, the vector pEGFP-C2 (Clontech, Franklin Lakes, NJ), and pCI-neo (Promega) were digested respectively with the Eco RI and Bam HI enzymes and were ligated at 16°C overnight and transformed into the competent cells of E. coli Top10 strain (Invitrogen). After being sequenced, the resultant plasmids pEGFP-C2-F3 and pCI-neo-F3 were purified respectively using the Bioscience Plasmid Purified Kit (Marligen, Biosciences, Ijamsville, MD) and transfected into PK-15 cells. Briefly, when PK-15 cells reached 90–95% confluency, the cells were transfected respectively with the plasmids pEGFP-C2-F3 and pCI-neo-F3 using the lipofectamine™ 2000 reagent (1 mg/mL; Life Technology, Grand Island, NY) according to the manufacturer's protocol. Mock-transfected cells with pEGFP-C2 and pCI-neo vectors were included as controls. From 5 to 60 h post-transfection, the PK-15 cells were observed. At 48 h post-transfection, the PK-15 cells were detected by using the above-mentioned IHC with MAbs to PCV2-ORF3-encoding protein.
RT-PCR and Northern blot analysis of ORF3 transcription
To identify the mRNA of ORF3 transcription, total cell RNAs were prepared from PCV2-infected PK-15 cells at 10 to 60 h post-inoculation (h.p.i.) and RT-PCR was performed. Briefly, between 10 and 60 h.p.i., the cells were lysed and the RNAs were extracted with Trizol according to the manufacturer's instructions (Gibco). The resulting RNA samples were incubated with DNase I (Promega) for 60 min at 37°C to remove any contaminating viral DNA. Reverse transcription was carried out with 1 μg of total RNA using ORF3 specific reverse primer R357 following the RevertAid First Strand cDNA Synthesis Kit (MBI Fermentas, Burlington, Canada). PCR was performed with the primers F671 and R357 by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and polymerization at 72°C for 1 min, with a final extension at 72°C for 10 min. PCR products were analyzed on a 1.2% agarose gel and identified by DNA sequencing. The RNA without reverse transcription was used as a control in order to avoid DNA contamination. Northern blot analysis was performed as described elsewhere.(26) Briefly, the RNA samples were subjected to electrophoresis in 2.2 M formaldehyde gel and transferred onto a nylon membrane (Amersham). The transferred RNA was further hybridized with 32P-labeled ORF3 DNA probe in 3.2 M tetramethylammonium chloride solution containing 1% sodium-dodecyl sulfate overnight at 60°C. Finally, the membrane was washed twice in 0.1×SSC containing 0.5% SDS for 30 min at 60°C and then exposed to X-ray films.
Expressing detection of ORF3-encoding protein in PCV2 replication
Briefly, the monolayer of PK-15 cells was inoculated with PCV2 isolate HZ0201 as described previously.(19) Between 5 and 60 h.p.i., the monolayer of PK-15 cells was screened using the above-mentioned IHC with MAbs to PCV2-ORF3-encoding protein to detect the cellular expression of the putative ORF3-encoding protein during PCV2 replication.
Construction of PCV2-mORF3 and PCV2-normal infectious DNA clones
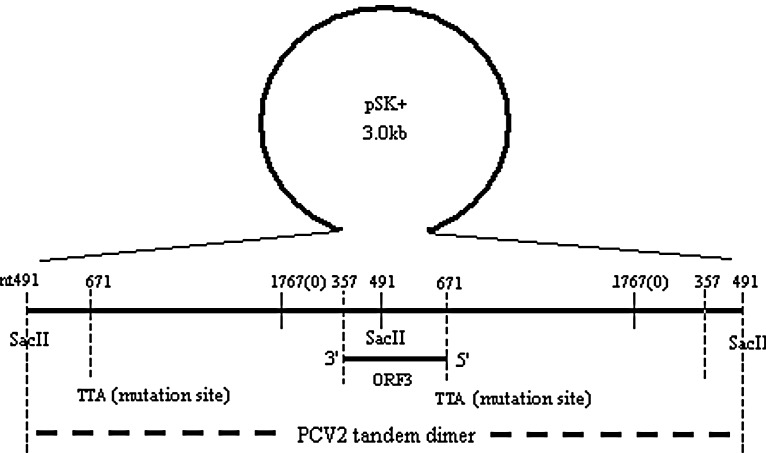
To generate the infectious DNA clone lacking ORF3 (Fig. 2), the initiation codon of ORF3 was mutated to ATT from ATG by designing the specific primers. Briefly, a modified 190 bp fragment (nt 491 to 680) was amplified with the primers F-sac2 containing the Sac II site and R-658 (Table 1) with the genomic DNA from the PCV2 strain HZ0201. Primers F661 and R-sac2 containing the Sac II site (Table 1) generated another subgenomic modified DNA fragment of 1603 bp (nt 661 to 491). Then a 1767 bp fragment of the complete PCV2 genome (nt 491 to 491) without ORF3 was amplified by PCR from the 190 bp and 1603 bp fragments using the primers F-sac2 and R-sac2. The PCR product was digested further with the Sac α enzyme and cloned into pBluescript SK+(pSK+) vector (Stratagene, La Jolla, CA). The competent cells of E. coli Top 10 strain were used for transformation. Recombinant plasmids containing the full-length PCV2 genome without ORF3 were isolated with the Bioscience Plasmid Purified Kit (Marligen) and were verified by restriction enzyme digestion and sequencing. The full-length PCV2 genomes were excised from the recombinant pSK+ vector with the Sac II digestion and dimerized at 37°C for 30 min to produce tandem dimmers. The tandem dimmers were subsequently cloned into the digested pSK+ vector, named pSK-PCV2-mORF3. The resultant plasmid pSK-PCV2-mORF3 was purified and transfected into PK-15 cells using the above-mentioned method. As a positive control, the construction of PCV2 infectious DNA clone (pSK-PCV2-normal) was generated with the primers F-sac2 and R-sac2 as described by Fenaux and colleagues.(27,28) At 48 h post-transfection, the above-mentioned IHC was applied to detect the presence of PCV2 virions with swine anti-PCV2 serum. In addition, PCV2-infected and PCV-free PK-15 cells were also used as controls.
FIG. 2.
Construction of PCV2 infectious DNA clone lacking ORF3. Linear PCV2 genome without ORF3 was amplified by mutating initiation codon ATG of ORF3 to ATT with designing the specific primers. The PCV2 infectious DNA clone was generated by ligating two full-length linear PCV2 genome without ORF3 in tandem into vector pSK+.
Results
Expression and purification of GST-ORF3 protein expressed in E. coli
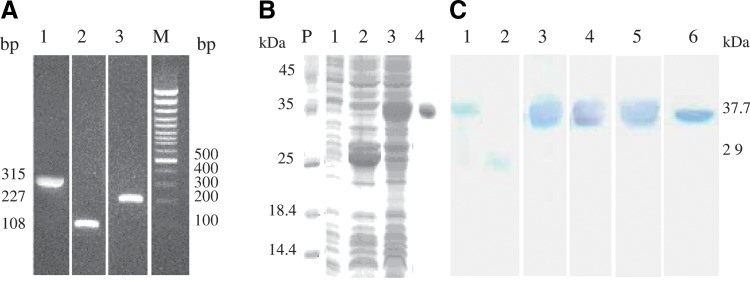
According to Kane's report,(29) 12 rare codons of E. coli were found within PCV2-ORF3 DNA sequence. As shown in Figure 1 and Table 1, to express ORF3-encoding protein in E. coli efficiently, nine bases of six rare codons within ORF3 DNA sequence for E. coli were mutated artificially by the designing specific primers and two DNA fragments of 108 bp and 227 bp were obtained by PCR. Subsequently, the two fragments generated an entirely rebuilt ORF3 DNA sequence of 315 bp by PCR technique (Fig. 3A). The recombinant pGEX-4T-1-F3 was constructed by inserting the ORF3 DNA fragment of 315 bp into the multiple cloning site (MCS) of pGEX-4T-1vector containing the glutathione S-transferase (GST) gene. After induction with IPTG, the GST-ORF3 protein recognized with rabbit anti-GST polyclonal antibody, an approximate molecular weight of 37.7 kDa, was expressed in the E. coli BL21 strain containing the pGEX-4T-1-F3 plasmid (Fig. 3B,C). Furthermore, to purify the expressed GST-ORF3 protein, the supernatant of the bacterial lysates containing GST-ORF3 protein was loaded to HiTrap affinity column. Unfortunately, the collected elution was not reacted with rabbit anti-GST polyclonal antibody in Western blot assay, indicating that there was no GST-ORF3 protein in the elution. Subsequently, after separating on 15% SDS-PAGE gel and cutting off the specific protein band within the gel, the purified GST-ORF3 protein was obtained and the yield of the purified protein was about 1 mg/L bacterial culture, as assessed by the Bradford method (Fig. 3B).
FIG. 3.
PCR, SDS-PAGE, and Western blot analyses of in vitro expression of ORF3-encoding protein. (A) PCR amplification of rebuilt ORF3. Lane 1, 315 bp fragment of the rebuilt ORF3; lane 2, 108 bp fragment; lane 3, 227 bp fragment; lane M, 100 bp marker. (B) SDS-PAGE analysis of ORF3-encoding protein expressed in E. coli. Lane P, protein marker; lane 1, cell lysates of E. coli strain BL21; lane 2, cell lysates of E. coli strain BL21 transformed with the vector pGEX-4T-1; lane 3, cell lysates of E. coli strain BL21 transformed with the recombinant plasmid pGEX-4T-1-F3; lane 4, purified GST-ORF3 protein expressed in E. coli strain BL21. (C) Western blot analyses of GST-ORF3 protein and MAbs against ORF3-encoding protein. Lane 1, GST-ORF3 protein recognized with rabbit anti-GST polyclonal antibody; lane 2, positive control reacted with rabbit anti-GST polyclonal antibody; lanes 3–6, MAbs 1H3, 3B1, 3D12, and 3G7 against PCV2-ORF3-encoding protein strongly reacted with GST-ORF3 protein expressed in E. coli.
Production and isotypes of MAbs against PCV2 ORF3-encoding protein
The supernatant of hybridomas secreting MAb to ORF3-encoding protein was detected by an indirect ELISA using antigens (i.e., GST-ORF3 protein, recombinant Cap protein of PCV2, the purified PCV2 and PCV1 virions, and lysates of E. coli BL21 strain containing the parental pGEX-4T-1 vector, PK-15 cell, and RPMI 1640 medium. Only the positive hybridomas reacting strongly with GST-ORF3 protein were further selected and cloned. Four hybridoma cell lines to ORF3-encoding protein of PCV2 were selected and characterized and then designated MAbs 1H3, 3B1, 3D12, and 3G7 (Table 2). All four MAbs were found to belong to the IgG1 subclass, with kappa light chains.
Table 2.
Optical Densities for Reactivity of MAbs with Purified ORF3 Protein of PCV2 in ELISA
| |
|
|
OD490b at MAb (supernatant) dilution |
|||||
|---|---|---|---|---|---|---|---|---|
| MAb | Isotype | Light chain | 1:10 | 1:20 | 1:40 | 1:80 | 1:160 | 1:320 |
| 3B1 | 1.889 | 1.561 | 1.321 | 1.025 | 0.654 | 0.324 | ||
| 3D12 | 2.869 | 2.310 | 1.641 | 1.214 | 0.894 | 0.650 | ||
| 3G7 | 1.521 | 1.204 | 0.856 | 0.652 | 0.456 | 0.213 | ||
| 1H3 | IgG1 | κ | 2.521 | 2.103 | 1.412 | 1.102 | 0.752 | 0.501 |
| Anti-IBDVa | 0.098 | 0.085 | 0.086 | 0.090 | 0.072c | 0.076d | ||
MAb from an irrelevant virus, infectious bursal disease virus.
Optical density at 490 nm.
Plate background.
Conjugated background.
Reactivity, specificity, and VN activity of MAbs against ORF3-encoding protein
The ability of each MAb was detected using Western blot assay to recognize a linear epitope on the ORF3-encoding protein. Bacterial lysates containing GST-ORF3 protein, purified GST-ORF3 protein, purified PCV2 and PCV1 virions, lysates of E. coli BL21 strain and E. coli BL21 containing the parental pGEX-4T-1 vector, and PK-15 cells were separated by SDS-PAGE and transferred onto nitrocellulose membranes. In Western blot analysis, MAbs 1H3, 3B1, 3D12, and 3G7 reacted strongly and specifically with the GST-ORF3 protein (Fig. 3C), and no signal was detected with the BL21 strain and pGEX-4T-1, the purified PCV2 and PCV1 virions, and PK-15 cells. In addition, after all MAbs were neutralized with PCV2 virions, PCV2 virions were still replicated in the infected PK-15 cell, showing that the infectivity of PCV2 virions could not be neutralized.
Expression of ORF3 for PCV2 in PK-15 cells
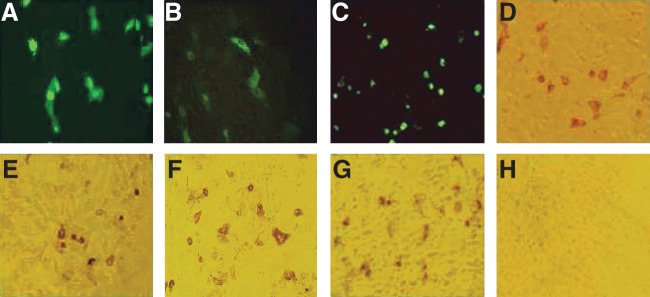
To express ORF3-encoding protein in eukaryotic cells, the ORF3 sequence of PCV2 was cloned respectively into the MCS of the plasmids pEGFP-C2 and pCI-neo. At 8–9 h post-transfection, GFP-ORF3 fusion protein began to present in the cytoplasm of PK-15 cells. Later, the cells presenting the fluorescence increased gradually, and the expressed GFP-ORF3 fusion protein was found in the cytoplasm, or in the nucleus, or in both the cytoplasm and nucleus (Fig. 4). Moreover, the GFP-ORF3 fusion protein expressed in the PK-15 cell could be recognized specifically with the MAbs 1H3, 3B1, 3D12, and 3G7 in immunohistochemical analysis (data not shown). Similarly, the entire ORF3-encoding protein was achieved by expression in the PK-15 cells transfected with pCI-neo-F3 vector by IHC analysis with the MAbs 1H3, 3B1, 3D12, and 3G7 (Fig. 4), showing that ORF3-encoding protein expressed in PK-15 cells possess the binding epitopes of the MAbs for the ORF3-encoding protein expressed in E. coli. In addition, most transfected PK-15 cells with fluorescence began to shrink and segment after 48 h post-transfection (Fig. 4). In contrast, no signals were detected in PK-15 cells, and PK-15 cells transfected respectively with pEGFP-C2 and pCI-neo vectors (data not shown).
FIG. 4.
PK-15 cells transfected with the vectors pEGFP-C2-F3 and pCI-noe-F3. (A) PK-15 cells transfected with pEGFP-C2 vector. (B) GFP-ORF3 fusion protein was expressed in PK-15 cells at 9 h post-transfection. (C) PK-15 cells expressing GFP-ORF3 fusion protein shrank and segmented at 48 h post-transfection. (D–G) PK-15 cells transfected with the recombinant plasmid pCI-neo-F3 was recognized specifically by MAbs 1H3, 3B1, 3D12, and 3G7 in IHC analysis. (H) PK-15 cells transfected with the vector pCI-neo in IHC analysis.
Intracellular transcription and expression of ORF3 for PCV2
To confirm whether the mRNA of ORF3 was transcribed during the replication of PCV2 virions, total RNAs extracted from PCV2-infected PK-15 cells at 10 to 60 h.p.i. were subjected to RT-PCR using ORF3 specific primers. As shown in Figure 5, findings revealed that the RT-PCR product of 315 bp in length was determined at 12 to 60 h.p.i. Sequencing analysis further showed that the nucleotide sequence of the RT-PCR product was 100% identified to ORF3 of PCV2. In the previous studies, Cheung and colleagues21 considered that the RNA transcripts were detected at 18 h.p.i., while we found that the mRNA of ORF3 was detected at 12 h.p.i., indicating that the transcription of ORF3 appeared in the early stage of PCV2 replication. In Northern blot assay, the transferred RNA was hybridized weakly with a 32P-labeled probe of the PCV2-ORF3 sequence (data not shown). However, in the analysis of the intracellular ORF3-encoding protein expression, no positive signals were detected by IHC assay with MAbs to ORF3-encoding protein in PK-15 cells infected with the mixture of PCV2 virions and MAbs to ORF3-encoding protein at 5 to 60 h.p.i.
FIG. 5.
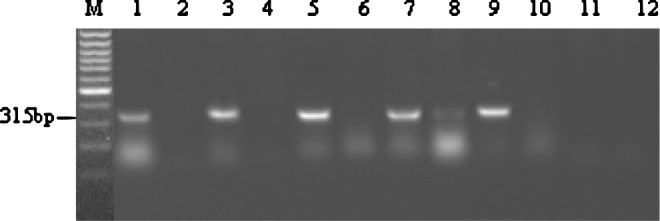
RT-PCR analysis of ORF3 mRNA transcription in PCV2-infected PK-15 cells. Lanes 1, 3, 5, 7, and 9, RT-PCR products of ORF3 were amplified at 12, 24, 36, 48, and 60 h.p.i. from the total RNA of PCV2-infected PK-15 cells. Lanes 2, 4, 6, 8, and 10, PCR products were amplified at 12, 24, 36, 48, and 60 h.p.i. from the total RNA of PCV2-infected PK-15 cells to avoid any DNA contamination as a negative control; lane 11, RT-PCR product of PK-15 cells; lane 12, PCR product of PK-15 cells.
Role of putative ORF3 in replication of PCV2 virions
The IHC reactivities of swine PCV2 antiserum with clones pSK–PCV2-mORF3 and pSK-PCV2-normal after transfection are shown in Figure 6. At 48 h post-transfection, the positive signals were obtained with swine PCV2 antiserum in PK-15 cells transfected with pSK-PCV2-normal. In contrast, no positive signals appeared in the cells transfected with pSK-PCV2-mORF3 vector and the PCV2-free PK-15 cells, indicating that the knock-out of ORF3 inhibited the replication of PCV2 virions and verifying that transcription and translation of the putative ORF3 play pivotal roles in the replication of PCV2 virions. In addition, within cells transfected with PCV2 clone pSK-PCV2-normal, the positive signals appeared predominantly as small, dense, granular intranuclear inclusion bodies resembling the prereplication sites reported in herpes simplex viruses.(30)
FIG. 6.
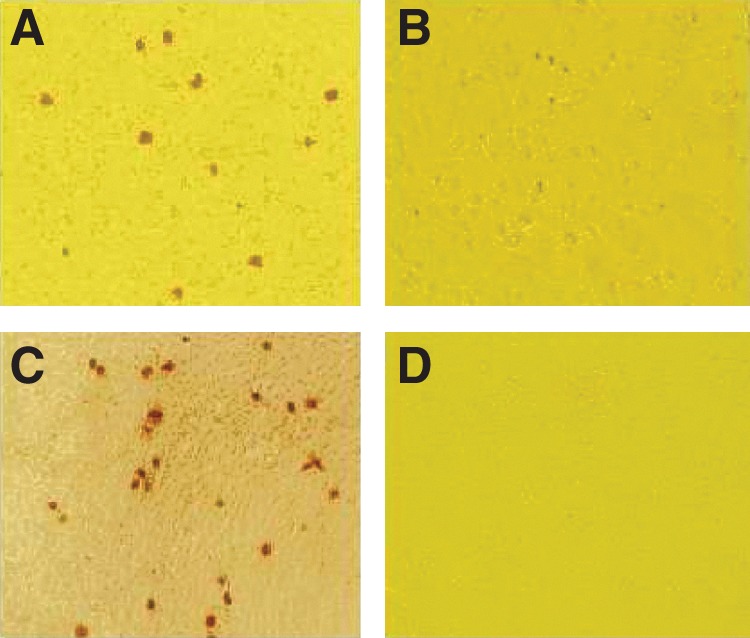
IHC reactivity with swine PCV2 antiserum in PK-15 cells transfected with PCV2 infectious DNA clones pSK-PCV2-mORF3 and pSK-PCV2-normal. (A) PK-15 cells transfected with pSK-PCV2-normal was reacted with swine PCV2 antiserum. (B) PK-15 cells transfected with pSK-PCV2-mORF3 could not be recognized by swine PCV2 antiserum. (C) PK-15 cells infected with PCV2 virions were recognized by swine PCV2 antiserum. (D) PK-15 cells.
Discussion
In this report we describe how the PCV2-ORF3-encoding protein, as a GST-fusion protein with an approximate molecular weight of 37.7 kDa (Fig. 3B), was obtained for expression using pGEX-4T-1 vector in E. coli, with some optimization of DNA sequence. In this experiment, we also constructed the recombinant vector containing ORF3 DNA sequence without modifications using the vectors pET, pBAD His/B, and pET-30a, as well as pGEX-4T-1. Interestingly, we found that no expression of ORF3-encoding protein was observed. Sequence analysis revealed that the DNA sequence of ORF3 contains 12 rare condons for E. coli, which may be an important factor in the failure of the ORF3-encoding protein to express in E. coli, as previous reports demonstrated that some rare condons of the inserted foreign gene for E. coli were disadvantageous for a foreign gene expression.(29) Even if the ORF3 DNA sequence was optimized, with the exception of the pGEX-4T-1 vector, the vectors pET, pBAD His/B, and pET-30a failed to express the ORF3-encoding protein in E. coli. Therefore, we concluded that the pGEX-4T-1 vector and the E. coli BL21 strain was a suitable prokaryotic expression system for ORF3 of PCV2.
As for purification of the GST-ORF3 protein, although the supernatant of bacterial lysates were verified containing the soluble GST-ORF3 protein by SDS-PAGE and Western blot assay, but GST-ORF3 protein expressed in E. coli failed to purify with a commercial HiTrap affinity column, and only the purified 37.7 kDa-GST-ORF3 protein was obtained by “cutting off” the gel. The reason for this observation was not clear. It is possible that the ORF3 moiety interfered with the proper folding of GST and therefore reduced dramatically the interaction of GST with glutathione, indicating that GST conformations within the GST-ORF3 protein were changed.(31)
The MAbs 1H3, 3B1, 3D12, and 3G7 generated in this study could recognize specifically GST-ORF3 protein expressed in E. coli and ORF3-encoding protein expressed in PK-15 cells in Western blot and IHC assays, indicating that all MAbs to ORF3-encoding protein expressed in E. coli possessed the binding epitopes of natural ORF3-encoding protein expressed in PK-15 cells. In the VN assay, we further observed that PCV2 virions still replicated in PK-15 cells after inoculation with a mixture of PCV2 and MAb, raising the question of whether these MAbs were non-neutralizing for the epitopes of ORF3-encoding protein or ORF3-encoding protein was a non-structural protein for PCV2 virions. In general, the MAbs against ORF3-encoding protein of PCV2 generated in this study should be a valuable and useful tool for in-depth studies of molecular biology and structure of PCV2 ORF3-encoding protein and for virus-cell interaction studies.
In the analyses of mRNA transcription and expression for ORF3-encoding protein, we found that the RT-PCR product was amplified successfully from the total RNAs extracted from the PCV2-infected PK-15 cells (Fig. 5), and that the weaker positive signal was observed after the total cellular RNA was hybridized with a 32P-labeled probe of ORF3 DNA sequence, confirming that ORF3 was transcribed in PCV2 replication. Previous reports proposed that 9 PCV2-specific RNAs were detected during productive infection in PK-15 cells,(21,25,32) including the viral capsid protein RNA (CR), a cluster of five Rep-associated RNAs (Rep, Rep’, Rep3a, Rep3b, Rep3c), and three NS-associated RNAs (NS515, NS672, and NS0). Members of the Rep-associated RNA cluster all share common 5′ and 3′ nt sequences. The three NS-associated RNAs are probably transcribed from three different promoters present inside ORF1, independent from the Rep promoter. However, all of the Rep-associated RNAs and NS-associated RNAs are transcribed in the same clockwise orientation as the Rep’ RNA. In this experiment, compared with the above-mentioned nine RNAs, the RNA of ORF3 we discovered is anticlockwise inside ORF1, and the specific reverse primer was used. Therefore, we confirmed that the RNA of ORF3 was novel in PCV2 replication. Unfortunately, although mRNA of ORF3 was monitored (Fig. 5), none of the MAbs detected the ORF3-encoding protein in purified PCV2 virions and PK-15 cells infected with PCV2 as we expected. This could be due to the amount of ORF3-encoding protein being too low to detect in PCV2 replication.
In conclusion, in the experiment using PCV2 infectious DNA clones, we found that the new progeny PCV2 antigens were not observed in PK-15 cells transfected with the plasmid pSK-PCV2-mORF3 after 48 h post-transfection, with the exception of PK-15 cells transfected with the clone pSK-PCV2-normal with ORF3. Therefore, we concluded that the putative ORF3 protein played a pivotal role in the process of PCV2 replication. But we are unclear about whether the ORF3-encoding protein is a replicase. Furthermore, when the ORF3-encoding protein was expressed in PK-15 cells, we also investigated whether or not the fluorescence cells expressing ORF3-encoding protein shrunk and segmented after 48 h post-transfection compared to PK-15 cells transfected with the vector pEGFP-C2. This phenomenon resembled the apoptosis of VP3 apoptin of chicken anemia virus. Therefore, we wondered whether the ORF3-encoding protein also plays an important role in PCV2 pathogenesis. Fenaux and colleagues(28) reported previously that a chimeric PCV1-2 virus was attenuated when compared to normal PCV2, suggesting that another protein important to the virulence of PCV2 besides ORF2 still exists.
Acknowledgments
This work was supported by a grant (30370052) from the National Natural Science Foundation of China.
Author Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.McNulty M. Dale J. Lukert P. Mankertz A. Randles J. Todd D. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press; San Diego: 2000. Virus taxonomy: classification, nomenclature of viruses; pp. 299–303. [Google Scholar]
- 2.Fauquet MM. Maniloff J. Desselberger U. Ball LA. Virus Taxonomys: VIIIth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; Amseterdam: 2005. [Google Scholar]
- 3.Tischer I. Gelderblom H. Vettermann W. Koch MA. A very small porcine virus with circular single-stranded-DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- 4.Todd D. Niagro FD. Ritchie BW. Curran W. Allan GM. Lukert PD. Latimer KS. Steffens WL., 3rd McNulty MS. Comparison of three animal viruses with circular single-stranded DNA genomes. Arch Virol. 1991;117:129–135. doi: 10.1007/BF01310498. [DOI] [PubMed] [Google Scholar]
- 5.Todd D. Weston JH. Soike D. Smyth JA. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology. 2001;286:354–362. doi: 10.1006/viro.2001.0985. [DOI] [PubMed] [Google Scholar]
- 6.Woods LW. Latimer KS. Barr BC. Niagro FD. Campagnoli RP. Nordhausen RW. Castro AE. Circovirus-like infection in a pigeon. J Vet Diagn Invest. 1993;5:609–612. doi: 10.1177/104063879300500417. [DOI] [PubMed] [Google Scholar]
- 7.Allan GM. McNeilly F. Cassidy JP. Reilly GA. Adair B. Ellis WA. McNulty MS. Pathogenesis of porcine circovirus; experimental infections of colostrum deprived piglets and examination of pig foetal material. Vet Microbiol. 1995;44:49–64. doi: 10.1016/0378-1135(94)00136-k. [DOI] [PubMed] [Google Scholar]
- 8.Tischer I. Rasch R. Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A. 1974;226:153–167. [PubMed] [Google Scholar]
- 9.Tischer I. Mields W. Wolff D. Vagt M. Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. 1986;91:271–276. doi: 10.1007/BF01314286. [DOI] [PubMed] [Google Scholar]
- 10.Harding JCS. Clark EG. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS) Swine Health Prod. 1997;5:201–203. [Google Scholar]
- 11.Clark EG. Proceedings of the American Association of Swine Practitioners. American Association of Swine Practitioners; Quebec City, Canada: 1997. Post-weaning multisystemic wasting syndrome; pp. 499–501. [Google Scholar]
- 12.Morozov I. Sirinarumitr T. Sorden SD. Halbur PG. Morgan MK. Yoon KJ. Paul PS. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J Clin Microbiol. 1998;36:2535–2541. doi: 10.1128/jcm.36.9.2535-2541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allan G. Meehan B. Todd D. Kennedy S. McNeilly F. Ellis J. Clark EG. Harding J. Espuna E. Botner A. Charreyre C. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet Rec. 1998;142:467–468. [PubMed] [Google Scholar]
- 14.Allan GM. Mc Neilly F. Meehan BM. Kennedy S. Mackie DP. Ellis JA. Clark EG. Espuna E. Saubi N. Riera P. Botner A. Charreyre CE. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol. 1999;66:115–123. doi: 10.1016/s0378-1135(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 15.Ellis J. Hassard L. Clark E. Harding J. Allan G. Willson P. Strokappe J. Martin K. McNeilly F. Meehan B. Todd D. Haines D. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can Vet J. 1998;39:44–51. [PMC free article] [PubMed] [Google Scholar]
- 16.Meehan BM. McNeilly F. Todd D. Kennedy S. Jewhurst VA. Ellis JA. Hassard LE. Clark EG. Haines DM. Allan GM. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998;79:2171–2179. doi: 10.1099/0022-1317-79-9-2171. [DOI] [PubMed] [Google Scholar]
- 17.Hamel AL. Lin LL. Nayar GPS. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J Virol. 1998;72:5262–5267. doi: 10.1128/jvi.72.6.5262-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meehan BM. Creelan JL. McNulty MS. Todd D. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J Gen Virol. 1997;78:221–227. doi: 10.1099/0022-1317-78-1-221. [DOI] [PubMed] [Google Scholar]
- 19.Zhou JY. Chen QX. Ye JX. Shen HG. Chen TF. Shang SB. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet Res Commun. 2006;30:205–220. doi: 10.1007/s11259-006-3203-x. [DOI] [PubMed] [Google Scholar]
- 20.Mankertz A. Hillenbrand B. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology. 2001;279:429–438. doi: 10.1006/viro.2000.0730. [DOI] [PubMed] [Google Scholar]
- 21.Cheung AK. Transcriptional analysis of porcine circovirus type 2. Virology. 2003;305:168–180. doi: 10.1006/viro.2002.1733. [DOI] [PubMed] [Google Scholar]
- 22.Liu J. Chen I. Kwang J. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J Virol. 2005;79:8262–8274. doi: 10.1128/JVI.79.13.8262-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J. Chen I. Du Q. Chua H. Kwang J. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J Virol. 2006;80:5065–5073. doi: 10.1128/JVI.80.10.5065-5073.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou JY. Shang SB. Gong H. Chen QX. Wu JX. Shen HG. Chen TF. Guo JQ. In vitro expression, monoclonal antibody and bioactivity for capsid protein of porcine circovirus type II without nuclear localization signal. J Biotechnol. 2005;118:201–211. doi: 10.1016/j.jbiotec.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Cheung AK. Bolin SR. Kinetics of porcine circovirus type 2 replication. Arch Virol. 2002;147:43–58. doi: 10.1007/s705-002-8302-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhou JY. Wu JX. Cheng LQ. Zheng XJ. Gong H. Shang SB. Zhou EM. Expression of immunogenic S1 glycoprotein of infectious bronchitis virus in transgenic potatoes. J Virol. 2003;77:9090–9093. doi: 10.1128/JVI.77.16.9090-9093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenaux M. Halbur PG. Haqshenas G. Royer R. Thomas P. Nawagitgul P. Gill M. Toth TE. Meng XJ. Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J Virol. 2002;76:541–551. doi: 10.1128/JVI.76.2.541-551.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenaux M. Opriessnig T. Halbur PG. Elvinger F. Meng XJ. A chimeric porcine circovirus (PCV) with the immunogenic capsid gene of the pathogenic PCV type 2 (PCV2) cloned into the genomic backbone of the nonpathogenic PCV1 induces protective immunity against PCV2 infection in pigs. J Virol. 2004;78:6297–6303. doi: 10.1128/JVI.78.12.6297-6303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- 30.Whitley RJ. Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q. Willson P. Attoh-Poku S. Babiuk LA. Bacterial expression of an immunologically reactive PCV2 ORF2 fusion protein. Protein Expr Purif. 2001;21:115–120. doi: 10.1006/prep.2000.1356. [DOI] [PubMed] [Google Scholar]
- 32.Cheung AK. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology. 2003;310:41–49. doi: 10.1016/s0042-6822(03)00096-5. [DOI] [PubMed] [Google Scholar]