Abstract
Erythropoietin (EPO) is a neuroprotective cytokine in models of ischemic and nervous system injury, where it reduces neuronal apoptosis and inflammatory cytokines and increases neurogenesis and angiogenesis. EPO also improves cognition in healthy volunteers and schizophrenic patients. We studied the effect of EPO administration on the gene-expression profile in the ischemic cortex of rats after cerebral ischemia at early time points (2 and 6 h). EPO treatment up-regulated genes already increased by ischemia. Hierarchical clustering and analysis of overrepresented functional categories identified genes implicated in synaptic plasticity—Arc, BDNF, Egr1, and Egr2, of which Egr2 was the most significantly regulated. Up-regulation of Arc, BDNF, Dusp5, Egr1, Egr2, Egr4, and Nr4a3 was confirmed by quantitative PCR. We investigated the up-regulation of Egr2/Krox20 further because of its role in neuronal plasticity. Its elevation by EPO was confirmed in an independent in vivo experiment of cerebral ischemia in rats. Using the rat neuroblastoma B104, we found that wild-type cells that do not express EPO receptor (EPOR) do not respond to EPO by inducing Egr2. However, EPOR-expressing B104 cells induce Egr2 early upon incubation with EPO, indicating that Egr2 induction is a direct effect of EPO and that EPOR mediates this effect. Because these changes occur in vivo before decreased inflammatory cytokines or neuronal apoptosis is evident, these findings provide a molecular mechanism for the neuroreparative effects of cytokines and suggest a mechanism of neuroprotection by which promotion of a plastic phenotype results in decreased inflammation and neuronal death.
Keywords: microarrays, ischemia-reperfusion injury, neurotrophins, early genes, neuronal cells
Since our first report (1), several studies have documented the neuroprotective effect of erythropoietin (EPO) in models of ischemic and traumatic brain injury (reviewed in refs. 2–4) and the role of endogenous EPO in ischemic preconditioning (5). Multiple mechanisms can account for the action of EPO, including inhibition of neuronal apoptosis (6) and decreased neuroinflammation (7, 8). EPO also activates repair, in particular through promotion of neurogenesis, oligodendrogenesis, and angiogenesis (9, 10), as well as mobilization of endothelial progenitor cells (11). It also improves cognition, long-term potentiation (LTP), and synaptic plasticity (4, 12–14).
However, the early effects of EPO responsible for its neuroprotective activities are not understood, and there even is debate whether the classical EPO receptor (EPOR) alone mediates these effects or an additional tissue-protective coreceptor is required (15–18).
In the study presented here, we investigated the effect of EPO on the gene-expression profile of the brain using the rat model of cerebral ischemia induced by middle cerebral artery occlusion (MCAO) with which we performed most of the studies on EPO. To identify early events induced by EPO, experiments were carried out at the time points 2 and 6 h post-MCAO, when ischemic damage is not yet detected by histology. The results obtained show that the early effects of EPO are on genes important for neuronal synaptic plasticity, particularly early growth response 2 (Egr2). In vitro experiments using a neuronal cell line show that EPOR is necessary for EPO induction of Egr2, clearly demonstrating that EPOR is implicated in the effects of EPO on cells of the nervous system and not just in its erythropoietic activity on erythroid precursors. These results strengthen the evidence of EPO as a tissue-reparative cytokine.
Results
Identification of EPO-Regulated Genes.
Three groups of rats were studied: 12 sham-operated rats (S), 12 ischemic rats undergoing MCAO with saline treatment (I), and 12 ischemic rats undergoing MCAO with EPO treatment (IE). Six rats per group were killed 2 h after MCAO, and six rats were killed at 6 h after MCAO, obtaining six experimental groups (S, I, and IE at 2 h and 6 h). Microarray analysis was performed in the ischemic cortex to identify genes differentially expressed in the EPO-treated groups. For microarray analysis, RNA samples were pooled from six rats to obtain three biological replicates per group. Each replicate was obtained from two rats. Each rat contributed to only one pool.
With a cutoff of P < 0.01 and of a fold-change of 2 (corresponding to a log base 2 change of 1), strikingly at 6 h EPO induced the expression of only one gene, Egr2. At 2 h one gene (Olr792_predicted) was up-regulated, and one (LOC683790) was down-regulated, but their absolute expression level was very low (just above the 4.2 expression threshold).
Because we intended to use the microarray analysis only as a first discovery step, and we intended to validate and pursue any difference of interest by quantitative PCR (qPCR), we decided to lower the stringency to P < 0.05/1.5-fold to see if there was a discernible pattern in the transcripts affected by EPO. At this stringency, as shown in Table 1, EPO regulated 1.4% and 2.2% (at 2 h and 6 h, respectively) of the transcripts affected by ischemia, but only 0.09% and 0.2%, respectively, of those unaffected by ischemia.
Table 1.
Summary of transcriptional changes by ischemia or ischemia+EPO at 2 and 6 h
| Change | Total | Up by EPO | Down by EPO |
| 2 h | |||
| Up by ischemia | 385 | 0 | 5 |
| Down by ischemia | 240 | 4 | 0 |
| Unaffected | 39,707 | 7 | 28 |
| 6 h | |||
| Up by ischemia | 1,115 | 25 | 4 |
| Down by ischemia | 442 | 9 | 1 |
| Unaffected | 37,677 | 52 | 19 |
Expression changes at P < 0.05, 1.5-fold, were considered. Ischemia was compared with sham-operated rats, and EPO-treated ischemic rats were compared with ischemia alone.
At 2 h, EPO up-regulated 11 transcripts (three genes) and down-regulated 33 transcripts (10 genes) (Table S1); neither manual screening nor functional classification analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) could identify any functional cluster. At 6 h (Table 2; see Table S2 for the full list), EPO up-regulated 86 transcripts (29 genes), and down-regulated 24 transcripts (13 genes).
Table 2.
Genes significantly changed by EPO in ischemic cortex at 6 h and relative change in ischemic versus sham
| Gene symbol | Accession number | Ischemia+EPO vs. ischemia |
Ischemia vs. sham |
||
| Fold change | P value | Fold change | P value | ||
| Up-regulated | |||||
| RGD1304775_predicted | XM_237151 | 2.80 | 0.0469 | ns | — |
| RGD1310265_predicted | XM_001070727 | 2.38 | 0.0423 | ns | — |
| Krt14 | D63774 | 2.32 | 0.0377 | ns | — |
| Slc10a1 | NM_017047 | 2.28 | 0.0273 | ns | — |
| LOC679379 | XM_001055377 | 2.23 | 0.0468 | ns | — |
| ENSRNOT00000014809 | ENSRNOT00000014809 | 2.02 | 0.0229 | ns | — |
| Ces5 | XM_341636 | 1.62 | 0.0479 | ns | — |
| RGD1563378_predicted | XM_228994 | 1.60 | 0.0342 | ns | — |
| Olr1461_predicted | NM_001000022 | 1.46 | 0.0476 | ns | — |
| BDNF* | NM_012513 | 1.03 | 0.0467 | 1.49 | 0.0026 |
| Dusp5 | NM_133578 | 1.01 | 0.0333 | 1.42 | 0.0018 |
| Egr2 | NM_053633 | 1.01 | 0.0077 | 1.42 | 0.0089 |
| Olr372_predicted | NM_001001048 | 1.00 | 0.0153 | ns | — |
| Arc* | NM_019361 | 0.92 | 0.0467 | 1.57 | 0.0008 |
| Fosl2 | NM_012954 | 0.86 | 0.0099 | ns | — |
| Mas1 | NM_012757 | 0.85 | 0.0188 | ns | — |
| Egr4 | NM_019137 | 0.85 | 0.0253 | 1.08 | 0.0066 |
| LOC684624 | XM_001070871 | 0.84 | 0.0472 | ns | — |
| Rem2 | NM_022685 | 0.75 | 0.0183 | 1.29 | 0.0192 |
| Olr1678_predicted | NM_001000893 | 0.73 | 0.0395 | ns | — |
| Prssl1 | NM_001003956 | 0.68 | 0.0057 | ns | — |
| XM_224859 | XM_224859 | 0.66 | 0.0169 | ns | — |
| RGD1311223_predicted | XM_345971 | 0.65 | 0.0300 | ns | — |
| Nr4a3* | DQ268830 | 0.64 | 0.0089 | ns | — |
| Cdkl3 | NM_021772 | 0.61 | 0.0274 | ns | — |
| Egr1* | NM_012551 | 0.60 | 0.0270 | ns | — |
| RGD1562685_predicted | XM_231463 | 0.60 | 0.0099 | ns | — |
| Angptl4 | NM_199115 | 0.59 | 0.0285 | 1.02 | 0.0123 |
| Ccl7* | NM_001007612 | 0.59 | 0.0228 | 2.84 | 0.0008 |
| Down-regulated | |||||
| Olr750_predicted | NM_001000366 | −1.56 | 0.0321 | ns | — |
| Atp7a | NM_052803 | −1.21 | 0.0159 | ns | — |
| Trem1_predicted | XM_217336 | −1.03 | 0.0243 | 1.67 | 0.0057 |
| Olr1630_predicted | NM_001000092 | −0.97 | 0.0240 | ns | — |
| RGD1307937 | NM_001013877 | −0.82 | 0.0265 | ns | — |
| RGD1310352 | XM_220404 | −0.71 | 0.0141 | ns | — |
| Zfp606 | XM_218283 | −0.67 | 0.0429 | ns | — |
| Cxcl2 | NM_053647 | −0.65 | 0.0369 | 6.49 | 1.4E-07 |
| LOC679115 | XM_001054757 | −0.60 | 0.0235 | ns | — |
| RGD1310980_predicted | XM_343381 | −0.60 | 0.0358 | ns | — |
| LOC680443 | XM_001057208 | −0.60 | 0.0115 | ns | — |
| Rnf24_predicted | XM_342522 | −0.59 | 0.0243 | ns | — |
| Crispld1_predicted | XM_237258 | −0.59 | 0.0491 | ns | — |
Only genes with a functional annotation changed more than 1.5-fold, P < 0.05, in ischemia+EPO vs. ischemia are included. Fold change is expressed as log base 2 ratio and is the average of triplicate samples. ns, not significant.
*When genes are identified by different probes/replicates, the average of the gene expression level (gProcessed Signal) of all the different probes/replicates has been calculated. For these genes, the numbers of significantly different transcripts/total transcripts were Ccl7, 2/2; Nr4a3, 2/3; BDNF, 6/11; Arc 7/10; Egr1, 8/10. All replicates were considered when calculating statistical significance. A list of all significantly up-regulated transcripts, including unmapped probe IDs and replicates, is presented in Table S2.
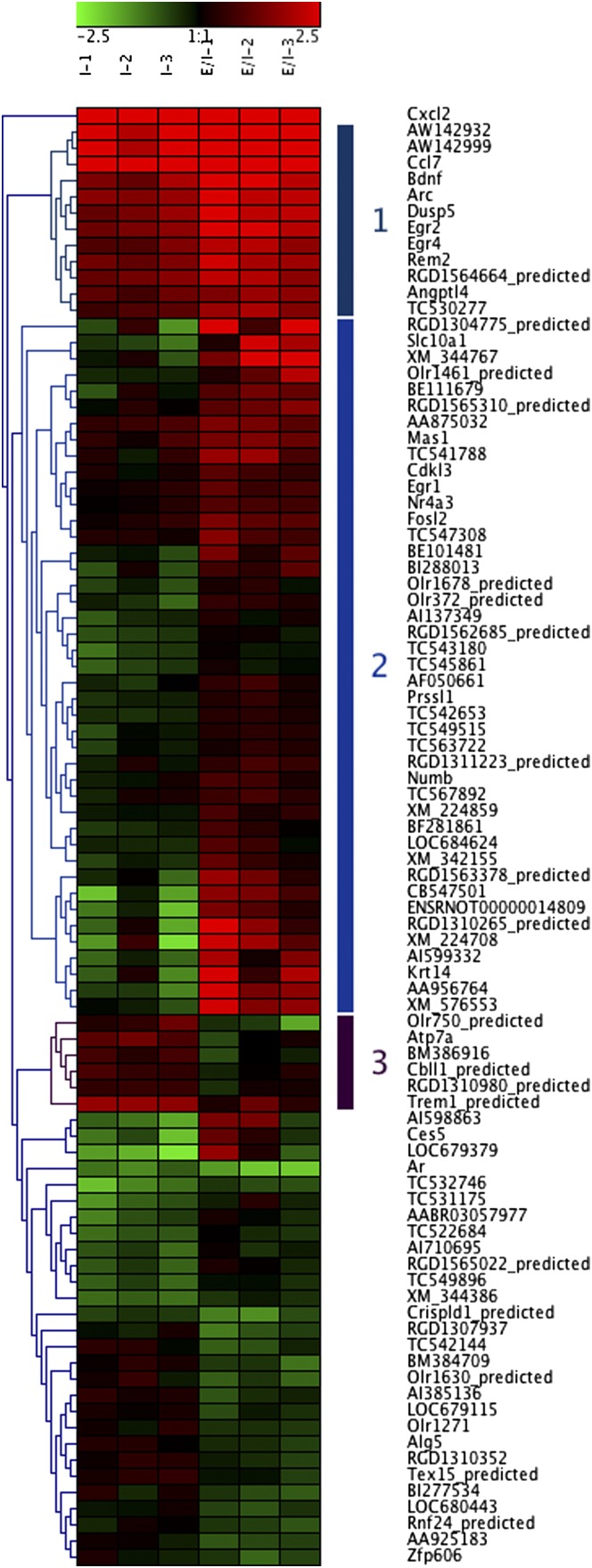
Hierarchical cluster analysis was performed on the genes regulated by EPO at 6 h (Fig. 1). Transcripts in cluster 1 include Arc, BDNF, Ccl7, Dusp5, Egr2, and Egr4, which already were up-regulated markedly by ischemia and were up-regulated further by EPO. Transcripts in cluster 2 (including Egr1, Fosl2, and Nr4a3) were not regulated significantly by ischemia at 6 h but were up-regulated by ischemia+EPO. Cluster 3 included a few genes whose up-regulation by ischemia was inhibited by EPO, such as Trem1 and Atp7a.
Fig. 1.
Cluster analysis. Hierarchical clustering and heat map of the differentially expressed transcripts identified by comparing ischemia+EPO (IE) vs. ischemia alone (I) and setting a threshold of 1.5-fold change, P < 0.05. Each sample (pooled RNA from two rats) represents the expression change compared with the mean of three samples from sham-operated rats. Red indicates an increase and green indicates a decrease in expression compared with sham controls. Average linkage clustering analysis was performed using Genesis software.
We then used DAVID to identify overrepresented (enriched) functional categories among the EPO–up-regulated genes. The top ranking categories were “regulation of neuronal synaptic plasticity,” “behavior,” and “learning or memory” (Table 3), comprising genes in clusters 1 and 2 from Fig. 1. No enriched functional categories were identified analyzing the transcripts down-regulated by EPO.
Table 3.
Functional categories enriched in EPO–up-regulated genes
| Category | Fold enrichment | Gene symbols | P value |
| Regulation of neuronal synaptic plasticity | 44.9 | BDNF, Egr1, Egr2, Arc | 7.9E-05 |
| Behavior | 7.8 | BDNF, Egr1, Egr2, fosl-2, Nr4a3, CCl7 | 6.6E-04 |
| Learning or memory | 16.7 | BDNF, Egr1, Egr2, fosl-2 | 1.5E-03 |
| Zinc finger | 45.3 | Egr1, Egr2, Egr4 | 1.7E-03 |
DAVID Functional Annotation Chart analysis showing the overrepresented (enriched) categories among the genes up-regulated by EPO in rat ischemic cortex at 6 h. The four top categories are shown. Reported are the fold enrichment, the list of the gene symbols, and the significance of the enrichment (P value).
Because we and others had reported that EPO decreased neuroinflammation at later times [24 h after MCAO or later (7, 8, 19, 20)], we were surprised that no inflammatory cytokines or their receptors were among the transcripts down-regulated by EPO. In fact, in agreement with previous studies, ischemia markedly induced several inflammatory genes, including Il1b, Il6, Tnf, and many other cytokines, but their induction was unaffected by EPO (Dataset S1). The only genes with the Gene Ontology (GO) database description “inflammatory response” or “immune response” whose expression was down-regulated significantly by EPO were Cxcl2 and Trem1, whereas Ccl7 was up-regulated (Table 2 and Dataset S1). Interestingly, Ccl7 is a chemokine but also belongs to the GO category “behavior.” Likewise, because a previous study on PC-12 cells treated with EPO for 24 h reported an up-regulation of antiapoptotic Bad, Bax, and Bcl-xL (21), we specifically looked for genes related to apoptosis. None of them was affected by EPO, even when transcripts with low (below 4.2) expression levels were taken into account, as can be seen from Dataset S2 that lists all genes with “apoptosis” or “cell death” in the GO.
Validation of Microarray Data by PCR.
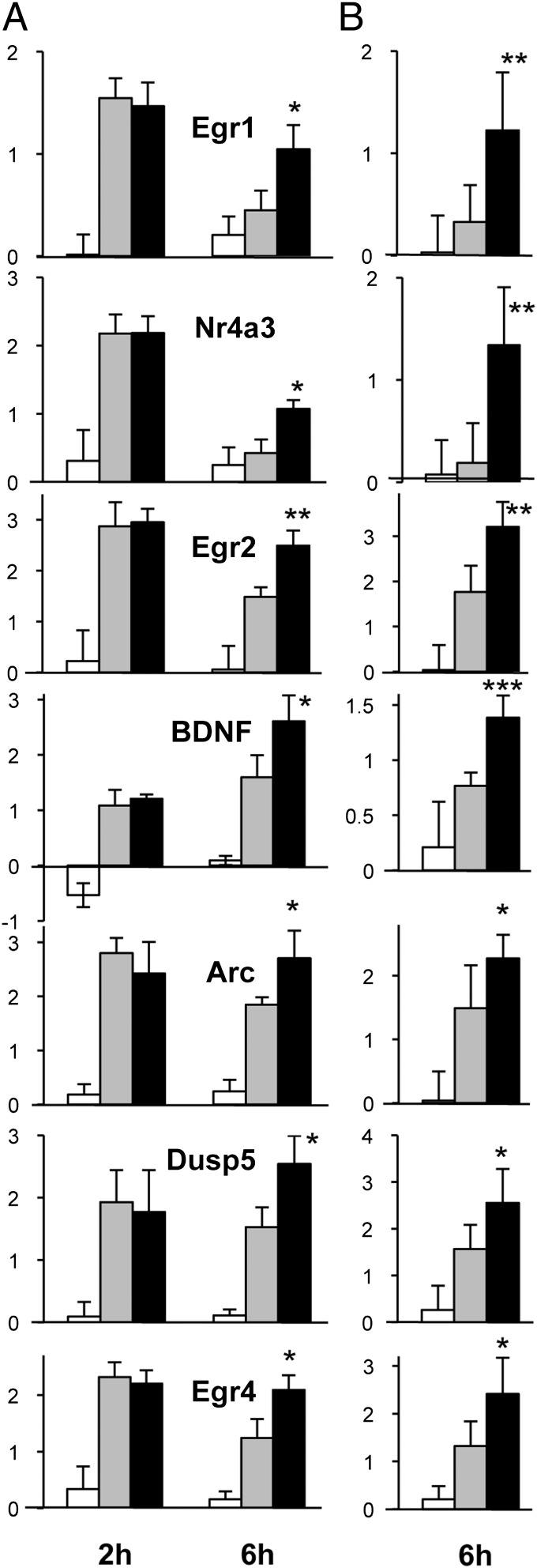
Selected genes among those significantly up-regulated by EPO at 6 h were validated by qPCR. In this case, unlike the microarray experiment, samples were not pooled, and qPCR analysis was performed on six individual rats per group. We also looked in the dataset for their expression at 2 h. Fig. 2 reports the expression data from the microarrays at 2 h and 6 h (Fig. 2A) and PCR validation at 6 h (Fig. 2B). Both Egr1 and Nr4a3, belonging to cluster 2, were strongly induced by ischemia at 2 h but returned to the control level by 6 h. The effect of EPO on these genes was to maintain and prolong their otherwise transient induction by ischemia. All the other genes (Egr2, BDNF, Arc, Dusp5, and Egr4), belonging to cluster 1 were induced by ischemia at 2 h and 6 h and were up-regulated further by EPO at 6 h. All the results obtained by microarrays at 6 h were confirmed by qPCR (Fig. 2B).
Fig. 2.
PCR validation of the microarray data. Results for seven genes are shown, comparing (A) expression data from microarrays at 2 h and 6 h (white bars represent sham surgery; gray bars represent ischemia; black bars represent ischemia+EPO) and (B) respective results from qPCR analysis at 6 h. Results are expressed as log base 2 ratio vs. one of the sham samples at 6 h. Microarray results are the average ± SD of triplicate samples (each sample was obtained by pooling two rat cortices). qPCR results are the average ± SD of six independent, nonpooled samples assayed in duplicate. ***P < 0.001; **P < 0.01; *P < 0.05 vs. ischemia by Student’s t test.
EPO-Induced Egr2 mRNA Expression in Neuronal Cells.
Our findings on genes involved in synaptic plasticity support the importance of EPO in neurorepair. Because only Egr2 was identified with the highest stringency analysis (fold-change of 2 and P < 0.01), we first sought to reproduce its induction by EPO in vivo in a second, independent cerebral ischemia experiment carried out exactly as the one used for microarray analysis (6 h after MCAO, six rats per group; three groups: sham, ischemia, and ischemia+EPO). Egr2, measured by qPCR, was induced significantly in ischemic compared with sham-operated rats (log base 2 expression ratio ± SD of I vs. S: 1.5 ± 1, P < 0.05) and was up-regulated further by EPO in ischemic animals (log base 2 ratio ± SD, IE vs. I: 0.8 ± 0.5, P < 0.05). Thus, EPO increased Egr2 in ischemic animals 1.8-fold (log base 2 ratio = 0.8), confirming the results of the first experiment.
Because the microarray experiment did not include a group of rats treated with EPO in the absence of ischemia, we wondered whether EPO directly induced expression of Egr2 or only up-regulated Egr2 induced by cerebral ischemia. For this purpose, we treated healthy rats with the same dose of EPO (50 µg/kg i.p.) and measured the expression of Egr2 in the brain at 2 h and 6 h. The results showed that EPO did not affect Egr2 expression, compared with that in rats injected with saline alone, at any time point [Egr2 mRNA levels, log base 2 expression ratio ± SD, EPO vs. no EPO, n = 6; at 2 h: −0.5 ± 0.3, nonsignificant (ns); at 6 h: 0.3 ± 0.9, ns]. Therefore, the in vivo effect of EPO on Egr2 was to modulate its induction by ischemia.
Because change in gene expression in the brain can take place in several cell populations, we investigated in vitro the effect of EPO on Egr2 in neuronal cells using the rat neuronal cell line B104. Serum-deprived cells were treated with 80 ng/mL EPO, and Egr2 mRNA expression was measured 1, 3, and 5 h later. In our experiments, EPO did not affect Egr2 expression in wild-type B104 cells, as measured by qPCR (log base 2 expression ratio ± SD, EPO vs. no EPO, n = 3; at 1 h: 0.01 ± 0.06, ns; at 3 h: −0.12 ± 0.09, ns; at 5 h: 0.19 ± 0.06, ns). However, we found that these cells do not express detectable EPOR by qPCR (fluorescence threshold cycle for EPOR amplification was >38). On the other hand, EPOR is up-regulated in brain injury and ischemia (22). We therefore overexpressed EPOR in B104 cells. As shown in Fig. 3, EPO induced Egr2 mRNA at 1 h by about 10-fold; then the levels decreased but still were up-regulated (1.6-fold) at 3 h and returned to control level at 5 h. Of note, EPOR-expressing cells showed functional EPOR signaling in terms of autophosphorylation upon EPO treatment (see Fig. S2).
Fig. 3.
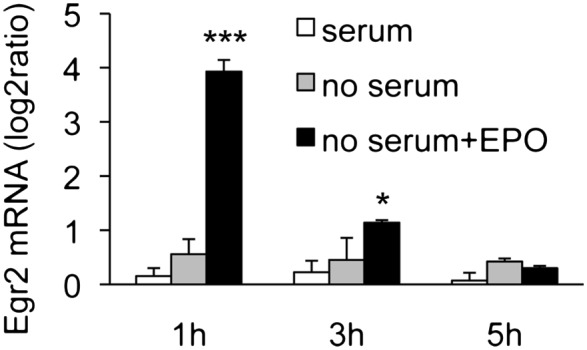
EPO induces Egr2 mRNA in rat B104-EPOR neuronal cells in vitro. Cells were plated in 24-well plates at 120,000 cells/mL in complete medium. After overnight incubation, the cells were deprived of serum for 4 h and then were stimulated with EPO (80 ng/mL) for the indicated time. Egr2 mRNA was measured by qPCR, using GAPDH as a housekeeping gene. Results represent the change in expression level vs. one of the control (serum) samples, expressed as log base 2 ratio, and are the mean ± SD of triplicate samples assayed in duplicate. ***P < 0.001; *P < 0.05 vs. no serum by Student’s t test.
Discussion
Overall, the main transcriptional effect of EPO at early time points was to regulate genes whose expression already was affected by ischemia. For the majority of transcripts, EPO amplified or prolonged the effect of ischemia, which was particularly evident at 6 h, as shown in Table 1, suggesting that EPO potentiates protective or reparative pathways already activated by ischemic injury. In particular, Egr2 and other genes implicated in synaptic plasticity were up-regulated, or their induction by ischemia was prolonged. Among these genes was BDNF, thus confirming reports of its induction by EPO in stroke and experimental autoimmune encephalomyelitis (EAE) (9, 23). Although a previous study reported the induction of Egr1 by EPO in erythroid cells (24), the effect of EPO on Egr2 was not investigated in that study.
We were surprised to find no effect of EPO on genes related to inflammation or apoptosis, because a previous microarray study in a mouse model of neonatal brain hypoxia/ischemia showed inhibition of these pathways (25). However, the time point used in that study was 24 h or longer and thus may reflect the lesser damage in EPO-treated animals (25). Unlike Juul et al. (25) who observed, at 24 h or later, an overall normalizing effect of EPO on genes up- or down-regulated by ischemia, we found that at earlier time points EPO amplified responses to ischemia. Of note, the lack of inhibitory effect of EPO on the expression of inflammatory cytokines (with the exception of Cxcl2) was observed even if these genes were markedly induced at the time points analyzed (Dataset S1). This lack of effect probably indicates that EPO does not inhibit the early triggering of the inflammatory response and is in agreement with our earlier report that EPO, although decreasing inflammatory cytokines 24 h after stroke (8) or in EAE (26), did not have any direct effect on the production of inflammatory cytokines by macrophages or glial cells (8). Therefore, the decreased neuroinflammation and the decreased expression of Il6, Tnf, and Ccl2 observed at 24 h may be secondary to neuroprotection/neurorepair (8).
Interestingly, EPO did not affect brain Egr2 expression in the absence of cerebral ischemia, thus strengthening the hypothesis of Brines and Cerami (27) that tissue injury “primes” cells for response to EPO. For instance, TNF could produce such a priming, as reported with primary neurons (22), and TNF is induced early in cerebral ischemia, along with most inflammatory cytokines (28) (see also Dataset S1). Thus, although EPO did not reduce inflammatory genes at these early time points, one might speculate that neuroinflammation might be a factor that primes the brain for the tissue-protective action of EPO.
Induction of Egr2 may be important for many central actions of EPO. Egr2, also known as “Krox20,” is part of the Kruppel-like zinc finger transcription factor family, which also includes Egr1, Egr3, and Egr4 and has several functions that might be important in the pharmacodynamics of EPO in neurological diseases. Egr genes are induced by neuronal activity and brain injury, stimuli that cause synaptic plasticity (reviewed in refs. 29 and 30). The most thoroughly studied member of the family is Egr1, whose role in synaptic plasticity associated with learning and memory is well documented (30, 31). Egr2 is induced by neuronal activity (29), but less is known about its specific role. However, Egr2 was clearly shown to mediate stabilization and maintenance of LTP (32, 33) and cognitive functions associated with attention (34) in models in which Egr1 was induced only transiently (33) or was not induced (34). Therefore, different members of the Egr family might mediate different cognitive functions associated with neuronal plasticity. The finding that EPO preferentially induces Egr2 in cerebral ischemia might highlight a specific pathway through which EPO induces functional recovery in stroke and improves cognitive functions in diseases such as schizophrenia (35) and multiple sclerosis (12).
Although the molecular mechanisms that link Egr induction to long-term effects mediating neuronal plasticity are unknown, Egr1 and Egr3 can regulate directly activity-regulated cytoskeleton-associated protein (Arc) (36), a plasticity-associated gene involved in the maintenance, but not in the induction, of LTP and consolidation of long-term memory (37). Arc can be induced as an early gene, similar to the Egr, but also through a protein synthesis-dependent mechanism mediated by Egr3 (36). Interestingly, Arc also is among the genes induced by EPO in our model.
Previous studies have shown that several early genes are up-regulated in cerebral ischemia. Although the most studied are fos/jun family members, zinc finger transcription factors, including Egr2, also are induced in the brain after permanent (38) or transient (39) ischemia. Studies addressing the role of fos/jun in neurotoxicity/neuroprotection have produced apparently contradictory evidence. In particular, fos/jun members are implicated in neuronal apoptosis (40), but when c-fos is inhibited in vivo with antisense oligonucleotides, cerebral ischemia-induced brain damage is increased (41), and ischemia-induced NGF is inhibited (42), suggesting a protective function. There are no studies investigating the role of Egr2 in stroke by blocking its expression, but indirect evidence for a protective role of Egr in stroke is provided by a study showing that these genes are expressed preferentially in surviving neurons compared with neurons committed to die (38). Furthermore, a study carried out at 6 h after ischemia identified Egr1, Egr2, Egr4, and Nr4a3 among the neuroprotective genes up-regulated by hypothermia in a model of hypothermia-induced neuroprotection in experimental stroke in rats (43). Likewise, Egr2 protects osteoclasts and T cells from apoptosis (44, 45) and therefore might contribute to the well-known antiapoptotic effect of EPO (46).
Further studies in which Egr2 in the CNS is inhibited by either conditional knockout or antisense oligonucleotides will be necessary, and are feasible, to investigate the relevance of Egr2, and possibly other genes reported here, in the neuroprotective effect of EPO.
Methods
Animals and Treatments.
All experimental procedures were performed in accordance with the European Communities Council Directive #86/609 for care of laboratory animals and in agreement with national regulations on animal research in Italy and Turkey. Surgery was carried out on male Crl:CD (SD)BR rats weighing 250–285 g. Recombinant human EPO (rhEPO) (Creative Dynamics) was given i.p. at the dose of 50 µg/kg 1 h after MCAO, as described previously (1, 8). Ischemic rats received either rhEPO or saline (vehicle). Animals were killed 2 or 6 h after MCAO, the brains were removed, and the ipsilateral cortex was frozen in liquid nitrogen.
RNA Extraction.
Tissue was homogenized in TRIzol (Invitrogen, Life Technologies). Total RNA was extracted from the homogenates using silica spin columns provided with the TRIzol Plus RNA Purification kit, following the manufacturer’s instructions (Invitrogen). RNA quality and concentration were determined using a NanoDrop ND-1000 (NanoDrop Technologies) and an Agilent 2100 Bioanalyzer (Agilent Technologies). The RNA from the six rats in each group was pooled to obtain three biological replicates per group, each replicate containing RNA pooled from two rats. This pooling strategy was designed to limit the number of microarrays. It has been shown that, provided multiple pools are analyzed for each group and each pool is a biological replicate, pooling RNA samples for microarray analysis can decrease variability, improve accuracy, and increase power (47, 48). In total, 18 arrays were done: three sham (S), three ischemic+saline (I), and three ischemic+EPO (IE) samples at each time point (2 and 6 h).
Microarrays.
For each sample, 1 µg of total RNA spiked with 10 viral polyA transcript controls (Agilent) was converted to double-stranded cDNA in a reverse-transcription reaction. Subsequently the sample was converted to antisense cRNA, amplified, and labeled with Cyanine 3-CTP (Cy3) in an in vitro transcription reaction according to the manufacturer’s protocol (Agilent). Purified and labeled cRNA (28 pmol of Cy3-labeled cRNA) was hybridized on Rat Whole Genome array (ID 014879; Agilent) followed by manual washing, according to the manufacturer’s procedures. To assess the raw probe signal intensities, arrays were scanned using the Agilent DNA MicroArray Scanner with SureScan High-Resolution Technology, and probe signals were quantified using Agilent’s Feature Extraction software (version 9.1.1.1).
Microarray Data Analysis.
Raw data from the microarray experiments have been deposited at the Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo under accession no. GSE33725. Normalized data were analyzed using GeneSpring GX software (Agilent). We then filtered out transcripts with very low expression, because these transcripts may be a confounding factor in the analysis and introduce noise (49). Based on the frequency distribution of the transcript expression levels (gProcessed Signal) shown in Fig. S1, we filtered out all genes whose average gene expression was <18.4 (log base 2 < 4.2) (49), thereby removing 12.8% of the genes. Transcript expression between the experimental groups was compared by Student’s t test done on the log base 2 of the gProcessed Signal. Fold change in the expression was calculated as the ratio between the average of the gProcessed Signals of the various groups and where indicated was expressed as log base 2 ratio. Functional annotation and biological term enrichment analysis was done by using the DAVID database (50).
PCR Validation.
Reverse transcription and real-time qPCR were carried out as reported (22) on RNA from each individual rat, without pooling. PCR reactions were run on the MX3000 PCR machine (Stratagene, Agilent), using Taqman gene expression assays (Applied Biosystems, Life Technologies) and Brilliant II qPCR master mix (Stratagene). Gene expression was quantified using the ΔΔCt method, following Applied Biosystems guidelines. Genes expression was considered undetectable when the threshold cycle for fluorescence detection was >38. Results were normalized to GAPDH expression (housekeeping gene) and expressed as log base 2 of the relative gene expression (ratio) vs. one of the sham samples at 6 h, chosen as the calibrator (51).
Cell Culture.
The rat neuroblastoma B104 cell line and the genetically modified B104-EPOR cell line were cultured in DMEM (PAA Laboratories) supplemented with 10% (vol/vol) FBS (Invitrogen). When treated with EPO, cells were switched to medium without serum with 5 µg/mL insulin, 5 µg/mL transferrin, and 5 ng/mL selenium (Sigma-Aldrich). Total RNA was extracted with TRIzol (Invitrogen), and reverse transcription and qPCR were done as above.
Generation of Genetically Modified B104-EPOR Cells.
B104 cells were genetically modified to express EPOR (B104-EPOR) constitutively. Production of lentivector particles, gene transfer, and cloning of B104 cells were performed as described (52). Expression and EPO-induced activation of EPOR in transduced cells is shown in Fig. S2. A more detailed description of the techniques used for molecular cloning and to detect expression and activation of EPOR in B104-EPOR cells is included in SI Methods.
Supplementary Material
Acknowledgments
We thank Joke Allemeersch (Flanders Institute for Biotechnology) for help with analyzing the data and Sara Triulzi (Mario Negri Institute) for help with animal experiments. This work was supported by the Interuniversity Poles of Attraction Program-Belgian Science Policy Grant P5/12 and by the European Regional Development Fund Grant Trans Channel Neuroscience Network (TC2N) (to P. Vandenabeele and P.G.). P.G. is supported by the Brighton and Sussex Medical School and the R. M. Phillips Charitable Trust. P. Vandenabeele is holder of a Methusalem grant (BOF09/01M00709) from the Flemish Government. Y.C. and A.A. are supported by the National Multiple Sclerosis Society-USA (Promise 2010).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO), http://www.ncbi.nlm.nih.gov/geo (accession no. GSE33725).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200554109/-/DCSupplemental.
References
- 1.Brines ML, et al. Erythropoietin crosses the blood-brain barrier to protect against experimental brain injury. Proc Natl Acad Sci USA. 2000;97:10526–10531. doi: 10.1073/pnas.97.19.10526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brines M, Cerami A. Emerging biological roles for erythropoietin in the nervous system. Nat Rev Neurosci. 2005;6:484–494. doi: 10.1038/nrn1687. [DOI] [PubMed] [Google Scholar]
- 3.Byts N, Sirén AL. Erythropoietin: A multimodal neuroprotective agent. Exp Transl Stroke Med. 2009;1:4. doi: 10.1186/2040-7378-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargin D, Friedrichs H, El-Kordi A, Ehrenreich H. Erythropoietin as neuroprotective and neuroregenerative treatment strategy: Comprehensive overview of 12 years of preclinical and clinical research. Best Pract Res Clin Anaesthesiol. 2010;24:573–594. doi: 10.1016/j.bpa.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Pacary E, Petit E, Bernaudin M. Erythropoietin, a cytoprotective and regenerative cytokine, and the hypoxic brain. Neurodegener Dis. 2006;3:87–93. doi: 10.1159/000092098. [DOI] [PubMed] [Google Scholar]
- 6.Sirén AL, et al. Erythropoietin prevents neuronal apoptosis after cerebral ischemia and metabolic stress. Proc Natl Acad Sci USA. 2001;98:4044–4049. doi: 10.1073/pnas.051606598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa P, et al. Reduced functional deficits, neuroinflammation, and secondary tissue damage after treatment of stroke by nonerythropoietic erythropoietin derivatives. J Cereb Blood Flow Metab. 2007;27:552–563. doi: 10.1038/sj.jcbfm.9600370. [DOI] [PubMed] [Google Scholar]
- 8.Villa P, et al. Erythropoietin selectively attenuates cytokine production and inflammation in cerebral ischemia by targeting neuronal apoptosis. J Exp Med. 2003;198:971–975. doi: 10.1084/jem.20021067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Zhang Z, Wang Y, Zhang R, Chopp M. Treatment of stroke with erythropoietin enhances neurogenesis and angiogenesis and improves neurological function in rats. Stroke. 2004;35:1732–1737. doi: 10.1161/01.STR.0000132196.49028.a4. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, et al. Erythropoietin amplifies stroke-induced oligodendrogenesis in the rat. PLoS ONE. 2010;5:e11016. doi: 10.1371/journal.pone.0011016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahlmann FH, et al. Erythropoietin regulates endothelial progenitor cells. Blood. 2004;103:921–926. doi: 10.1182/blood-2003-04-1284. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenreich H, et al. Exploring recombinant human erythropoietin in chronic progressive multiple sclerosis. Brain. 2007;130:2577–2588. doi: 10.1093/brain/awm203. [DOI] [PubMed] [Google Scholar]
- 13.Adamcio B, et al. Erythropoietin enhances hippocampal long-term potentiation and memory. BMC Biol. 2008;6:37. doi: 10.1186/1741-7007-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrenreich H, Bartels C, Sargin D, Stawicki S, Krampe H. Recombinant human erythropoietin in the treatment of human brain disease: Focus on cognition. J Ren Nutr. 2008;18:146–153. doi: 10.1053/j.jrn.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 15.Brines M, et al. Erythropoietin mediates tissue protection through an erythropoietin and common beta-subunit heteroreceptor. Proc Natl Acad Sci USA. 2004;101:14907–14912. doi: 10.1073/pnas.0406491101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Um M, Gross AW, Lodish HF. A “classical” homodimeric erythropoietin receptor is essential for the antiapoptotic effects of erythropoietin on differentiated neuroblastoma SH-SY5Y and pheochromocytoma PC-12 cells. Cell Signal. 2007;19:634–645. doi: 10.1016/j.cellsig.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Brines M, Cerami A. Erythropoietin-mediated tissue protection: Reducing collateral damage from the primary injury response. J Intern Med. 2008;264:405–432. doi: 10.1111/j.1365-2796.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- 18.Ghezzi P, et al. Erythropoietin: Not just about erythropoiesis. Lancet. 2010;375:2142. doi: 10.1016/S0140-6736(10)60992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Calvert JW, Zhang JH. Neonatal hypoxia/ischemia is associated with decreased inflammatory mediators after erythropoietin administration. Stroke. 2005;36:1672–1678. doi: 10.1161/01.STR.0000173406.04891.8c. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Post-ischemic treatment with erythropoietin or carbamylated erythropoietin reduces infarction and improves neurological outcome in a rat model of focal cerebral ischemia. Br J Pharmacol. 2007;151:1377–1384. doi: 10.1038/sj.bjp.0707285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renzi MJ, et al. Erythropoietin induces changes in gene expression in PC-12 cells. Brain Res Mol Brain Res. 2002;104:86–95. doi: 10.1016/s0169-328x(02)00323-6. [DOI] [PubMed] [Google Scholar]
- 22.Taoufik E, et al. TNF receptor I sensitizes neurons to erythropoietin- and VEGF-mediated neuroprotection after ischemic and excitotoxic injury. Proc Natl Acad Sci USA. 2008;105:6185–6190. doi: 10.1073/pnas.0801447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Erythropoietin treatment improves neurological functional recovery in EAE mice. Brain Res. 2005;1034:34–39. doi: 10.1016/j.brainres.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Fang J, et al. EPO modulation of cell-cycle regulatory genes, and cell division, in primary bone marrow erythroblasts. Blood. 2007;110:2361–2370. doi: 10.1182/blood-2006-12-063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juul SE, et al. Microarray analysis of high-dose recombinant erythropoietin treatment of unilateral brain injury in neonatal mouse hippocampus. Pediatr Res. 2009;65:485–492. doi: 10.1203/PDR.0b013e31819d90c8. [DOI] [PubMed] [Google Scholar]
- 26.Savino C, et al. Delayed administration of erythropoietin and its non-erythropoietic derivatives ameliorates chronic murine autoimmune encephalomyelitis. J Neuroimmunol. 2006;172:27–37. doi: 10.1016/j.jneuroim.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 27.Brines M, Cerami A. The receptor that tames the innate immune response. Mol Med. 2012;18:486–496. doi: 10.2119/molmed.2011.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meistrell ME, 3rd, et al. Tumor necrosis factor is a brain damaging cytokine in cerebral ischemia. Shock. 1997;8:341–348. [PubMed] [Google Scholar]
- 29.O’Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM. The EGR family of transcription-regulatory factors: Progress at the interface of molecular and systems neuroscience. Trends Neurosci. 1999;22:167–173. doi: 10.1016/s0166-2236(98)01343-5. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Cadahía B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol. 2011;89:61–73. doi: 10.1139/O10-138. [DOI] [PubMed] [Google Scholar]
- 31.Worley PF, et al. Thresholds for synaptic activation of transcription factors in hippocampus: Correlation with long-term enhancement. J Neurosci. 1993;13:4776–4786. doi: 10.1523/JNEUROSCI.13-11-04776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inokuchi K, Murayama A, Ozawa F. mRNA differential display reveals Krox-20 as a neural plasticity-regulated gene in the rat hippocampus. Biochem Biophys Res Commun. 1996;221:430–436. doi: 10.1006/bbrc.1996.0612. [DOI] [PubMed] [Google Scholar]
- 33.Williams J, et al. Krox20 may play a key role in the stabilization of long-term potentiation. Brain Res Mol Brain Res. 1995;28:87–93. doi: 10.1016/0169-328x(94)00187-j. [DOI] [PubMed] [Google Scholar]
- 34.DeSteno DA, Schmauss C. Induction of early growth response gene 2 expression in the forebrain of mice performing an attention-set-shifting task. Neuroscience. 2008;152:417–428. doi: 10.1016/j.neuroscience.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehrenreich H, et al. Improvement of cognitive functions in chronic schizophrenic patients by recombinant human erythropoietin. Mol Psychiatry. 2007;12:206–220. doi: 10.1038/sj.mp.4001907. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzowski JF, et al. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honkaniemi J, Sharp FR. Global ischemia induces immediate-early genes encoding zinc finger transcription factors. J Cereb Blood Flow Metab. 1996;16:557–565. doi: 10.1097/00004647-199607000-00005. [DOI] [PubMed] [Google Scholar]
- 39.An G, Lin TN, Liu JS, Hsu CY. Induction of Krox-20 expression after focal cerebral ischemia. Biochem Biophys Res Commun. 1992;188:1104–1110. doi: 10.1016/0006-291x(92)91345-q. [DOI] [PubMed] [Google Scholar]
- 40.Ham J, et al. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Widmayer MA, Zhang B, Cui JK, Baskin DS. Suppression of post-ischemic-induced fos protein expression by an antisense oligonucleotide to c-fos mRNA leads to increased tissue damage. Brain Res. 1999;832:112–117. doi: 10.1016/s0006-8993(99)01459-6. [DOI] [PubMed] [Google Scholar]
- 42.Cui JK, Hsu CY, Liu PK. Suppression of postischemic hippocampal nerve growth factor expression by a c-fos antisense oligodeoxynucleotide. J Neurosci. 1999;19:1335–1344. doi: 10.1523/JNEUROSCI.19-04-01335.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohta H, Terao Y, Shintani Y, Kiyota Y. Therapeutic time window of post-ischemic mild hypothermia and the gene expression associated with the neuroprotection in rat focal cerebral ischemia. Neurosci Res. 2007;57:424–433. doi: 10.1016/j.neures.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Bradley EW, Ruan MM, Oursler MJ. Novel pro-survival functions of the Kruppel-like transcription factor Egr2 in promotion of macrophage colony-stimulating factor-mediated osteoclast survival downstream of the MEK/ERK pathway. J Biol Chem. 2008;283:8055–8064. doi: 10.1074/jbc.M709500200. [DOI] [PubMed] [Google Scholar]
- 45.Lawson VJ, Weston K, Maurice D. Early growth response 2 regulates the survival of thymocytes during positive selection. Eur J Immunol. 2010;40:232–241. doi: 10.1002/eji.200939567. [DOI] [PubMed] [Google Scholar]
- 46.Ghezzi P, Brines M. Erythropoietin as an antiapoptotic, tissue-protective cytokine. Cell Death Differ. 2004;11(Suppl 1):S37–S44. doi: 10.1038/sj.cdd.4401450. [DOI] [PubMed] [Google Scholar]
- 47.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: From disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 49.Servant N, et al. EMA - A R package for Easy Microarray data analysis. BMC Res Notes. 2010;3:277. doi: 10.1186/1756-0500-3-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Annenkov A, et al. A chimeric receptor of the insulin-like growth factor receptor type 1 (IGFR1) and a single chain antibody specific to myelin oligodendrocyte glycoprotein activates the IGF1R signalling cascade in CG4 oligodendrocyte progenitors. Biochim Biophys Acta. 2011;1813:1428–1437. doi: 10.1016/j.bbamcr.2011.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.