Abstract
Heart wall myofibers wind as helices around the ventricles, strengthening them in a manner analogous to the reinforcement of concrete cylindrical columns by spiral steel cables [Richart FE, et al. (1929) Univ of Illinois, Eng Exp Stn Bull 190]. A multitude of such fibers, arranged smoothly and regularly, contract and relax as an integrated functional unit as the heart beats. To orchestrate this motion, fiber tangling must be avoided and pumping should be efficient. Current models of myofiber orientation across the heart wall suggest groupings into sheets or bands, but the precise geometry of bundles of myofibers is unknown. Here we show that this arrangement takes the form of a special minimal surface, the generalized helicoid [Blair DE, Vanstone JR (1978) Minimal Submanifolds and Geodesics 13–16], closing the gap between individual myofibers and their collective wall structure. The model holds across species, with a smooth variation in its three curvature parameters within the myocardial wall providing tight fits to diffusion magnetic resonance images from the rat, the dog, and the human. Mathematically it explains how myofibers are bundled in the heart wall while economizing fiber length and optimizing ventricular ejection volume as they contract. The generalized helicoid provides a unique foundation for analyzing the fibrous composite of the heart wall and should therefore find applications in heart tissue engineering and in the study of heart muscle diseases.
Keywords: myocardium, myofiber geometry, diffusion tensor MRI
Histological studies of the mammalian heart (1–4) corroborate the finding that individual myofibers in the left ventricle (LV) are aligned to form helical curves (Fig. 1B). Several formal analyses (1, 2, 5–7) support the view that this alignment is mechanically optimal. Moving beyond considerations of individual fibers has proved difficult. An advantage of certain fibrous composites, such as those in plant cell walls, bone, insect cuticle, and fiberglass, is that their fiber geometries offer efficient reinforcement (8) by equalizing stiffness in all directions parallel to the plane in which fibers lie. Our approach has been to retain the mathematical precision available for the analysis of individual fibers while switching scales to that of the composite arrangement. Anatomical studies reveal that at such scales, fibers remain almost parallel locally (9), as illustrated in Fig. 1C with the colors depicting changes in orientation. We show that they are packed together to achieve this organization, while maintaining their helical form (4), via a unique structural arrangement in which they bundle into a special surface: a generalized helicoid (10–14). Because this object is a minimal surface (15, 16), it generalizes the geodesic properties of the individual helices (1, 2, 5–7) to the more global scale of the ventricular wall. We also show that this minimal surface structure can be maintained as the heart beats, with simulations revealing the power of using the proper mathematical coordinates. Previous models (1, 2, 5–7) apply to selected regions of the LV myocardium but exclude the apex. They describe the orientation of individual fibers but not volumetric bundles of them. The arrangement of myofibers in generalized helicoids characterizes their orientation throughout the heart wall.
Fig. 1.
Fiber geometry in the left ventricle of a rat. (A) An axial slice (gray) with both a single helical fiber passing through a voxel (B) and a bundle of fibers passing through neighboring locations (C–E). (B) The geometry of a single fiber is characterized locally by its rate of bending in the osculating tangent-normal (TN) plane (curvature) and out of it (torsion). The TN plane is shown in blue. (C) The geometry of a bundle of fibers is more complex. Here we show a narrow slice of fibers arranged across the thickness of the wall, emerging from the slice in A. The colors are used to visualize changes in orientation. (D) More abstractly, we now show fibers passing through a plane of voxels in green, and those in a neighboring plane in a cubic lattice in blue. The placement of a local coordinate frame allows fiber bundle geometry to be characterized by curvature measures in directions T, N, and B. (E) Comparing the model to data: The orientations corresponding to GHM fitting are overlaid in color on the principal eigenvector direction e1 (shown with gray to black shading in the direction from below to above the slice plane), for an axial slice of the rat heart DT-MRI data (compare with C).
Model
Setting up the right local coordinate frame is critical to developing the generalized helicoid model (GHM). Fig. 1D depicts a myofiber passing through a particular location (voxel) in a rectangular grid in three-dimensional Euclidean space, with its tangent vector lying in the plane of the page, along with fibers passing through neighboring voxels. An orthogonal coordinate frame is placed using the fiber orientation for tangent vector T, the in-page direction for the binormal vector B, and their cross-product for the normal vector N. The differential geometry of the collection of myofibers can now be characterized by specifying the rates of change of fiber orientation for displacements in the directions of T, N, and B.
The GHM expresses fiber orientation in terms of these curvatures by prescribing an orientation function  given by
given by
 |
at each point (x,y,z) within the LV wall with respect to a local coordinate frame (17, 18). Here θ(x,y,z) represents the orientation in the x–y plane with respect to the x axis, which is aligned with the local fiber direction, and with the z axis taken to be the component of the heart wall orthogonal to it (following heart myofiber geometry literature, refs. 19 and 20). This choice ensures that the reference frame rotates smoothly and consistently throughout the LV myocardium. Because it is small (21, 22) we do not directly model the component of fiber orientation out of the x–y plane in the local neighborhood of (x,y,z).
Fig. 2 illustrates the effects of the parameters of the GHM, with the orientation θ shown by a unit length vector field in the x–y plane and with fibers abstracted by streamline traces in θ (23). The KT parameter causes bending in the direction tangential to a fiber (Fig. 2A). With positive KN added, the fibers fan out. Finally, KB defines angular change in the z direction. In this example, KB has the effect of creating rotated copies of the same streamlines in planes parallel to the x–y plane (Fig. 2C). Mathematically the GHM’s scalar parameters KT, KN, and KB correspond locally to the three curvatures of the myofiber bundle, describing the amount that the fiber orientation changes for displacements in the directions T, N, and B, respectively (17, 18). As such it provides coordinates for describing the geometry of a bundle of myofibers in three dimensions. In contrast, a helix (1, 2, 5–7) only describes the geometry of a single fiber viewed as a one-dimensional curve.
Fig. 2.
The effect of varying the parameters KT,KN, and KB of the GHM. The x, y, and z axes of its reference frame are shown in red, green, and blue, and correspond to the directions T, N, and B. A slice of the unit length vector field specified by θ(x,y,z) in the x–y plane is shown in black. In each panel a single “fiber” is traced in red using a forward Euler approach (23) in the direction T, with neighboring fiber traces shown in black or gray. In A and B, fibers are only shown in the x–y plane because with KB = 0, θ is only a function of (x,y). (A) With only tangential curvature KT the fibers bend but remain locally parallel to one another in the x–y plane. (B) With positive normal curvature KN added the fibers fan away from one another in the x–y plane. (C) The effect of adding KB is to cause the fibers to rotate in planes parallel to the x–y plane. Here traced fibers are shown in six planes, one in the x–y plane (red), two above it (black), and three below it (gray). Notice the similarity between this and the rotation of fiber orientations from epicardium to endocardium in Fig. 1E.
A fundamental property of the GHM, as shown in ref. 17 and in the Appendix, is that (x,y,z,θ(x,y,z)) is a minimal surface when embedded in  ; i.e., its mean curvature, given by the trace of the shape operator, vanishes everywhere. A more familiar example of a minimal surface is the bubble that results from dipping a wire frame into a soapy film; that is, the minimal surface required to enclose a given volume of air. Blair and Vanstone show in ref. 10 that complete ruled minimal n-dimensional hypersurfaces
; i.e., its mean curvature, given by the trace of the shape operator, vanishes everywhere. A more familiar example of a minimal surface is the bubble that results from dipping a wire frame into a soapy film; that is, the minimal surface required to enclose a given volume of air. Blair and Vanstone show in ref. 10 that complete ruled minimal n-dimensional hypersurfaces  in
in  are products of
are products of 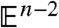 and a helicoid in
and a helicoid in 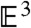 . As such, in
. As such, in  , a generalized helicoid is the product of a helicoid in
, a generalized helicoid is the product of a helicoid in  and the real line. Barbosa et al. (11) and Thas (13, 14) independently derive the parametric equations of such minimal generalized helicoids. There is a relationship between these results and those on generalized helical curves. In ref. 24, Hayden proves that, in Riemannian spaces of n dimensions, with n even, at least one of the curvatures of a generalized helix must vanish. One can deduce from this result that, in
and the real line. Barbosa et al. (11) and Thas (13, 14) independently derive the parametric equations of such minimal generalized helicoids. There is a relationship between these results and those on generalized helical curves. In ref. 24, Hayden proves that, in Riemannian spaces of n dimensions, with n even, at least one of the curvatures of a generalized helix must vanish. One can deduce from this result that, in  , a generalized helix can have non-null curvature and torsion, but its higher-order curvatures must be null. Thus, in
, a generalized helix can have non-null curvature and torsion, but its higher-order curvatures must be null. Thus, in  , a generalized helix must be the ordinary 3D helix with a linear coordinate function describing its fourth dimension.
, a generalized helix must be the ordinary 3D helix with a linear coordinate function describing its fourth dimension.
Results
We have tested our model against data from diffusion tensor (DT) MRI from three different mammals: rat, dog, and human. DT-MRI provides estimates of myofiber orientation over the full myocardial volume at high spatial resolution by measuring the orientation dependence of the Brownian motion of water molecules (25). Diffusion in the myocardium is anisotropic due to its fibrous structure. Several studies (26–28) show that the principal eigenvector e1 of a DT is locally aligned with myofiber orientation at the spatial scale of a typical image voxel. The use of DT-MRI has advantages over earlier dissection studies which were typically restricted to a small set of locations (3) and are therefore difficult to reproduce.
Returning to Fig. 1D, let T and B represent the x and z axis directions at each voxel. Applying GHM fitting to the fiber bundle shown in Fig. 1C, one expects a large binormal curvature KB, capturing the variation of fiber angle in the direction perpendicular to the heart wall. In contrast, the curvatures KT and KN along and across fibers, respectively, appear to be small. Our results confirm this trend throughout the LV myocardium, consistent with the anatomical observation that fibers are almost parallel locally but turn as one penetrates the heart wall (1–3, 26, 29). Fig. 1E compares our model to the data by overlaying the orientations corresponding to the best fit GHMs in color on the DT-MRI based fiber directions (gray to black) in an axial slice of rat LV myocardium.
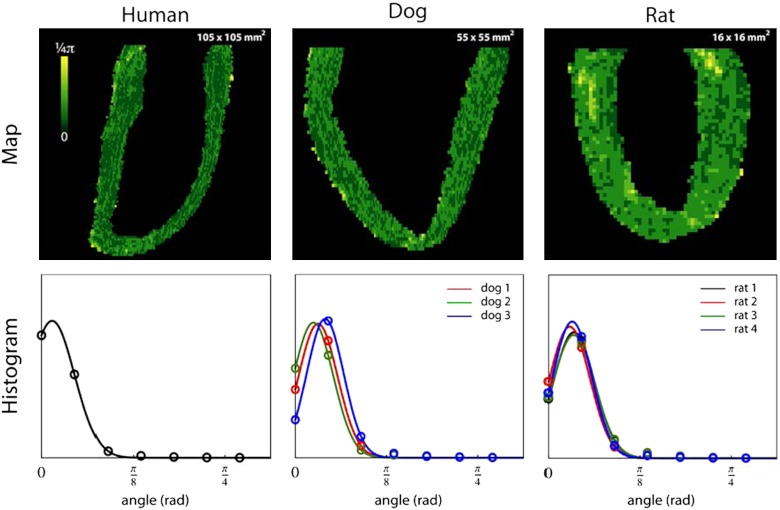
In Fig. 3, we plot histograms of the distributions of KB, KN, and KT for one human, three dog, and four rat DT-MRI datasets. The human, dog, and rat datasets have spatial resolutions of 0.4297 × 0.4297 × 1.0, 0.3125 × 0.3125 × 0.8, and 0.25 × 0.25 × 0.25 mm3, respectively. To perform a cross-species comparison, all curvature parameters were normalized based on the maximum epicardial cross-sectional diameter (MD). The histograms were then normalized with respect to the number of voxels contained in the dataset. The histogram of normalized KB values for each dataset peaks around -0.15 to -0.22 rad/mm, revealing a remarkable consistency of design across these three species. The mean KB value for each dataset is also an order of magnitude larger than the mean KN and KT values, which are both centered at zero. The rightmost column of Fig. 3 depicts the spatial variation of the KB parameter in a long-axis slice from a single dataset of each species, showing it to be homogeneous throughout the LV myocardial wall, including the apex, an area excluded by other models (1, 2, 5–7). GHM fits for transmural penetrations at locations near the base, the equator, and the apex of a rat heart dataset are shown in Fig. 4. Table 1 and Fig. 5 provide a quantitative and qualitative assessment of the GHM fitting angular errors over all the heart datasets. A von Mises distribution (30) fit shows these errors to be consistently low, with the μ parameter ranging from 2.63° (0.046 rad) to 7.63° (0.128 rad), and to have concentration κ higher than 100 (or equivalently, low variance). Locations of high error coincide with dataset boundaries, or are close to the base or insertions of the right ventricle.
Fig. 3.
Normalized KB, KN, and KT histograms for GHM fits of the human, dog, and rat heart DT-MRI datasets. The right-most column shows the spatial variation of the normalized KB parameter in a long-axis slice from one dataset of each species, with the color map ranging from -0.7 to 0.7 rad/mm. The plots show KB to be consistent across species as well as homogenous in the heart wall, and KN and KT to be close to zero. Locations where KB has a different sign and value (red arrows) reveal interesting anatomy near the insertions of the right ventricle and at the base close to the valves and the atria.
Fig. 4.
GHM fits at three locations in a rat heart dataset. (A) Transmural penetrations in red at the apex, equator, and base (counterclockwise from bottom). (B) The GHM fit at the base. (C) The GHM fit at the apex. (D) The GHM fit at the equator. In B–D, the measured DT-MRI fiber orientations are shown in pink with extrapolated fibers based on the GHM fits in blue. For each penetration the model smoothly interpolates the measured fiber orientations via its curvature parameter KB.
Table 1.
Quantitative assessment of the GHM fits for all heart DT-MRI datasets
| * | KB, rad/mm | KN, rad/mm | KT, rad/mm | μ, rad | κ(-) |
| Human | -0.17 ± 0.18 | 0.003 ± 0.085 | -0.005 ± 0.081 | 0.046 ± 0.002 | 108 ± 8 |
| Dog 1 | -0.15 ± 0.16 | 0.001 ± 0.101 | -0.001 ± 0.101 | 0.098 ± 0.002 | 141 ± 9 |
| Dog 2 | -0.17 ± 0.15 | 0.004 ± 0.096 | -0.011 ± 0.090 | 0.078 ± 0.001 | 136 ± 9 |
| Dog 3 | -0.17 ± 0.16 | 0.006 ± 0.099 | -0.011 ± 0.096 | 0.128 ± 0.003 | 157 ± 7 |
| Rat 1 | -0.19 ± 0.14 | 0.008 ± 0.053 | 0.003 ± 0.053 | 0.109 ± 0.003 | 128 ± 9 |
| Rat 2 | -0.21 ± 0.12 | 0.008 ± 0.045 | 0.005 ± 0.043 | 0.090 ± 0.002 | 132 ± 7 |
| Rat 3 | -0.22 ± 0.11 | 0.008 ± 0.043 | 0.006 ± 0.049 | 0.107 ± 0.003 | 121 ± 9 |
| Rat 4 | -0.20 ± 0.14 | 0.008 ± 0.053 | 0.000 ± 0.053 | 0.101 ± 0.001 | 144 ± 6 |
*All values reported as mean ± standard deviation. A neighborhood of 5 × 5 × 5 about the central voxel v is used for each fit. Errors are measured only for the voxels closest to v. KB, KN, and KT are species-normalized values.
Fig. 5.
Statistical analysis of the angular error of fit β. (Upper) The spatial variation of β in a long-axis slice from one dataset of each species. Locations of higher error in the rat dataset coincide with anatomically interesting regions, where KB has a different sign (red arrows in Fig. 3). (Lower) A statistical analysis obtained by fitting a von Mises distribution (30) (solid lines), with the μ and κ parameters shown in Table 1, to the angular errors from discretized GHM fits (open circles).
In a second experiment, we assess the effect of increasing neighborhood size (N = 3, 5, 7, 9) on GHM fits for the human heart dataset. Here we measure error for all voxels within the associated spherical neighborhood, not just the closest voxel neighbors. The von Mises analysis, presented in Table 2, shows that, although the error increases slightly, the larger number of error samples now leads to much tighter von Mises fits, with the concentration parameter κ increasing by a factor of 4 or more. In interpreting these results, it is important to recall that because the mathematical framework we follow is motivated by Cartan’s moving frame construction, we adapt the local coordinate frame to the object under consideration. When this frame is made too large, the tangent plane approximation to the heart wall becomes less accurate.
Table 2.
Quantitative assessment of the effect of increasing neighborhood size on the GHM fit for the human heart dataset
| Neighborhood* | μ, rad | κ(-) |
| 3 × 3 × 3 | 0.125 ± 0.00077 | 610 ± 23 |
| 5 × 5 × 5 | 0.145 ± 0.00156 | 478 ± 33 |
| 7 × 7 × 7 | 0.162 ± 0.00198 | 444 ± 37 |
| 9 × 9 × 9 | 0.173 ± 0.00213 | 430 ± 38 |
*All values reported as mean ± standard deviation. The errors are now measured for all voxels within the sphere of maximal radius inscribed in the neighborhood.
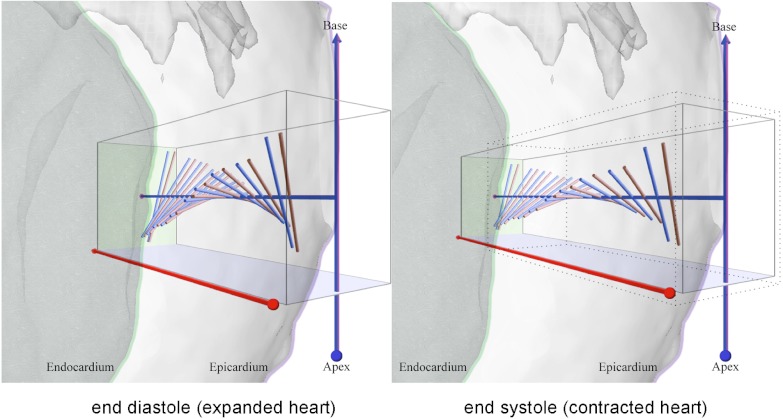
The changes implied by a beating heart, including fiber shortening and wall thickening (31), reveal another surprising aspect of the curvature parameter KB. Chen et al. (26) have reported DT-MRI measurements of fiber orientation along transmural penetrations from epicardium to endocardium on rat hearts fixed at end systole and end diastole. Their results show the heart wall to thicken significantly as myofibers contract, but the total change in fiber orientation from outer wall to inner wall to be quantitatively preserved. Using parameters consistent with their findings and our GHM fits for a transmural sampling at a location near the equator of the dataset in Fig. 4, Fig. 6 and Movie S1 show how this rearrangement of fiber geometry is predicted simply by a decrease in KB. Remarkably, throughout the contraction process, the minimal surface structure provided by the GHM is preserved.
Fig. 6.
A simulation of fiber contraction for an equatorial penetration of the rat heart in Fig. 4. To situate the anatomy the epicardium and endocardium are overlaid as transparent gray surfaces, with their contours shown in pink and green, respectively. The left subfigure shows the DT-MRI fiber orientations prior to contraction in pink with extrapolated fibers based on GHM fits and their corresponding KB curvatures in blue. The simulation (left to right) shows the effect of decreasing the length of each fiber by 14.2%, increasing their radii by 8.0% to preserve fiber volume and increasing the wall thickness by 41.9%, as reported in the findings of Chen et al. (26). As a result, in the right subfigure, the endocardium patch (green rectangle) has been displaced in the direction of the ventricular chamber. The dotted box in the right panel is a copy of the box in the left panel to illustrate changes to scale. By reducing the description of myofiber geometry to the single curvature parameter KB, the fiber rearrangement after contraction in the right subfigure is explained by simply decreasing it, while preserving the GHM structure. The full cycle from end diastole to end systole and back is visualized in Movie S1.
Discussion
Our bundling of heart wall myofibers into generalized helicoids adds significant dimensions to current knowledge of LV myofiber structure and function. First, this organization shows that myofiber orientation as a function of position is a minimal surface (15, 16) throughout the myocardium. Whereas previous models have considered only the geometry of individual (one-dimensional) fibers (1, 2, 6, 7) or their groupings in (two-dimensional) sheets (9, 19, 26, 28), our analysis of fiber orientation applies in three dimensions across the entire ventricular volume. The property that a minimal surface is locally area minimizing (15, 16) generalizes current understanding that individual helical fibers, being geodesic (1, 2, 6, 7), are length minimizing. Other properties of minimal surfaces are also inherited by the generalized helicoid organization. For example, for a harmonic minimal surface defined on a plane, it can be shown that the second variation of its area depends only on the component of a variation normal to it (15, 16). This property suggests that shortening fibers along their length in the direction given by θ could contribute to an efficient shrinkage of volume by yielding a displacement in the direction normal to the heart wall. It has in fact been established that a helical orientation of myofibers is essential to achieve a transmurally homogeneous workload for all myocytes within the healthy myocardium (32–34). More globally, the helicoidal myofiber architecture induces torsion of the left ventricle during contraction. Second, our GHM fits with small KT and KN but high KB formalize Neville’s stacked helicoids model (8) and Bouligand’s generalized twisted model (35). These two models have been used as qualitative descriptions of helicoidal arrangements in a variety of biological and man-made fibrous composites (8, 35), but without a formal mathematical underpinning. Furthermore, they have never before been applied to the heart wall. Our formalization along with our experimental fits to DT-MRI data reveal an additional purpose of the generalized helicoid organization, which is that it allows for wall stiffness to be equalized in the plane approximately tangent to the heart wall (8, p. 95), giving it mechanical strength.
In tissue bioengineering, understanding the basic structural properties of heart wall myofibers is fundamental. Their higher-order structural arrangement in generalized helicoids is likely to find application in the design of scaffolds for artificial heart muscle growth (36), in the study of myocardial infarction or other heart pathologies that rearrange fiber geometry (37, 38), and in understanding the relationship between individual fibers and their laminar organization into sheets (9, 19, 26, 28).
Materials and Methods
Diffusion Tensor Magnetic Resonance Imaging.
DT-MRI data for the dog and human hearts were obtained from The Center for Cardiovascular Bioinformatics and Modeling (http://www.ccbm.jhu.edu/research/DTMRIDS.php) at Johns Hopkins University. The institutional animal care committee at the Eindhoven University of Technology approved the following procedures for the acquisition of the rat heart datasets. Male Wistar rats were sedated by 3% isoflurane in medical air. For each specimen, the skin and ribs were cut to expose the heart. The left ventricular wall was penetrated at the apex with an 18 gauge perfusion needle. The vena cava inferior was cut and the vascular bed was perfused with 10,000 units of heparin L-1 in 100 mL of PBS. Subsequently, the perfusate was switched to 100 mL 4% phosphate buffered paraformaldehyde to induce tissue fixation. After all muscular contractions ceased, the heart was excised, rinsed thoroughly with PBS, and stored overnight in PBS at 4 °C.
DT-MRI measurements were then obtained on a 6.3 T horizontal-bore MRI scanner (Oxford Instruments) equipped with a 12-cm inner diameter providing a maximum gradient strength of 400 mT/m and a quadrature driven birdcage coil with an inner diameter of 32 mm (RAPID Biomedical). Each heart was placed in a plastic tube filled with Fomblin (Fens) for susceptibility matching, with medical gauze used to immobilize the specimen against mechanical vibrations. The left ventricular long axis was visually aligned with the centerline of the magnet bore. Diffusion-weighted images were collected at room temperature using a three-dimensional spin-echo sequence with unipolar diffusion sensitizing pulsed field gradients. The field of view was 32 × 16 × 16 mm3, with matrix dimensions 128 × 64 × 64, yielding 250 × 250 × 250 μm3 isotropic voxels (echo time 25 ms, repetition time 1,000 ms, 1 signal average). Pulsed field gradients were applied in 10 directions (39), with a diffusion weighting b value of 900 s/mm2. One additional measurement was performed without diffusion weighting. Paravision 4.0 was used to reconstruct the diffusion tensor which was diagonalized to obtain the three eigenvalues (λ1, λ2, and λ3) and eigenvectors (e1, e2, and e3) for each voxel. The principal eigenvector corresponds to the local orientation of myofibers (27, 28).
Generalized Helicoid Model Fitting.
In each dataset, the left ventricle was manually segmented by an expert. A GHM fit was obtained at each voxel v by searching over KT, KN, and KB values in the range [-0.7,…,0.7] rad/mm, in increments of 0.1 rad/mm for the human and dog datasets (a total of 15 × 15 × 15 = 3,375 {KT,KN,KB} triplets) and a range of [-2.75,…,2.75] rad/mm, in increments of 0.25 rad/mm, for the rat datasets (a total of 23 × 23 × 23 = 12,167 {KT,KN,KB} triplets). For each triplet, the GHM equation was applied within a local neighborhood N of size 5 × 5 × 5 voxels with origin at v to provide a 3D orientation α at each voxel v∈N. This neighborhood size is about the largest that can be used for the rat heart datasets, where we have approximately 10 voxels from outer wall to inner wall. In our implementation, for storage reasons, we discretized a GHM’s orientation α to the closest direction among 100 uniformly distributed samples on the hemisphere. The angle between α and the fiber direction, indicated by the DT principal eigenvector e1 at v was then computed, and this angular difference was averaged over all v∈N to provide a goodness-of-fit measure for the model. We note that, due to the discretization of the GHM’s orientation, even a perfect fit would result in an average error of about 5.4°. The goodness-of-fits measure was used to determine the best-fitting set of {KT,KN,KB} parameter values. In order to control for overall heart size differences, the {KT,KN,KB} parameters were normalized with respect to the MD of the human heart, such that, for example, KBnorm = KB × MD/MDhuman. For each fit, the x axis of the GHM frame at v was aligned with e1, with its z axis aligned with the projection of the local normal vector to the heart wall onto the plane orthogonal to e1. The heart wall normal direction was estimated using the local gradient to a Euclidean distance function (40) within the heart wall. In this manner, the frame rotated smoothly throughout the heart wall and allowed the z axis to be associated with the penetration direction used to define a fiber angle in the literature (19, 20).
Quantitative Assessment of GHM Fitting Errors.
The angular error of fit β at v was computed as the average angular difference between data and model direction, within the six nearest neighbors of v. The calculated β values were placed in bins corresponding to the inherent discretization of the data directions and were statistically analyzed by obtaining a least squares fit of a von Mises probability density function (30):
 |
Here μ is a measure of the amount of angular error β with κ a measure of its concentration at that amount—i.e., 1/κ is analogous to the variance of a normal distribution. We report the mean and standard deviation of KT,KN, and KB and the μ and κ parameters for each dataset in Table 1, with an associated quantitative and qualitative assessment in Fig. 5. We carry out an additional experiment to assess the effect of increasing neighborhood size (N = 3, 5, 7, 9) on the human heart dataset, measuring the error for all voxels within the associated spherical neighborhood. The μ and κ parameters resulting from a von Mises fit are presented in Table 2.
Appendix: The GHM Model for θ Is a Minimal Surface
A minimal surface is one whose mean curvature vanishes everywhere (15, 16). The mean curvature is given by the trace of the shape operator, which is the differential dN expressed in terms of the local coordinates of the tangent space  at a point p, of the surface normal vector N. We begin by expressing (x,y,z,θ) in the parametric form h(u,v,w) = (x(u,v,w),y(u,v,w),z(u,v,w),θ(u,v,w)), where θ is a three-dimensional surface in four-dimensional Euclidean space
at a point p, of the surface normal vector N. We begin by expressing (x,y,z,θ) in the parametric form h(u,v,w) = (x(u,v,w),y(u,v,w),z(u,v,w),θ(u,v,w)), where θ is a three-dimensional surface in four-dimensional Euclidean space  . We rearrange the GHM expression for θ as
. We rearrange the GHM expression for θ as
 |
[1] |
and let z(u,v,w) = w and θ(u,v,w) = v to get 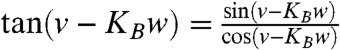 . Multiplying the numerator and the denominator of the right-hand side of this equation by the parameter u and requiring equality with the right-hand side of [1] leads to the parametric equations
. Multiplying the numerator and the denominator of the right-hand side of this equation by the parameter u and requiring equality with the right-hand side of [1] leads to the parametric equations
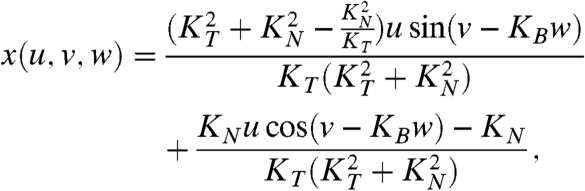 |
[2] |
 |
[3] |
| [4] |
An expression for the normal vector N at a given point of the surface is found by solving for the null space of JT, where J is the Jacobian matrix of parametrization h. The solution space is a line in  , and so N is taken as the direction vector of that line:
, and so N is taken as the direction vector of that line:
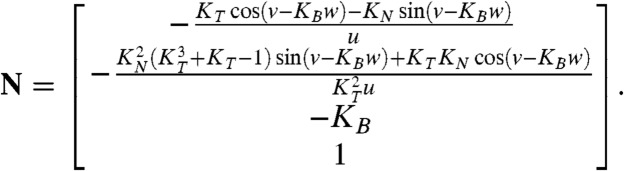 |
[5] |
Projected in the local coordinates of the tangent space  at p, which are given by ∂h/∂u = (∂x/∂u,∂y/∂u,∂z/∂u,∂θ/∂u), ∂h/∂v, and ∂h/∂w, dN becomes
at p, which are given by ∂h/∂u = (∂x/∂u,∂y/∂u,∂z/∂u,∂θ/∂u), ∂h/∂v, and ∂h/∂w, dN becomes
 |
[6] |
where  denotes the Euclidean dot product in
denotes the Euclidean dot product in  . With the expressions for h = (x,y,z,θ) provided in [2–4] and with the expression for the normal vector N provided in [5], one can verify that
. With the expressions for h = (x,y,z,θ) provided in [2–4] and with the expression for the normal vector N provided in [5], one can verify that 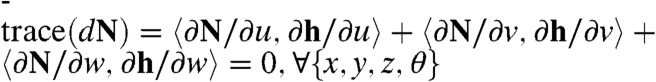 , which completes the proof.
, which completes the proof.
Supplementary Material
Acknowledgments.
We thank Patrick A. Helm and Raimond L. Winslow at the Center for Cardiovascular Bioinformatics and Modeling, and Elliot McVeigh at the National Institutes of Health (NIH), for provision of the human and dog DT-MRI data; G. B. Pike for helpful discussions; and I. Siddiqi and O. Siddiqi for comments on the manuscript. P.S. is supported by the NIH. G.J.S. is supported by the Dutch Technology Foundation, the applied science division of the Netherlands Organization for Scientific Research and the Technology Program of the Ministry of Economic Affairs (Grants 07952 and 10191). S.W.Z. is supported by the NIH (National Institute on Alcohol Abuse and Alcoholism) and the National Science Foundation. K.S. is supported by the Natural Sciences and Engineering Research Council of Canada, Fonds de recherche du Québec—Nature et technologies, and McGill University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data analyzed in this paper is archived at the following location: http://www.cim.mcgill.ca/~shape/dtmri; name: heart; password: helicoid. This database combines human and dog diffusion MRI datasets that are publicly available at http://www.ccbm.jhu.edu/research/DTMRIDS.php with rat diffusion MRI datasets that were collected at the Eindhoven University of Technology.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120785109/-/DCSupplemental.
References
- 1.Streeter DD, Powers WD, Alison Ross M, Torrent-Guasp F. Cardiovascular System Dynamics. Cambridge, MA: MIT Press; 1978. Three-dimensional fiber orientation in the mammalian heart; pp. 73–84. [Google Scholar]
- 2.Streeter DD. Gross morphology and fiber geometry of the heart. In: Berne RM, Sperelakis N, editors. Handbook of Physiology, Section 2. The Heart. New York: Williams and Wilkins; 1979. pp. 61–112. [Google Scholar]
- 3.Streeter DD, Spotnitz HM, Patel DP, Ross J, Sonnenblick EH. Fiber orientation in the canine left ventricle during diastole and systole. Circ Res. 1969;24:339–347. doi: 10.1161/01.res.24.3.339. [DOI] [PubMed] [Google Scholar]
- 4.Streeter DD, Hanna WT. Engineering mechanics for successive states in canine left ventricular myocardium. II. Fiber angle and sarcomere length. Circ Res. 1973;33:656–664. doi: 10.1161/01.res.33.6.656. [DOI] [PubMed] [Google Scholar]
- 5.Peskin CS. Mathematical Aspects of Heart Physiology. New York: New York University; 1975. (Courant Institute of Mathematical Sciences Lecture Notes). [Google Scholar]
- 6.Peskin CS. Fiber architecture of the left ventricular wall: An asymptotic analysis. Commun Pur Appl Math. 1989;42:79–113. [Google Scholar]
- 7.Horowitz A, Perl M, Sideman S. Geodesics as a mechanically optimal fiber geometry for the left ventricle. Basic Res Cardiol. 1993;88(Suppl 2):67–74. [PubMed] [Google Scholar]
- 8.Neville AC. Biology of Fibrous Composites: Development Beyond the Cell Membrane. Cambridge, UK: Cambridge Univ Press; 1993. [Google Scholar]
- 9.Gilbert S, Benson A, Li P, Holden A. Regional localisation of left ventricular sheet structure: Integration with current models of cardiac fibre, sheet and band structure. Eur J Cardiothorac Surg. 2007;32:231–249. doi: 10.1016/j.ejcts.2007.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Blair DE, Vanstone JR. Minimal Submanifolds and Geodesics. Tokyo: Kaigai Publ; 1978. A generalization of the helicoid; pp. 13–16. [Google Scholar]
- 11.Barbosa JM, Dajczer M, Jorge LP. Minimal ruled submanifolds in spaces of constant curvature. Indiana U Math J. 1984;33:531–547. [Google Scholar]
- 12.Dillen F. Ruled submanifolds of finite type. Proc Am Math Soc. 1992;114:795–798. [Google Scholar]
- 13.Thas C. Technical Report, Seminar of Higher Geometry. Belgium: State University of Ghent; 1979. Minimal generalized ruled surfaces in the Euclidean space Em. [Google Scholar]
- 14.Thas C. A property of the Veronese surface. J Geom. 1988;32:157–168. [Google Scholar]
- 15.Nitsche JJ. Lectures on Minimal Surfaces: Volume 1, Introduction, Fundamentals, Geometry and Basic Boundary Value Problems. Cambridge, UK: Cambridge Univ Press; 1989. [Google Scholar]
- 16.Osserman R. A Survey of Minimal Surfaces. Mineola, NY: Dover; 2002. [Google Scholar]
- 17.Savadjiev P. Montreal: McGill Univ; 2009. Perceptual Organisation in Diffusion MRI: Curves and Streamline Flows; p. 143. PhD thesis. [Google Scholar]
- 18.Savadjiev P, Zucker SW, Siddiqi K. On the differential geometry of 3D flow patterns: Generalized helicoids and diffusion MRI analysis; Proceedings of the IEEE 11th International Conference on Computer Vision; 2007. 10.1109/ICCV.2007.4409086. [Google Scholar]
- 19.LeGrice IJ, et al. Laminar structure of the heart: Ventricular myocyte arrangement and connective tissue architecture in the dog. Am J Physiol Heart and Circ Physiol. 1995;269:H571–H582. doi: 10.1152/ajpheart.1995.269.2.H571. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen PM, LeGrice IJ, Smaill BH, Hunter PJ. Mathematical model of geometry and fibrous structure of the heart. Am J Physiol Heart and Circ Physiol. 1991;260:H1365–1378. doi: 10.1152/ajpheart.1991.260.4.H1365. [DOI] [PubMed] [Google Scholar]
- 21.Geerts L, Bovendeerd P, Nicolay K, Arts T. Characterization of the normal cardiac myofiber field in goat measured with MR-diffusion tensor imaging. Am J Physiol Heart and Circ Physiol. 2002;283:H139–145. doi: 10.1152/ajpheart.00968.2001. [DOI] [PubMed] [Google Scholar]
- 22.LeGrice IJ, Hunter PJ, Smaill BH. Laminar structure of the heart: A mathematical model. Am J Physiol Heart and Circ Physiol. 1997;272:H2466–H2476. doi: 10.1152/ajpheart.1997.272.5.H2466. [DOI] [PubMed] [Google Scholar]
- 23.Piuze E, Kry PG, Siddiqi K. Generalized helicoids for modeling hair geometry. Comput Graph Forum. 2011;30:247–256. [Google Scholar]
- 24.Hayden HA. On a generalized helix in a Riemannian n-space. Proc Lond Math Soc. 1931;s2-32:337–345. [Google Scholar]
- 25.Jones DK, editor. Diffusion MRI: Theory, Methods, and Applications. New York: Oxford Univ Press; 2010. [Google Scholar]
- 26.Chen J, et al. Regional ventricular wall thickening reflects changes in cardiac fiber and sheet structure during contraction: Quantification with diffusion tensor MRI. Am J Physiol Heart and Circ Physiol. 2005;289:H1898–H1907. doi: 10.1152/ajpheart.00041.2005. [DOI] [PubMed] [Google Scholar]
- 27.Hsu EW, Muzikant AL, Matulevicius SA, Penland RC, Henriquez CS. Magnetic resonance myocardial fiber-orientation mapping with direct histological correlation. Am J Physiol Heart and Circ Physiol. 1998;274:H1627–H1634. doi: 10.1152/ajpheart.1998.274.5.H1627. [DOI] [PubMed] [Google Scholar]
- 28.Tseng W-YI, Wedeen VJ, Reese TG, Smith RN, Halpern EF. Diffusion tensor MRI of myocardial fibers and sheets: Correspondence with visible cut-face texture. J Magn Reson Imaging. 2003;17:31–42. doi: 10.1002/jmri.10223. [DOI] [PubMed] [Google Scholar]
- 29.Coghlan C, Hoffman J. Leonardo da Vinci’s flights of the mind must continue: Cardiac architecture and the fundamental relation of form and function revisited. Eur J Cardiothorac Surg. 2006;29(Suppl 1):S4–17. doi: 10.1016/j.ejcts.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Fisher RA. Dispersion on a sphere. Proc R Soc London Ser A. 1953;217:295–305. [Google Scholar]
- 31.Spotnitz HM. Macro design, structure, and mechanics of the left ventricle. J Thorac Cadiovasc Surg. 2000;119:1053–1077. doi: 10.1016/S0022-5223(00)70106-1. [DOI] [PubMed] [Google Scholar]
- 32.Aelen FW, et al. Relation between torsion and cross-sectional area change in the human left ventricle. J Biomech. 1997;30:207–212. doi: 10.1016/s0021-9290(96)00147-9. [DOI] [PubMed] [Google Scholar]
- 33.Arts T, Veenstra PC, Reneman RS. Torsion of the left ventricle during the ejection phase in the intact dog. Cardiovasc Res. 1984;18:183–193. doi: 10.1093/cvr/18.3.183. [DOI] [PubMed] [Google Scholar]
- 34.Lumens J, Delhaas T, Arts T, Cowan BR, Young AA. Impaired subendocardial contractile myofiber function in asymptomatic aged humans, as detected using MRI. Am J Physiol Heart Circ Physiol. 2006;291:H1573–H1579. doi: 10.1152/ajpheart.00074.2006. [DOI] [PubMed] [Google Scholar]
- 35.Bouligand Y. Twisted fibrous arrangements in biological materials and cholesteric mesophases. Tissue Cell. 1972;4:189–217. doi: 10.1016/s0040-8166(72)80042-9. [DOI] [PubMed] [Google Scholar]
- 36.Jawad H, Lyon AR, Harding SE, Ali NN, Boccaccini AR. Myocardial tissue engineering. Br Med Bull. 2008;87:31–47. doi: 10.1093/bmb/ldn026. [DOI] [PubMed] [Google Scholar]
- 37.Tseng W-YI, Dou J, Reese TG, Wedeen VJ. Imaging myocardial fiber disarray and intramural strain hypokinesis in hypertrophic cardiomyopathy with MRI. J Magn Reson Imaging. 2006;23:1–8. doi: 10.1002/jmri.20473. [DOI] [PubMed] [Google Scholar]
- 38.Strijkers GJ, et al. Diffusion tensor imaging of left ventricular remodeling in response to myocardial infarction in the mouse. NMR Biomed. 2009;22:182–190. doi: 10.1002/nbm.1299. [DOI] [PubMed] [Google Scholar]
- 39.Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Res Med. 1999;42:515–525. [PubMed] [Google Scholar]
- 40.Borgefors G. Distance transformations in arbitrary dimensions. Comput Vision Graph. 1984;27:321–345. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.