Abstract
Drosophila ovarian germ cells require Sex-lethal (Sxl) to exit from the stem cell state and to enter the differentiation pathway. Sxl encodes a female-specific RNA binding protein and in somatic cells serves as the developmental switch gene for somatic sex determination and X-chromosome dosage compensation. None of the known Sxl target genes are required for germline differentiation, leaving open the question of how Sxl promotes the transition from stem cell to committed daughter cell. We address the mechanism by which Sxl regulates this transition through the identification of nanos as one of its target genes. Previous studies have shown that Nanos protein is necessary for GSC self-renewal and is rapidly down-regulated in the daughter cells fated to differentiate in the adult ovary. We find that this dynamic expression pattern is limited to female germ cells and is under Sxl control. In the absence of Sxl, or in male germ cells, Nanos protein is continuously expressed. Furthermore, this female-specific expression pattern is dependent on the presence of canonical Sxl binding sites located in the nanos 3′ untranslated region. These results, combined with the observation that nanos RNA associates with the Sxl protein in ovarian extracts and loss and gain of function studies, suggest that Sxl enables the switch from germline stem cell to committed daughter cell by posttranscriptional down-regulation of nanos expression. These findings connect sexual identity to the stem cell self-renewal/differentiation decision and highlight the importance of posttranscriptional gene regulatory networks in controlling stem cell behavior.
In adults, tissue homeostasis depends on a stable population of stem cells that have the capacity to give rise to both self-renewing and differentiating daughter cells. In Drosophila, the continuous supply of gametes throughout adulthood is accomplished by a stem cell–based system, the analysis of which has proved to be a powerful model for understanding the mechanisms that specify the choice between self-renewal and differentiation (1, 2). Much work has been done to understand how stem cell maintenance is governed by signals provided by the local microenvironment as well as to identify the cell-intrinsic factors required for self-renewal and the prevention of precocious differentiation. However, much less is known about the intrinsic machinery that enables daughter differentiation.
In the adult Drosophila ovary, the germline stem cells (GSCs) reside in a somatic niche, the microenvironment located at the anterior end of the each germarium. The niche maintains GSC fate by stimulating the Bmp signaling cascade that directly represses transcription of the differentiation promoting gene bag-of-marbles (bam) in the GSCs (3–6). When the GSC divides, the distal daughter cell moves out of the niche where it no longer receives (or responds to) the Bmp signals, bam is expressed, and the cell differentiates into a cystoblast (CB) (7). Interestingly, there have been reports of cells that coexpress bam with one or more of the GSC specific markers, suggesting that the cell fated to differentiate first passes through an intermediate stage that transitions, without dividing, to a mature CB cell (3, 7–11). Our recent studies uncovered a key role for Sex-lethal (Sxl) in the GSC-to-CB transition (12). In the adult, germ cells that lack Sxl protein can adopt a GSC fate; however, instead of then entering the differentiation pathway, the mutant GSC progeny are blocked at a stage that resembles an immature CB cell that coexpresses Bam protein and a set of GSC-specific markers. This GSC/CB cell switch defect is accompanied by continued proliferation and the formation of an ovarian tumor (12–14). Although these studies clearly show that germ cells require Sxl to transition from a stem cell to a fully committed daughter cell, the mechanism by which this occurs is not known.
Previous studies have shown that Sxl encodes a female-specific RNA binding protein that orchestrates sex-specific development and behavior by modulating the expression of a set of downstream target genes (15, 16). Sxl controls the sex determination and dosage compensation pathways by regulating the expression of transformer and male-specific-lethal-2 genes, but neither of these genes has a role in the germline (17–19). Notch transcripts are also subject to Sxl regulation, but only in a subset of somatic cells (20–22). Additional candidate genes have been identified by bioinformatic approaches, but their biological relevance to germline differentiation has yet to be established (23, 24). Thus, the Sxl target genes that mediate the GSC/CB cell fate switch remain to be discovered.
Here we identify nanos as a Sxl target gene in the adult germline, a conserved translational repressor that is necessary for maintaining a stable population of GSCs in the adult ovary (25–27). In the absence of nanos, all germ cells enter the differentiation pathway; therefore, it has been hypothesized that nanos maintains GSCs by repressing the translation of a set of as-yet-unidentified differentiation-promoting genes. nanos is then down-regulated, allowing the GSC/CB cell fate switch to occur. In germ cells, as in other cell types, nanos expression is regulated at the posttranscriptional level (28). Although studies have shown that nanos repression in CBs is dependent on bam function, the relationship is genetic, and the mechanism by which this occurs has not been established (28). We find nanos repression is under Sxl control, as it is not down-regulated in germ cells lacking Sxl protein. We further show that nanos RNA associates with the Sxl protein. More importantly nanos repression in CBs is dependent on the presence of Sxl binding sites located in the nanos 3′ UTR. These data, together with genetic epistasis experiments, support a model in which Sxl promotes the GSC/CB cell switch by down-regulation, and highlight the importance of posttranscriptional regulatory networks in controlling stem cell behavior.
Results
Nanos Expression Pattern in Differentiating Germ Cells Is Sexually Dimorphic.
In ovaries, the Nanos protein expression pattern is highly dynamic, changing as germ cells begin to differentiate (28). Using fully functional tagged transgenes to follow the expression of Bam and Nanos, we confirmed the previously reported findings. Our data show that Nanos protein accumulates in the GSCs located at the anterior tip of the germarium (Fig. 1A, Region 1). When the GSC divides, one cell remains at the tip and retains its GSC identity, whereas the other daughter moves away from the tip and initiates a differentiation program which includes significant accumulation of the Bam protein and rapid down-regulation of the Nanos protein (Fig. 1A, Region 2). This daughter cell undergoes four mitotic divisions to form an interconnected 16-cell cyst (Fig. S1D). As the cells cease to divide, the relationship between Nanos and Bam expression inverts, culminating with high levels of Nanos, as Bam expression is extinguished in the 16-cell cyst (Fig. 1A, Region 3). In sharp contrast to the dynamic expression pattern observed in the ovary, Nanos is expressed in both the testicular GSCs and their Bam-expressing daughters (Fig. 1B). The sexually dimorphic expression pattern of Nanos suggests a regulatory mechanism involving a sex-specific factor.
Fig. 1.
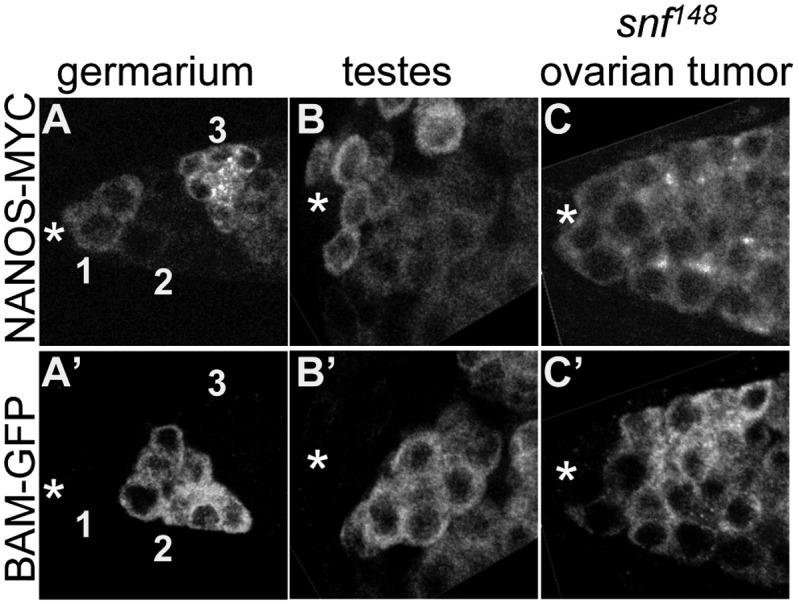
Nanos and Bam protein expression in ovaries, testis, and snf148 ovarian tumors. (A–C) Confocal images of gonads from WT animals and tumorous ovaries from snf148/snf148 females carrying fully functional copies of a Bam-GFP fusion protein and a Nanos-Myc fusion protein costained for GFP and MYC. (A) In the ovary, Bam and Nanos protein are expressed in nonoverlapping domains in early germ cells. The GSCs are located adjacent to the somatic cap cells, marked with an asterisk. Domains are numbered as in D. (B) In the testis, Nanos and Bam are coexpressed in early germ cells, except for the presumptive GSCs located adjacent to the hub, marked with an asterisk. (C) In the tip of the snf148/snf148 ovarian tumor, Nanos and Bam are coexpressed, except for the presumptive GSCs located adjacent to the somatic cap cells marked with an asterisk. Images in A–C are the same magnification.
Female-Specific Nanos Down-Regulation Is Attenuated in Germ Cells Lacking Sxl Protein.
The observation that nanos regulation in the germline is female specific suggests that nanos might be under Sxl control. Sxl protein is female specific. In agreement with previous studies, we find that Sxl protein is detectable in ovaries but not in testis (12, 29). In the germline, Sxl accumulates to very high levels in the cytoplasm of Nanos expressing GSCs (Fig. S1, Region 1) and remains detectable in the mitotically differentiating germ cells (Fig. S1, Region 2). Similar to Bam expression, cytoplasmic Sxl expression is absent or severely reduced in the newly formed 16-cell cyst, which expresses high levels of Nanos (Fig. S1, Region 3). This dynamic expression pattern is consistent with the possibility that Sxl has a role in down-regulating Nanos.
To assess the potential of a Sxl-mediated mechanism, we asked whether mutations that disrupt Sxl expression in the germline affect nanos expression. The extant female-sterile Sxl alleles are not ideal for these studies, because they are not protein-null alleles and their mutant phenotype is reversible by a variety of factors including temperature, genetic background, and infection with the reproductive parasite Wolbachia (29–31). As an alternative strategy, we examined Nanos expression in the snf148 germ cell tumor. snf148 is a female-sterile allele of the general splicing factor encoded by the sans-fille locus, which we have previously shown is due to the specific elimination of Sxl protein expression in germ cells (12, 32). In sharp contrast to WT germ cells, snf148 mutant germ cells coexpress Nanos and Bam (Fig. 1C). In addition, the WT expression pattern is restored in snf148 mutant germ cells carrying a copy of P{otu::SxlcDNA}, a transgene that expresses a SxlcDNA under control of a germline-specific promoter (Fig. S2). These studies demonstrate that Sxl is required for the down-regulation of Nanos in Bam-expressing cells.
Sxl Forms a Complex with nanos RNA.
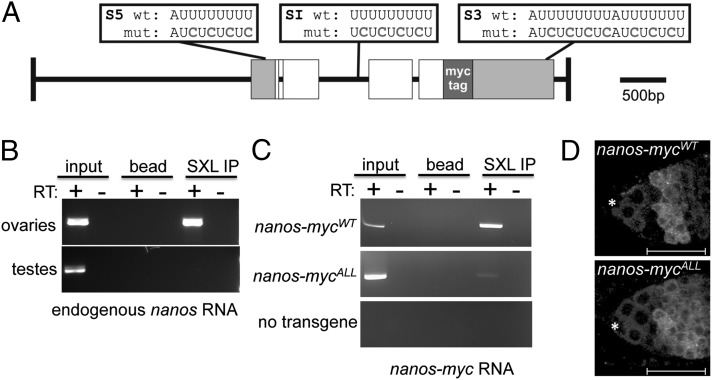
Sxl encodes an RNA binding protein which, in vitro, binds specifically to sequences that contain polyuridine runs of seven or more nucleotides (23, 33, 34). The nanos primary transcript has several such sequences located in its 5′ UTR, 3′ UTR, and within an intron (Fig. 2A); therefore, we asked whether nanos RNA is detectable in Sxl immunoprecipitates. The results of these RNA immunoprecipitation (IP) assays show that nanos RNA is detectable by RT-PCR in ovarian extracts, but not in control extracts made from testis that lack Sxl protein (Fig. 2B). Finding that Sxl is capable of associating with nanos RNA in ovarian extracts leads us to propose that Sxl modulates nanos expression directly.
Fig. 2.
The nanos gene is a Sxl target gene. (A) Schematic representation of the 5.8-kb genomic myc-tagged nanos constructs. Gray boxes indicate the 5′ and 3′ UTRs, white boxes represent the ORFs, and green box indicates the 6× MYC-tag inserted in frame. The location of the wild-type and mutated Sxl binding sites are shown above the diagram. (B) Sxl associates with nanos RNA. RNA IP assays were carried out in whole-cell extracts from ovaries or testis. The presence of nanos RNA in the IP pellet was detected by RT-PCR. (C) Mutagenesis of the S5, S1 and S3 binding sites impairs Sxl binding to nanos RNA. RNA IP assays were carried out in whole-cell extracts from ovaries carrying either the WT nanos-mycWT construct, the mutant nanos-mycALL construct, or no transgene. The presence of nanos-myc RNA in the IP pellet was detected by RT-PCR. (C) Confocal images of ovaries carrying either a WT or mutant myc-tagged nanos construct stained for Myc. The staining pattern of the nanos-mycWT parent construct resembles WT, whereas the mutant nanos-mycALL construct in which all of the S5, SI, and S3 binding sites are mutated is not down-regulated in the early differentiating germline cells. The location of the somatic cap cells is marked with an asterisk, and the regions are marked as in Fig. 1. Scale bars = 25 μm.
Sxl Binding Site in 3′ UTR Is Necessary for Nanos Down-Regulation.
If Sxl controls nanos expression by binding to its transcript, then mutating the predicted binding sites is predicted to abrogate Nanos down-regulation in Bam-expressing cells. To test this, we created a series of mutant myc-tagged nanos transgenes carrying U→C substitutions (Fig. 2A) expected to abolish Sxl binding based on in vitro studies (34). All constructs, including a WT control, were inserted into the same genomic location on the second chromosome to avoid differences in expression levels due to position effects (Materials and Methods).
In the first set of experiments, the consequence of simultaneously mutating all three Sxl binding sites was assessed. To determine whether mutating these sites would disrupt the Sxl/nanos RNA association in vivo, we immunoprecipiated Sxl complexes from ovarian extracts expressing either the mutant nanos-mycAll construct or the WT nanos-mycWT control. RT-PCR analysis revealed that although the nanos-mycWT mRNA was detectable in the immunoprecipitates, recovery of the nanos-mycAll mRNA was impaired (Fig. 2C). Finding that Sxl protein was still able to immunoprecipitate some nanos-mycALL RNA, however, suggests that Sxl remains tethered to the nanos RNA by another mechanism.
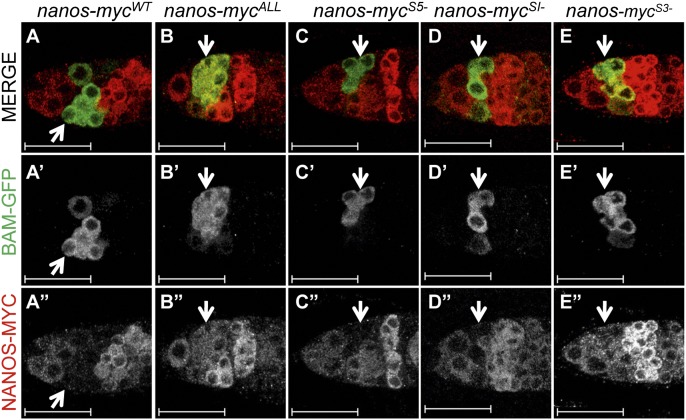
Nevertheless, examination of protein expression in the germaria revealed that the pattern of the mutant nanos-mycAll construct differed dramatically from the WT nanos-mycWT control (Fig. 2D). The protein expressed from the nanos-mycWT transgene showed the previously observed dynamic expression pattern: the protein is present in GSCs, falls below detectable levels in mitotically active cysts, and accumulates once more to high levels in 16-cell cysts. In striking contrast, the protein expressed from nanos-mycAll is expressed throughout the germarium, without any evidence of down-regulation in the mitotically active cells. To confirm that Nanos-mycAll protein is expressed in mitotically active cells, we re-examined expression in animals that also carry a copy of the fully functional Bam-GFP fusion protein. Costaining experiments reveal that although the Nanos-mycWT control is not detectable in Bam-GFP expressing cells, the mutant Nanos-mycAll protein and Bam-GFP are coexpressed (Fig. 3 A and B).
Fig. 3.
The nanos 3′ UTR Sxl binding site is essential for Nanos protein down-regulation. (A–E) Confocal images of ovaries from animals carrying a fully functional copy of a Bam-GFP fusion protein and one of the control or experimental Nanos-Myc fusion protein constructs costained for GFP and MYC. Arrows mark the location of the Bam-expressing cells. Scale bars = 25 μm.
In the second set of experiments, we assessed the consequences of mutating the Sxl binding sites individually. Of the three constructs, only nanos-mycS3−, which contains mutations in the 3′ UTR Sxl binding site, expresses a protein that shows significant coexpression with Bam-GFP (Fig. 3 C–E). Together these studies suggest that Sxl exerts its activity primarily through the poly(U) sequences located in the nanos 3′ UTR.
The Sxl binding site located in the 3′ UTR falls within a region that others have identified as nonessential for lowering Nanos protein levels in the germarium (28). In these experiments, expression of nanos transgenes deleted for nucleotides 500–800 of the 3′UTR, which removes all but the last nucleotide of the 3′ Sxl binding site, in the germarium was reported to be WT. Although the reason for this discrepancy remains unclear, one possibility is that the deletion construct removes other regulatory elements that negate the positive effects that we observed when specifically mutating the Sxl binding sites.
nanos Dysregulation Phenotype Is Sensitive to bam Gene Dosage.
Replacing the endogenous nanos 3′ UTR sequences with the tubulin 3′ UTR results in ectopic expression in the daughter cells fated to differentiate (28, 35), and a modest increase in the number of GSC-like cells in the germarium (35). To determine whether mutating the nanos Sxl binding sites is sufficient to delay or otherwise to interfere with the exit from the stem cell state, we followed the early stages of differentiation by the changing morphology of the fusome, labeled with Hu Li Tai Shao (Hts). The sphere-shaped fusome, also known as a spectrosome, is a distinguishing feature of both GSCs and CB cells (“round” in Fig. 4). Upon GSC and CB division, the spectrosome elongates and branches out to connect the two daughter cells. The presence of a short and linear fusome indicates either a dividing GSC that has not completed cytokinesis or a two-cell cyst (“short” in Fig. 4). As the two-cell cyst goes through three more rounds of cell division, the fusome develops into a branched structure that connects the cells within the multicellular cyst (“long” in Fig. 4).
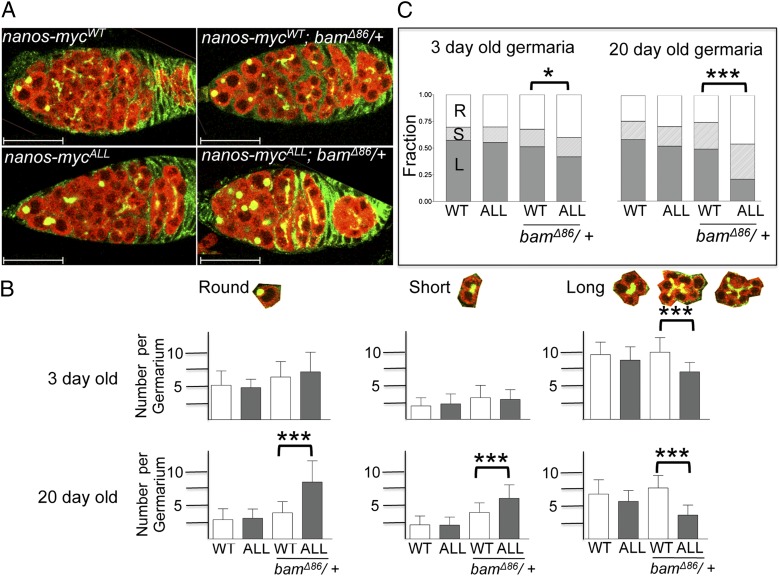
Fig. 4.
Impact of ectopic Nanos expression on germ cell differentiation. (A) Phenotypic impact of ectopic Nanos expression driven by two copies of the nanos-mycALL transgene is more pronounced in a bamΔ86/+ background. Confocal images of ovaries from 3-d-old animals stained for Vasa (red) to mark germ cells and 1B1 (green) to label the germ cell–specific spectrosomes and fusomes. 1B1 also labels somatic cell membranes. Scale bars = 25 μm. (B) Average number of spectrosomes/fusomes per germarium in different genetic backgrounds scored 3 and 20 d after eclosion. Spectrosomes/fusomes were classified as Round (R, spherical and not protruding into other cells), Short (S, extends to one adjacent cell), or Long (L, branches into two or more adjacent cells). Error bars represent SDs (n = 20 for each genotype/age group). Asterisks indicate significant differences by Student t test (***P < 0.001). (C) Relative distribution of the different types of spectrosome/fusomes structures per germarium. Asterisks indicate significant differences by the χ2 test (*P < 0.05, ***P < 0.001).
Interestingly, we find that ectopic nanos expression driven by a single copy of nanos-mycAll in a WT background has no apparent impact on germ cell differentiation. Even with two copies of the nanos-mycAll transgene, we do not observe a significant change in either the average number of spectrosome-containing cells or the relative distribution of cells with round/short/long structures compared with animals carrying two copies of the WT nanos-mycWT transgene (Fig. 4).
The failure to observe a significant phenotypic impact of expression driven by two copies of the nanos-mycAll suggests that eliminating Sxl regulation is, by itself, not sufficient to disrupt oogenesis. Prior work has suggested that bam is necessary for Nanos down-regulation (28). We therefore asked whether reducing bam dosage would reveal a mutant phenotype in 3-d-old ovaries. As illustrated in Fig. 4A and as quantitated in Fig. 4B, two copies of the nanos-mycALL transgene in a bam/+ heterozygous background results in a modest but consistent increase in the average number of spectrosome-containing cells (7.0 ± 2.9) compared with two copies of the WT transgene in a bam/+ heterozygous background (6.2 ± 2.2). The overall distribution of cells with round/short/long structures indicate that the increase in these cells is accompanied by a statistically significant loss of differentiated cysts (Fig. 4C).
Strikingly, the negative effect of nanos dysregulation on germ cell differentiation is more pronounced in aged flies. In the WT, the stem cell population decreases as the flies age (36), an effect that we observed in 20-d-old females expressing two copies of the WT transgene in a WT and bam/+ background (Fig. 4 B and C). This age-dependent loss, however, is reversed in age-matched flies expressing two copies of the nanos-mycAll transgene in a bam/+ background. The number of spectrosome containing cells increases over time (7.0 ± 2.9 → 8.3 ± 3.1; Fig. 4B), and the relative distribution of undifferentiated/differentiated cell types changes dramatically; more than 75% of all germ cells in the germarium fall into the GSC/CB or two-cell cyst category (Fig. 4C). Together with our earlier genetic studies showing that bam function depends on Sxl activity in the germline (12), these data lead us to conclude that Sxl and bam jointly control the entry into the differentiation pathway by lowering Nanos protein levels.
In the Absence of Sxl Protein in the Germline, nanos Is Essential for Tumor Growth.
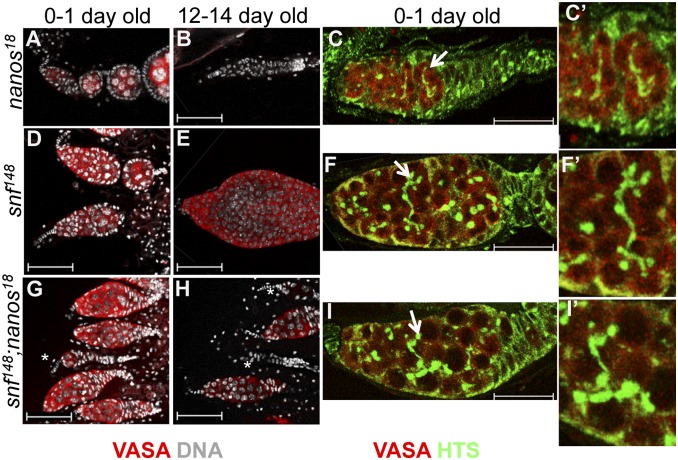
To further clarify the functional significance of Sxl-mediated nanos regulation, we asked whether dysregulated nanos expression is an essential component of the snf148 ovarian tumor phenotype. As reported previously (25), in young adults (0–1 d old), the majority of nanos18 mutant germaria produce gametes but become progressively agametic over time, presumably because the stem cell population is not maintained (Fig. 5 A and B). The snf148 ovarian tumor phenotype also changes with age, as tumor growth continues through adulthood (Fig. 5 D and E). Interestingly, tumor growth was strongly suppressed in double-mutant animals. In young animals, a significant fraction of all germaria (35%, n = 176) are agametic, a phenotype never observed in snf148 mutant animals (Fig. 5G). In older animals, the fraction of agametic germaria increases to 74% (n = 92), with a marked reduction in tumor size compared with similarly aged snf148 mutant animals (Fig. 5H). Notably, the majority of surviving double-mutant germ cells have abnormal fusome-like structures and fail to differentiate, indicating that they are more similar to snf than to nanos mutant germ cells (Fig. 5 C, F, and I). This result indicates that although nanos is required for proliferation of the tumorous germ cells, other factors must contribute to tumorigenesis.
Fig. 5.
snf148, nanos18 Double mutant analysis. Comparison of germaria from (A–C) nanos18/Df(3R) Exel6183, (D–F) snf148/snf148, and (G–I) double-mutant nanos18/Df(3R)Exel6183; snf148/snf148 females stained 0–1 d or 12–14 d after eclosion for Vasa, Hu Li Tai Shao (Hts) (C, F, and I), and DNA (A, B, D, E, G, H). Arrow in C, F, and I shows the region enlarged in C', F', and I'), illustrating the difference between the thin branching fusome structure seen in nanos18/Df(3R)Exel6183 and the abnormal long branching fusome structures visible in snf148/snf148 and double-mutant nanos18/Df(3R)Exel6183; snf148/snf148 females. Scale bars: 50 μm (A, B, D, E, G, and H); 25 μm (C, F, and I). Asterisks indicate germaria with few or no germ cells.
Discussion
Sxl has a pivotal role in the cell fate switch from a self-renewing GSC to a differentiation-competent CB (12). We address the underlying cellular mechanism by identifying nanos as a Sxl target gene. nanos is a conserved translational repressor that is thought to maintain GSC fate by silencing a set of as-yet-unidentified differentiation-promoting mRNAs. Cell fate switching occurs as one of the daughter cells initiates a differentiation program that includes significant accumulation of the Bam protein and rapid down-regulation of a set of GSC specific markers, including Nanos protein. Previous studies have shown that Nanos protein expression is dynamically regulated at the level of translation such that Nanos protein levels are high in GSCs but undetectable in all Bam-expressing daughter cells (28). We find that Sxl is responsible for this dynamic expression pattern. Moreover. regulation is direct. nanos RNA is bound by the Sxl protein in ovarian extracts, and nanos silencing is dependent on the presence of Sxl binding sites located in the nanos 3′ UTR. Together these studies point to a mechanism in which Sxl promotes the GSC/CB cell switch by lowering Nanos protein levels. The use of a female-specific factor to control nanos expression is consistent with earlier studies showing that the mechanism regulating GSC differentiation differs significantly between the sexes; for example, neither bam nor nanos is required for this process in males, although they are expressed (1). Thus, the Sxl-mediated posttranscriptional regulatory mechanism described here provides a direct link between sexual identity and execution of the appropriate differentiation pathway.
Sxl is expressed in both GSCs and their progeny, raising the question of how its role in Nanos down-regulation is restricted to Bam-expressing cells. bam Itself may confer cell-type specificity, as it too is required for lowering Nanos protein levels, although there are, as yet, no physical data to support direct regulation (28). Nevertheless, taken together with our genetic studies that reveal that bam requires Sxl activity to promote differentiation (12), these observations suggest that cell-type specificity could be achieved by a regulatory complex containing Sxl and Bam. Both Sxl and Bam have been shown to repress translation in other contexts (16, 37), further suggesting that the two proteins might function together to directly repress nanos translation. Biochemical studies will be required to test this model.
We previously showed that Sxl is required for germ cells to progress from a stem cell to a differentiation-competent CB fate, and that if this pathway is blocked, the mutant germ cells form a tumor (12). Although we show that inappropriate nanos expression is necessary for tumor growth, our genetic epistasis experiments indicate that nanos expression is not responsible for malignant transformation because the majority of surviving double-mutant germ cells continue to resemble a tumor cell. This conclusion is supported by studies carried out by us and others (28, 35) showing that forced expression of nanos in GSC progeny is not sufficient to block differentiation. Even in a genetically sensitized background where we observe an accumulation of extra stem cell–like germ cells, germ cells proceed through oogenesis. Therefore, these studies suggest that although forced expression of nanos is responsible for expanding the number of mutant germ cells, aberrant regulation of other genes and/or pathways under Sxl control elicit malignant transformation. The identification and analysis of additional Sxl targets genes will offer insights into how the failure to successfully exit the stem cell state is connected to the genesis of germ cell tumors.
Materials and Methods
Drosophila Strains.
The following mutant alleles and deficiencies were used in this study: snf148 (32), nos18 (also known as nosRC) (38), Df(3R)Exel6183 (39), and bamΔ86 (40). Transgenic lines published previously include P{bam-GFP} (4), P{nanos-myc} (41), and P{otu::SxlcDNA} (42). The lines constructed for this study include P{nanos-mycWT}, P{nanos-mycALL}, P{nanos-mycSI-}, P{nanos-mycS5-}, and P{nanos-mycS3-}. These five lines are structurally similar to the previously described P{nanos-myc} transgene (41) except that they were generated in the pACMAN vector and integrated into the attP40 docking site located on the second chromosome at 25C6. Genetic Services, Inc. carried out the injections and generated the transgenic flies. A complete description of the Drosophila strains used in this study is available from FlyBase (http://flybase.org).
Immunofluorescence and Image Analysis.
Ovaries were fixed and stained by standard methods. The following primary antibodies were used: mouse anti-HTS, 1:10 (1B1, Developmental Studies Hybridoma Bank); rat anti-VASA, 1:10 (Developmental Studies Hybridoma Bank); rabbit anti-GFP, 1:1,000 (#A1122, Invitrogen); rat anti-myc, 1:50 (#SC56633, Santa Cruz); mouse anti-myc, 1:50 (#SC40, 1:50, Santa Cruz); rabbit anti-myc, 1:50 (#SC789, Santa Cruz); mouse anti-SXL, 1:350 (m18, Developmental Studies Hybridoma Bank). Secondary antibodies coupled to FITC or Cy3 (Jackson ImmunoResearch) were used at 1:800; and secondary antibodies coupled to Alexa 488, 555, or 633 (Invitrogen) were used at 1:1,000. All images were acquired on an Zeis Axiophot microscope or on an inverted Leica DM IRE2 microscope with a Leica TCS SP2 AOBS filter-free UV/spectral confocal laser scanner.
RNA IP.
Using the conditions described previously (43, 44), RNA/protein complexes were immunoprecipitated from 550 μL crude extract in NET buffer (150 mM NaCL, 50 mM Tris, pH 7.5, 5 mM EDTA, and 0.05% Nonidet P-40) containing material from ∼50 ovaries or ∼100 testes from 0- to 2-d-old adult flies (Fig. 2B) or ∼100 ovaries from 0- to 2-d-old adult flies (Fig. 2D). Briefly, IPs were carried out with GammaBind Plus Sepharose beads (GE) beads pretreated with 20 μL mouse anti-Sxl antibodies (1:1 ratio of M104:M114; Developmental Studies Hybridoma Bank). After TRIzol purification, the precipitated RNA was resuspended in 20 μL nuclease-free water, and DNase treated. cDNA was synthesized from 8 μL of the RNA with random hexamers using the SuperScript First Strand Synthesis System (Invitrogen). The PCR reactions were performed in 25 μL total volume with either 1 μL (ovaries) or 2.5 μL (testes) of the RT reaction with the following primers: For endogenous nanos (Fig. 2B): forward CTGGAATTGCCGTACGCTTC and reverse GACATGCGACCGAGATCATC; for nanos-myc: forward CGATTTAAAGCTATGGAGCAAAA and reverse GACTTGGATTTGAGTGATCG. PCR conditions were as follows: 94 °C for 1 min; 30 cycles of 94 °C for 1 min, 55 °C for 1 min, and 72 °C for 1 min; and 72 °C for 10 min. Products were detected on a 2% agarose gel by staining with ethidium bromide.
Supplementary Material
Acknowledgments
We thank Y. Li, M. Buszczak, D. McKearin, R. Lehmann, Robin Wharton, the Developmental Studies Hybridoma Bank, and the Bloomington Stock Center for gifts of antibodies and/or fly stocks. We also thank T. Xie, M. L. Johnson, R. Conlon, H. Lou, A. Matthews, and J. McDonald for helpful discussions and comments on the manuscript. National Institutes of Health (NIH) Grant R01-GM61039 provided support for the initial stages of this work. NIH National Center for Research Resources shared instrumentation Grant RR-017980 that funded the confocal microscope used in this study, and NIH training Grant T32-HD07104 provided partial salary support for J.C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120473109/-/DCSupplemental.
References
- 1.Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: A decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamashita YM. Cell adhesion in regulation of asymmetric stem cell division. Curr Opin Cell Biol. 2010;22:605–610. doi: 10.1016/j.ceb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, McKearin DM. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr Biol. 2003;13:1786–1791. doi: 10.1016/j.cub.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- 5.Song X, et al. Bmp signals from niche cells directly repress transcription of a differentiation-promoting gene, bag of marbles, in germline stem cells in the Drosophila ovary. Development. 2004;131:1353–1364. doi: 10.1242/dev.01026. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, McKearin D. Gene circuitry controlling a stem cell niche. Curr Biol. 2005;15:179–184. doi: 10.1016/j.cub.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Ohlstein B, McKearin D. Ectopic expression of the Drosophila Bam protein eliminates oogenic germline stem cells. Development. 1997;124:3651–3662. doi: 10.1242/dev.124.18.3651. [DOI] [PubMed] [Google Scholar]
- 8.Gilboa L, Forbes A, Tazuke SI, Fuller MT, Lehmann R. Germ line stem cell differentiation in Drosophila requires gap junctions and proceeds via an intermediate state. Development. 2003;130:6625–6634. doi: 10.1242/dev.00853. [DOI] [PubMed] [Google Scholar]
- 9.Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci USA. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casanueva MO, Ferguson EL. Germline stem cell number in the Drosophila ovary is regulated by redundant mechanisms that control Dpp signaling. Development. 2004;131:1881–1890. doi: 10.1242/dev.01076. [DOI] [PubMed] [Google Scholar]
- 11.Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–178. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Chau J, Kulnane LS, Salz HK. Sex-lethal facilitates the transition from germline stem cell to committed daughter cell in the Drosophila ovary. Genetics. 2009;182:121–132. doi: 10.1534/genetics.109.100693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schüpbach T. Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex-lethal in Drosophila melanogaster. Genetics. 1985;109:529–548. doi: 10.1093/genetics/109.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashiyama K, Hayashi Y, Kobayashi S. Drosophila Sex lethal gene initiates female development in germline progenitors. Science. 2011;333:885–888. doi: 10.1126/science.1208146. [DOI] [PubMed] [Google Scholar]
- 15.Salz HK. Sex determination in insects: A binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21:395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salz HK, Erickson JW. Sex determination in Drosophila: The view from the top. Fly (Austin) 2010;4:60–70. doi: 10.4161/fly.4.1.11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh JL, Wieschaus E. Is sex determination in germ line and soma controlled by separate genetic mechanisms? Nature. 1978;272:249–251. doi: 10.1038/272249a0. [DOI] [PubMed] [Google Scholar]
- 18.Schüpbach T. Autosomal mutations that interfere with sex determination in somatic cells of Drosophila have no direct effect on the germline. Dev Biol. 1982;89:117–127. doi: 10.1016/0012-1606(82)90300-1. [DOI] [PubMed] [Google Scholar]
- 19.Bachiller D, Sánchez L. Mutations affecting dosage compensation in Drosophila melanogaster: Effects in the germline. Dev Biol. 1986;118:379–384. doi: 10.1016/0012-1606(86)90007-2. [DOI] [PubMed] [Google Scholar]
- 20.Penn JK, Schedl P. The master switch gene sex-lethal promotes female development by negatively regulating the N-signaling pathway. Dev Cell. 2007;12:275–286. doi: 10.1016/j.devcel.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suissa Y, et al. Hrp48 attenuates Sxl expression to allow for proper notch expression and signaling in wing development. Proc Natl Acad Sci USA. 2010;107:6930–6935. doi: 10.1073/pnas.0910570107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suissa Y, Ordan E, Deshpande G, Gerlitz O. Males and females: Creating differences while maintaining the similarities. Fly (Austin) 2011;5:25–28. doi: 10.1073/pnas.0910570107. [DOI] [PubMed] [Google Scholar]
- 23.Robida MD, Rahn A, Singh R. Genome-wide identification of alternatively spliced mRNA targets of specific RNA-binding proteins. PLoS ONE. 2007;2:e520. doi: 10.1371/journal.pone.0000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gawande B, Robida MD, Rahn A, Singh R. Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J. 2006;25:1263–1272. doi: 10.1038/sj.emboj.7601022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 26.Gilboa L, Lehmann R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development. 2004;131:4895–4905. doi: 10.1242/dev.01373. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Minor NT, Park JK, McKearin DM, Maines JZ. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc Natl Acad Sci USA. 2009;106:9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bopp D, Horabin JI, Lersch RA, Cline TW, Schedl P. Expression of the Sex-lethal gene is controlled at multiple levels during Drosophila oogenesis. Development. 1993;118:797–812. doi: 10.1242/dev.118.3.797. [DOI] [PubMed] [Google Scholar]
- 30.Salz HK, Cline TW, Schedl P. Functional changes associated with structural alterations induced by mobilization of a P element inserted in the Sex-lethal gene of Drosophila. Genetics. 1987;117:221–231. doi: 10.1093/genetics/117.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starr DJ, Cline TW. A host parasite interaction rescues Drosophila oogenesis defects. Nature. 2002;418:76–79. doi: 10.1038/nature00843. [DOI] [PubMed] [Google Scholar]
- 32.Nagengast AA, Stitzinger SM, Tseng C-H, Mount SM, Salz HK. Sex-lethal splicing autoregulation in vivo: Interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development. 2003;130:463–471. doi: 10.1242/dev.00274. [DOI] [PubMed] [Google Scholar]
- 33.Singh R, Valcárcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 34.Samuels ME, et al. RNA binding by Sxl proteins in vitro and in vivo. Mol Cell Biol. 1994;14:4975–4990. doi: 10.1128/mcb.14.7.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris RE, Pargett M, Sutcliffe C, Umulis D, Ashe HL. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev Cell. 2011;20:72–83. doi: 10.1016/j.devcel.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan L, et al. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007;1:458–469. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Shen R, Weng C, Yu J, Xie T. eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc Natl Acad Sci USA. 2009;106:11623–11628. doi: 10.1073/pnas.0903325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann R, Nüsslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 39.Parks AL, et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 40.McKearin DM, Spradling AC. bag-of-marbles: A Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4(12B):2242–2251. doi: 10.1101/gad.4.12b.2242. [DOI] [PubMed] [Google Scholar]
- 41.Verrotti AC, Wharton RP. Nanos interacts with Cup in the female germline of Drosophila. Development. 2000;127:5225–5232. doi: 10.1242/dev.127.23.5225. [DOI] [PubMed] [Google Scholar]
- 42.Hager JH, Cline TW. Induction of female Sex-lethal RNA splicing in male germ cells: Implications for Drosophila germline sex determination. Development. 1997;124:5033–5048. doi: 10.1242/dev.124.24.5033. [DOI] [PubMed] [Google Scholar]
- 43.Johnson ML, Nagengast AA, Salz HK. PPS, a large multidomain protein, functions with Sex-lethal to regulate alternative splicing in Drosophila. PLoS Genet. 2010;6:e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stitzinger SM, Conrad TR, Zachlin AM, Salz HK. Functional analysis of SNF, the Drosophila U1A/U2B” homolog: Identification of dispensable and indispensable motifs for both snRNP assembly and function in vivo. RNA. 1999;5:1440–1450. doi: 10.1017/s1355838299991306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.