Abstract
Gene targeting by zinc-finger nucleases in one-cell embryos provides an expedite mutagenesis approach in mice, rats, and rabbits. This technology has been recently used to create knockout and knockin mutants through the deletion or insertion of nucleotides. Here we apply zinc-finger nucleases in one-cell mouse embryos to generate disease-related mutants harboring single nucleotide or codon replacements. Using a gene-targeting vector or a synthetic oligodesoxynucleotide as template for homologous recombination, we introduced missense and silent mutations into the Rab38 gene, encoding a small GTPase that regulates intracellular vesicle trafficking. These results demonstrate the feasibility of seamless gene editing in one-cell embryos to create genetic disease models and establish synthetic oligodesoxynucleotides as a simplified mutagenesis tool.
Gene targeting in embryonic stem (ES) cells is routinely applied to modify the mouse genome and established the mouse as the most commonly used genetic animal model (1). Direct genome editing by zinc-finger nucleases (ZFN) in one-cell embryos has been recently established as an alternative mutagenesis approach in mice, rats, rabbits, and zebrafish (2–9). Such nucleases are designed to induce double-strand breaks (DSBs) at preselected genomic target sites (10–13). DSBs targeted to coding exons frequently undergo sequence deletions leading to gene knockout or allow the insertion (knockin) of DNA sequences from gene-targeting vectors via homologous recombination (HR). We and others recently reported the generation of knockout and knockin mutants at the Rosa26, Mdr1a, Pxr, and IgM loci by microinjection of ZFNs one-cell embryos of mice, rats, and rabbits (3, 5, 8).
Genetically modified mammals can serve as valuable models of human inherited disorders, but the majority of disease-associated alleles represent single nucleotide replacements that lead to missense, nonsense, or silent mutations (14, 15). The successful use of ZFNs to induce knockout and knockin alleles prompted us to further explore whether disease-related mutants harboring seamless nucleotide replacements can be created as well. Here we apply gene editing in one-cell mouse embryos to introduce nucleotide replacements into the Rab38 gene.
RAB38 is a member of the RAB small GTPase family that regulates intracellular vesicle trafficking (16, 17). In mice and rats, Rab38 is expressed in melanocytes, retinal pigment epithelial cells, alveolar pneumocytes, and platelets (17–20). In chocolate mutant mice, the Rab38 gene exhibits a nucleotide replacement within codon 19 (Rab38cht) (21). The resulting G19V amino acid substitution impairs the sorting of the tyrosinase-related protein 1 (TYRP1) into melanosomes, leading to impaired pigment production. Homozygous Rab38cht mice exhibit a chocolate-like brown coat color, ocular hypopigmentation, and alterations of the lung surfactants and alveolar structure (22–24). In the rat substrains Fawn-hooded and Cinnamon, a point mutation in the Ruby locus, encoding the Rab38 homolog, causes hypopigmentation, platelet storage pool deficiency, and lung pathology (19, 20, 25). These RAB38 deficient mutants are considered as phenotypic models of Hermansky-Pudlak syndrome (HPS) in humans, characterized by oculocutaneous albinism (OCA), progressive pulmonary fibrosis, and platelet storage disease (20, 26, 27). HPS defines a group of eight recessive disorders that arise from defects in the biogenesis of lysosome-related organelles, including melanosomes and platelet-dense granules (26, 27). The genes associated with HPS encode components of the biogenesis of lysosome-related organelles (BLOC-1,-2-3) and the AP3 protein complexes that play roles in intracellular protein trafficking and/or organelle distribution. In Drosophila, a synthetic lethal interaction between the BLOC-1 binding partner RAB11 and its paralog lightoid, the ortholog of mammalian Rab38/32, argues for a partially overlapping function of these RAB proteins (28, 29). Therefore, Rab38 is considered a HPS-related locus, though patients harboring mutations in Rab38 have not been identified (26, 27).
By employing ZFNs in one-cell embryos, we introduced missense and silent mutations into the first exon of Rab38 using a gene-targeting vector or a synthetic oligodesoxynucleotide (ODN) as repair templates. These results demonstrate the feasibility of seamless gene editing in one-cell mouse embryos to create genetic disease models and establish synthetic ODNs as a simplified tool for mutagenesis.
Results
Generation of Rab38 G19V Mutants by ZFNRab38 and Gene-Targeting Vector.
For HR with the Rab38 locus, we designed a gene-targeting vector that includes a glycine to valine replacement at codon 19 (G19V) in the first exon, flanked by homology regions of 0.9 kb and 2.8 kb (Fig. 1A). To create the G19V replacement, a T was placed at the second codon position as found in Rab38cht (chocolate) mutants (21), whereas a silent A replacement was made at the third position as an identifier for our targeted Rab38IDG-Cht allele (Fig. 1B). These replacements further generated a SexAI and erased a BsaJI site and enabled us to discriminate the targeted Rab38IDG-Cht and the Rab38+ (wild type) allele by restriction analysis. To stimulate HR at the Rab38 locus, we used a ZFN pair (ZFNRab38) with a specific cleavage site located 50 bp downstream of codon 19 (Fig. 1A). To avoid the processing of the targeting vector by ZFNRab38, the vector‘s ZFN binding sequences were modified by silent nucleotide replacements (Fig. 1D).
Fig. 1.
Generation of Rab38 G19V mutants by ZFNRab38 and gene-targeting vector. (A) The targeting vector is designed for the insertion of a G19V mutation into the first exon of Rab38. The structure of the Rab38+ locus, the targeted Rab38IDG-Cht allele, and the location of the ZFNRab38 binding sites of the Rab38 5′ probe and of PCR primers P-for and P-rev are shown. The positions of SexAI (S), BsaJI (B), and ApaLI (A) restriction sites and of SexAI restriction fragments are indicated. (B) Comparison of Rab38 codons 17–20 of the Rab38 wild type allele, (Rab38+) the natural Rab38 chocolate (Rab38cht) allele, and the targeting vector (IDG-Cht). Nucleotides and amino acids differing from the wild type sequence are shown in red. The presence of a thymidine at the second position of codon 19 creates a valine codon, erases a BsaJI, and generates a SexAI site. The IDG-Cht G19V replacement was marked with an additional silent replacement at the third position of codon 19. (C) PCR amplification of a 213 bp region covering the first exon of Rab38 from C57BL/6 (B6), homozygous Rab38cht (Cht/Cht), heterozygous Rab38cht (Cht/+) control mice and pup R1.1 derived from embryos injected with ZFNRab38 and targeting vector (upper image). PCR products were digested with BsaJI (lower image) to determine the Rab38 genotype of pup R1.1 as Rab38+/Rab38IDG-Cht. (D) Comparison of sequences within the first exon of the Rab38 wild type gene, the targeting vector (IDG-Cht) and of BsaJI resistant PCR products amplified with primers P-for and P-rev from genomic DNA of pups R1.1, R3.5 and R3.9. The position of codon 19 and the ZFNRab38 binding regions are indicated. Nucleotides differing from the wild type sequence (blue background) are shown on a yellow background. (E) Southern blot analysis of SexAI digested genomic DNA using the Rab38 5′ probe shows a 10.5 kb and a 6.0 kb fragment from the Rab38+ loci of two C57BL/6 (B6) control mice and an additional 2.1 kb band derived from the targeted Rab38IDG-Cht allele of pups R1.1, R3.5, and R3.9. Homozygous Rab38IDG-Cht mutants (#47, #86) show the 10.5 kb and 2.1 kb bands, but not the 6.0 kb band. (F) Comparison of the coat color of a wild type C57BL/6N mouse and a homozygous Rab38IDG-Cht (#47) mouse.
The targeting vector was microinjected together with ZFNRab38 mRNAs into (C57BL/6N × FVB)F1 mouse one-cell embryos. From these injections, we obtained 87 offspring that were first genotyped for the presence of the Rab38IDG-Cht allele by PCR analysis. For this purpose, a 213 bp region of the first exon of Rab38 was PCR-amplified from tail DNA and digested with BsaJI. PCR products derived from the Rab38+ locus of wild type controls can be fully digested with BsaJI into fragments of 153 bp and 60 bp (Fig. 1C, B6). In contrast, the PCR product derived from homozygous Rab38cht mice is entirely resistant to BsaJI digestion (Fig. 1C, Cht/Cht), while PCR products from heterozygous mutants are only partially digestible due to the presence of a mutant, BsaJI-resistant allele (Fig. 1C, Cht/+). Among the 87 offspring from microinjections, we identified three pups (male R1.1, male R3.5, and female R3.9) which showed a partial resistance of PCR products to BsaJI digestion, as predicted for the presence of a Rab38IDG-Cht allele (Fig. 1C, R1.1). These Rab38IDG-Cht alleles were further characterized by sequence analysis of BsaJI-resistant PCR products derived from the recombined loci. As shown in Fig. 1D, all three samples exhibit the targeted G19V mutation and the silent exchange at the third position of codon 19 that identifies the targeted mutation. The targeted allele of pups R1.1 and R3.5 further included all silent mutations within the ZFN binding sites, whereas in pup R3.9 HR was terminated between these regions (Fig. 1D). To confirm the integrity of the targeted alleles, we analyzed genomic DNA derived from the three PCR-positive pups by Southern blotting using a hybridization probe located upstream of the vector·s 5′-homology arm (Fig. 1A). Upon digestion with SexAI, the Rab38IDG-Cht allele is predicted to generate a 2.1 kb band instead of the 6.0 kb band derived from the wild type locus (Fig. 1A). This analysis showed that the pups R1.1, R3.5, and R3.9 all exhibit the indicative 2.1 kb band which corresponds to a successful HR event of the targeting vector with a Rab38+ allele (Fig. 1E).
Taken together, the PCR and Southern blot analysis confirmed the identity and integrity of the targeted Rab38IDG-Cht alleles in the founder mice R1.1, R3.5, and R3.9. These mutants were identified among 87 pups derived from embryo injections, corresponding to a recombination rate of 3.5% at Rab38. This efficiency is comparable to our previous results from the Rosa26 locus that showed a 2%–4% rate for the ZFN-mediated integration of reporter genes (8).
To establish breeding colonies of Rab38IDG-Cht mutants, the founders were mated and the resulting offspring were genotyped for the presence of the targeted allele by PCR and Southern blot analysis. Founder R1.1 transmitted the Rab38IDG-Cht allele to 8 of 22 pups (36%), founder R3.5 to 5 of 24 pups (21%) and founder R3.9 to 5 of 10 pups (50%) indicating that ZFNRab38 expression did not interfere with the founder’s fertility. Statistical analysis revealed that mutant offspring appear at a submendelian ratio (18 positive pups of 56, P = 0.01045), suggesting that likely the founder mice R1.1 and R3.5 represent a mosaic of heterozygous targeted and wild type cells. Heterozygous mutant offspring were further mated to generate homozygous Rab38IDG-Cht mice. Genotyping of these offspring revealed a genotype distribution of 30.9% homozygote Rab38IDG-Cht (Fig. 1E, #47, #86), 48.1% heterozygote Rab38IDG-Cht, and 21.0% Rab38+/+ pups. The homozygous carriers of the Rab38IDG-Cht mutation exhibited the expected chocolate-like coat color (Fig. 1F, #47), as known from the natural Rab38cht mutants (21). This result indicates the functionality of the G19V mutation of the targeted Rab38IDG-Cht allele and provides the first genetic model with a targeted codon replacement generated by ZFN-assisted HR in one-cell embryos.
Targeting of the Rab38 Gene by ZFNRab38 and a Synthetic Oligodesoxynucleotide.
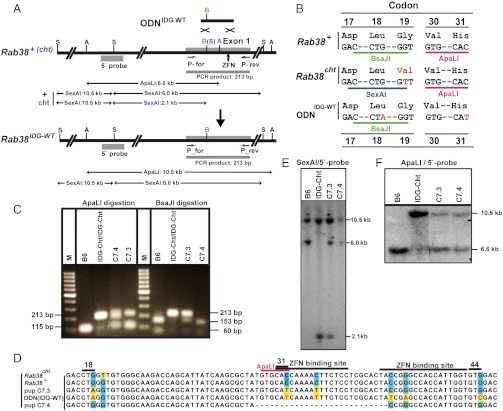
We further explored whether nucleotide replacements can be introduced into the Rab38 gene by the coinjection of ZFNRab38 and a single-stranded synthetic ODN instead of a plasmid-based gene-targeting vector. For this purpose, we used ODNIDG-WT corresponding to 144 nucleotides of the sense sequence of the first exon of Rab38 (Fig. 2A). ODNIDG-WT was designed to generate a Rab38IDG-WT allele by introduction of a silent nucleotide replacement within codon 18 (Fig. 2B). In addition, the ZFNRab38 recognition sequences were modified by silent replacements to prevent the potential processing of targeted alleles by ZFN nucleases (Fig. 2D). In addition, these replacements remove an ApaLI site from codon 30/31 (Fig. 2B). The ODNIDG-WT was coinjected with ZFNRab38 mRNAs into one-cell embryos derived from FVB female and Rab38cht (21) male mice. From these injections we obtained 60 pups that were genotyped for the presence of the Rab38IDG-WT allele by PCR amplification of a 213 bp region from the first exon of Rab38 (Fig. 2A). A fully recombined Rab38IDG-WT allele could be distinguished from the Rab38+ and Rab38cht alleles by the absence of the ApaLI site in the PCR fragment. PCR products from the Rab38+ and Rab38IDG-WT allele can be cleaved with BsaJI into 153 bp and 60 bp fragments (Fig. 2C, samples B6 and C7.4), whereas product from the Rab38cht allele is resistant to BsaJI.
Fig. 2.
Targeting of the Rab38 gene by ZFNRab38 and a synthetic oligodesoxynucleotide. (A) The targeting oligonucleotide ODNIDG-WT is designed for the introduction of a silent nucleotide replacement in codon 18 of a Rab38+ or Rab38cht allele. The structure of the Rab38+ and the Rab38cht (cht) locus, of the targeted Rab38IDG-WT allele, the location of the ZFNRab38 binding sites of the Rab38 5′-probe and of PCR primers P-for and P-rev are shown. The positions of SexAI (S), BsaJI (B), and ApaLI (A) restriction sites and of SexAI and ApaLI restriction fragments are indicated. The Rab38cht allele (cht) differs from Rab38+ (+) by the presence of a SexAI site instead of a BsaJI site in the first exon. (B) Comparison of Rab38 codons 17–19 and 30/31 of the Rab38 wild type allele (Rab38+), the Rab38cht allele and the targeting ODNIDG-WT. Nucleotides and amino acids differing from the wild type sequence are shown in red. For ODNIDG-WT, the presence of a thymidine at the third position of codon 31 erases an ApaLI site. (C) PCR amplification with primer pair P-for/P-rev of a 213 bp region covering the first exon of Rab38 from DNA from a C57BL/6 (B6) and a homozygous Rab38IDG-Cht (IDG-Cht/IDG-Cht) control mouse and pups C7.3 and C7.4 derived from Rab38+/Rab38cht embryos injected with ZFNRab38 and targeting ODNIDG-WT. The Rab38 genotype was determined by digestion of the PCR products with BsaJI, or ApaLI. PCR products derived from a Rab38+ allele (B6) or a targeted Rab38IDG-WT allele (recombined within codon 18) are digested by BsaJI into fragments of 153 bp and 60 bp; product from Rab38IDG-Cht alleles (IDG-Cht/IDG-Cht) is resistant to BsaJI digestion. Digestion with ApaLI of PCR products from Rab38+ and Rab38IDG-Cht alleles results into fragments of 115 bp and 98 bp that appear within a single band; product from a Rab38IDG-Cht allele (recombined within codon 30/31) is resistant to ApaLI digestion. The analysis of PCR products showed in both pups C7.3 and C7.4 the presence of an ApaLI resistant Rab38 allele. The more diffuse appearance of the ApaLI resistant PCR product of sample 7.4 is likely explained by the presence of a 27 bp deletion (see D) resulting in a reduced size of 186 bp and either an incomplete digestion or the presence of hybrid molecules formed with full-length wild type DNA strands. Pup C7.3 but not C7.4 harbors a BsaJI resistant Rab38cht allele. M: 100 bp size ladder. (D) Comparison of sequences within the first exon of the Rab38+ and Rab38cht alleles, the targeting ODNIDG-WT, and of ApaLI-resistant PCR products amplified with primers P-for and P-rev from genomic DNA of pups C7.3 and C7.4. The position of codon 18 and the ZFNRab38 binding regions are indicated. Nucleotides differing from the wild type sequence (blue background) are shown on a yellow background; nucleotide deletions are indicated by a dash. (E) Southern blot analysis of SexAI digested genomic DNA using the Rab38 5′-probe shows the presence of Rab38cht alleles (2.1 kb band) in a homozygous Rab38IDG-Cht control mouse (IDG-Cht) and pup C7.3 but its absence in pup C7.4. (F) Southern blot analysis of ApaLI-digested genomic DNA using the Rab38 5′-probe shows a 6.6 kb fragment from the Rab38+ alleles of a C57BL/6 control mouse (B6) and a 10.5 kb band for the Rab38IDG-Cht alleles from a homozygous Rab38IDG-Cht control (IDG-Cht/IDG-Cht). Both pups C7.3 and C7.4 exhibit a 6.6 kb band from an unmodified Rab38+ (C7.4) or Rab38cht C7.3) allele and a 10.5 kb band derived from an ApaLI resistant Rab38IDG-WT allele in pup C7.3 and a Rab38Δ allele in pup C7.4.
Interestingly, the PCR products from pup C7.3 showed partial resistance to ApaLI and BsaJI digestion (Fig. 2C). In contrast, PCR products from Pup C7.4 showed partial resistance to ApaLI but no resistance to BsaJI digestion (Fig. 2C). The sequence analysis of PCR products from pup C7.3 revealed a recombined Rab38IDG-WT allele that includes the nucleotide replacement within codon 18 and the upstream ZFN binding site, but not the replacements in the downstream ZFN binding site and in codon 44 (Fig. 2D). The Southern blot analysis of SexAI-digested DNA from pup C7.3 using the Rab38 5′-probe showed the presence of an unmodified Rab38cht allele as indicated by a 2.1 kb band (Fig. 2E). The genotype of pup C7.3 was further analyzed by digestion with ApaLI and confirmed the presence of a Rab38IDG-WT allele by an indicative 10.5 kb band and of the Rab38cht allele by a 6.6 kb band (Fig. 2F). Therefore, pup C7.3 exhibits a Rab38IDG-WT/Rab38cht genotype, suggesting that ODNIDG-WT recombined with the maternal Rab38+ allele, whereas the paternal Rab38cht allele was unaffected. For the Rab38IDG-WT allele HR occurred in the sequence covering codon 18 to 33, but was terminated between the oligonucleotide‘s silent replacements in the ZFN binding sites (Fig. 2D). Upon mating of the female founder C7.3 to wild type C57Bl/6 males, we obtained 13 pups, 4 of which harbored a Rab38IDG-WT/Rab38+ genotype as determined by PCR and Southern blot analysis (Fig. S1 A and B). The sequence analysis of PCR products derived from the germ line transmitted Rab38IDG-WT alleles showed their identity to the maternal allele of the founder C7.3 (Fig. S1C).
In contrast, the sequence and Southern blot analysis of DNA from pup C7.4 revealed the absence of a BsaJI-resistant, Rab38cht allele and the loss of the ApaLI site through a 27 bp deletion (Δ) within the ZFNRab38 binding site (Fig. 2D–F). This Rab38+/Rab38Δ genotype may arise by gene conversion from the maternal Rab38+ to the paternal Rab38cht allele, followed by a NHEJ repair-associated deletion that occurred in a second binding cycle of ZFNRab38 to the recombined allele.
Taken together, we obtained from 60 pups derived from microinjections one Rab38IDG-WT mutant (1.7%) that transmitted the modified allele to its offspring. This result provides proof-of-principle for the creation of single nucleotide replacements directly in one-cell embryos using ZFN technology and ODNs.
Discussion
ZFN-assisted gene editing in one-cell embryos provides a new, ES cell-independent paradigm to modify the genome of mammals and other vertebrates. The frequent loss of nucleotides associated with the NHEJ-mediated repair of induced DSBs provides an easy route for creating functional knockout alleles in the germline of mice, rats, rabbits, and zebrafish (2–9). The resulting loss-of-function mutants serve as valuable tools to decipher the essential role of targeted genes within the functional network that determines biological processes in vivo. Besides gene knockout, more specific questions on gene function require the creation of precisely targeted mutations, such as the replacement of codons for modeling disease alleles or the insertion of recombinase recognition sites to generate conditional alleles. This targeted mutagenesis approach requires the use of gene-targeting vectors which serve as template for the HR-mediated transfer of preplanned sequence modifications into the genome (1, 30). ZFN-assisted HR has been successfully used in mouse, rat, and rabbit models for the insertion of reporter genes (3, 5, 8), but the replacement of specific codons has yet not been reported.
By microinjection of ZFNRab38 and a gene-targeting vector into one-cell embryos, we were able to introduce a G19V chocolate mutation into the mouse Rab38 gene. Upon transfer of the injected embryos, we obtained three founder mice harboring the targeted mutation. Additional silent replacements, included into the ZFN-binding motifs of the targeting vector to prevent its nuclease processing, were cotransferred into the genome of two founders, but only partly transferred to the third founder. To avoid the cotransfer of such modifications in the future, we propose to include only one nucleotide replacement within the distant ZFN binding region. The three founders (two males, one female) transmitted the Rab38IDG-Cht allele to 32% of their offspring, indicating that ZFN manipulation does not interfere with fertility and that some of the founders were not fully heterozygous but mosaic for the targeted allele. As we noticed earlier, founders derived from one-cell embryo injections represent either fully heterozygous mutants that harbor the mutant allele in all body cells or mosaic mutants that bear the mutation in only part of the cells (8). Fully heterozygous founders are obtained if HR occurs in the pronucleus before genome replication, whereas later recombination at only one of the four parental chromatids results into mosaic heterozygotes. Further breeding of heterozygotes resulted in homozygous Rab38IDG-Cht mutants that recapitulated the coat color phenotype of the natural Rab38cht mutants.
Here we demonstrate that a mouse mutant harboring a preplanned codon replacement could be generated in a single step by gene targeting in one-cell embryos. As compared to the genetic manipulation of ES cells, gene targeting in one-cell embryos represents a straightforward approach that directly results in founder animals that can be used to establish a mutant colony. One-cell embryo gene targeting does not require the incorporation of selection marker genes into targeting vectors, which is an essential component of targeting vectors used for ES cell engineering. In addition, mutants established from ES cells require an additional breeding step for removal of the selection marker by Flp recombinase in order to avoid its interference with the function of the targeted gene (31, 32).
Typical gene-targeting vectors are plasmid-based constructs that include 4–10 kb homology regions derived from the target locus to guide the insertion of a desired mutation during the HR process (30). The construction of gene-targeting vectors is commonly a low-throughput and time-consuming task. In addition, the large size of these DNA constructs limits the number of molecules that can be introduced into embryos without eliciting toxicity. Both of these limitations are bypassed by the use of synthetic ODNs as sequence-specific repair templates. As recently shown in cultured cell lines, ZFN-mediated gene targeting is achieved with ODNs at a similar rate as compared to the use of classical targeting vectors (33). Our results with an ODN targeting the Rab38 gene indicate that these molecules also serve as repair templates in one-cell embryos, enabling nucleotide replacements. This simplified targeting approach expedites the generation of mutant alleles as compared to the time-consuming construction of plasmid vectors. Since ODNs are able to induce in vitro sequence insertions and deletions (33), we expect that such modifications will further expand the utility of ODN-directed gene targeting in embryos. As it was also shown that the inclusion of silent mutations into targeting ODNs is not mandatory (33), we expect that it will be possible to generate targeted alleles without any modification in the ZFN recognition sequence.
Taken together, we demonstrate that mutants harboring nucleotide replacements can be generated by gene editing in one-cell embryos. Furthermore, we show that a synthetic ODN serves as repair template. We consider these techniques useful for the generation of genetic disease models in rodents and other vertebrates to study the role of human disease-associated mutations, which mostly exhibit single nucleotide replacements (14, 15). As recently reported for rats and zebrafish, the limited availability of ZFNs is overcome by the use of modular TAL-based nucleases that enable to address any genomic target site (34, 35). These findings and our results of nucleotide specific gene editing open further prospects for modeling human disease alleles in vertebrates.
Material and Methods
Targeting Vector and Single-Stranded Oligodesoxynucleotide Design.
The targeting vector pRab38(IDG-Cht) comprised two homology regions encompassing 942 and 2788 bp of genomic sequence flanking exon 1 of the mouse Rab38 gene (Fig. 1A). For this purpose, the vector’s 5′- and 3′-homology arms were amplified from the genomic BAC clone RPCI-421G2 (derived from the C57BL/6J genome, Imagenes GmbH, Berlin) using the primer pair RabCht-1 (5′-CTCACCGCATTACCCTGGGCGTTGAAACCGAAGAAGACC-3′) and RabCht-2 (5′-GATAATGCTGGTCTTGCCCACtaCCAGGTCGCCGATC-3′), and the primer pair RabCht-3 (5′-GTGGGCAAGACCAGCATTATCAAGCGCTATGTGCAtCAAAAtTTCTCCTCGCACTAtCGaGCCACCATTGGTGTGGACTTCG-3′) and RabCht-4 (5′-GCTTATCGATACCGTCGACTCAGTTGACTACTACCCGTTC-3′). Primers RabCht-2 and -3 were selected such that they overlap by 21 bp within exon 1 of Rab38, immediately downstream of codon 19. Within the sequence of codon 19, primer RabCht-2 contained two nucleotide changes (small letters) that modify codon 19 from the wild type sequence GGT, coding for glycine, into GTA, coding for valine. This new chocolate mutation can be distinguished from the natural chocolate mutation, which exhibits only a single nucleotide exchange within codon 19 (GTT) coding for valine (21). The recognition region for the ZFN-Rab38-L and -R zinc-finger proteins is located 50 bp downstream of codon 19. For the construction of the targeting vector, each 18 bp ZFN recognition sequence in the 3′-homology region was further modified by the introduction of two silent nucleotide substitutions (small letters) that do not alter the Rab38 coding sequence (Fig. 1D), in order to avoid the potential processing of the targeting vector by the Rab38 specific ZFNs. To construct the complete pRab38(IDG-Cht) targeting vector, the PCR products representing the 5′- and 3′- homology arms were combined by fusion PCR using primers RabCht-1 and -4 for amplification. Since primer 1 includes an I-SceI restriction site and primer 4 a SalI restriction site, the I-SceI and SalI digested 3.7 kb PCR product was cloned into the backbone of the vector pRosa26.3-3 (8) that was opened with I-SceI and SalI. The integrity of the completed vector was confirmed by DNA sequencing.
The targeting oligodesoxynucleotide ODNIDG-WT for the second experiment, synthesized by Metabion (Martinsried, Germany), had a total length of 144 nucleotides including the IDG-WT mutation (Fig. 2B), and silent mutations within the ZFN recognition site (Fig. 2D). An additional silent mutation was included in codon 44 to differentiate between the Rab38+ allele and Rab38IDG-WT allele (Fig. 2D).
Injection of One-Cell Embryos.
The heterodimeric ZFNRab38 pair targets sequences (underlined) near to the site of the chocolate mutation (here WT sequence; bold) within the first exon of the mouse Rab38 gene: 5′-atgcagacacctcacaaggagcacctgtacaagctgctggtgatcggcgacctgggtgtgggcaagaccagcattatcaagcgctatgtgcaccaaaacttctcctcgcactaccgggccaccattggtgtggacttcgcgctgaaggtgctccactgggacccagagacggtggtgcgcttgcagctctgggacattgctg -3′. Expression constructs for these ZFNs were generated by gene synthesis (Genscript), followed by embedding of the reported ZFN recognition helix sequences (9) into a generic zinc-finger backbone based on the sequence of the ZIF268 protein (10, 36–38). For the preparation of ZFNRab38 mRNA, in vitro transcription from plasmid DNAs were performed as previously described (8). Each ZFNRab38 mRNA was diluted in injection buffer to a working concentration of 3 ng/μL for the first experiment and a working concentration of 7.5 ng/μL for the second. mRNAs were mixed and stored together with the targeting vector or ODN at -80 °C. The targeting vector pRab38(IDG-Cht) was used as a supercoiled plasmid. The vector was precipitated and dissolved in injection buffer to a working concentration of 15 ng/μL. The targeting oligodesoxynucleotide ODNIDG-WT was dissolved in water and diluted with injection buffer to a working concentration of 15 ng/μL. Mouse zygotes for the first experiment were obtained by mating of C57BL/6N males with superovulated FVB females (Charles River Germany) as described (8). In the second experiment, homozygous Rab38cht males (21), kindly provided by M. Seabra, were used instead of C57BL/6N. Zygotes were injected with a mixture of the targeting construct (15 ng/μL) and the ZFNRab38 mRNA (3 ng/μL each or 7.5 ng/μL each) in the same two-step procedure as described (8). Microinjections were performed into the larger (male) pronucleus of fertilized oocytes whenever possible. For those zygotes in which the pronuclei showed no obvious size difference, one of the pronuclei was randomly selected. Injected zygotes were transferred into pseudopregnant CD1 female mice, and viable adult mice were obtained. From the transfer of 428 zygotes, injected with the targeting vector pRab38(IDG-Cht), we obtained 87 newborns (20% recovery). The animals with correct homologous recombination events were mated, and the offspring were analyzed for germ line transmission. Whether the mutant allele was inherited at mendelian ratio was calculated with the exact binomial test using the R software (39). With the targeting oligodesoxynucleotide ODNIDG-WT, we obtained 60 mice from 180 transferred zygotes (33% recovery). All mice showed normal development and appeared healthy. Mice were handled according to institutional guidelines and housed in standard cages in a specific pathogen-free facility on a 12-h light/dark cycle with ad libitum access to food and water.
Preparation of Genomic DNA.
Genomic DNA was isolated from tail tips of mice derived from zygotes coinjected with ZFNRab38 mRNA and pRab38(IDG-Cht) vector or ODNIDG-WT, following the Wizard Genomic DNA Purification Kit (Promega) protocol. The obtained DNA pellet was dissolved in 100 μL 10 mM Tris-Cl, pH 8.5, incubated overnight at room temperature, and stored for further analysis at 4 °C.
PCR, Digestion, and Sequence Analysis.
To analyze the Rab38 alleles in the first round, we amplified the DNA from mice using the PCR primer pair P_for (5′-GGCCTCCAGGATGCAGACACC-3′) and P_rev (5′-CCAGCAATGTCCCAGAGCTGC-3′). Amplification was performed using Taq polymerase (Qiagen) in 25 μL reactions with 35 cycles of 94 °C–40 s, 58 °C—40 s, 72 °C–1 min. Afterwards, the PCR products were directly digested with 10 units of restriction enzyme in a volume of 30 μL and loaded on an 1.5% agarose gel. The undigested fragments, which showed the homologous recombined allele, were extracted with the Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sent for sequencing (GATC, Konstanz, Germany). The results were compared to the expected sequences using the Vector NTI software (Invitrogen).
Southern Blot Analysis.
Genomic DNA from positive recombined mice was digested overnight with the appropriate restriction enzyme. Southern blot analysis was performed as previously described (8), except for the washing step that was performed with 1x SSC at 70 °C (2 × 20 min.) using the Rab38 5′-probe, which was obtained by PCR amplification (P-Rab1: 5′-ctggaaactaaaattcaaggtgttatac-3′, P-Rab2: 5′-TATTCATTCACTTAACCATTTGTTC-3′) from C57BL/6N genomic DNA and purified with the Qiaquick PCR Purification Kit (Qiagen).
Supplementary Material
Acknowledgments.
We thank R. Kneuttinger, P. Kunath, A. Krause, A. Tasdemir, and S. Weidemann for excellent technical assistance; M. Seabra for Rab38cht mice; T. Faus-Kessler for statistical analysis; and B. Wefers and A. Pertek for reading the manuscript. This work was supported by the European Union within the EUCOMM project (Grant #LSHG-CT-2005-018931, to W.W.), by the German Ministry of Education and Research within the DIGTOP project (Grant #01GS0858, to W.W. and R.K.) of the NGFN-Plus program, and by the European Union Seventh Framework Programme (Grant #FP7/2007-2013, to O.O.) under grant agreement no. 251864.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121203109/-/DCSupplemental.
References
- 1.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.Carbery ID, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui X, et al. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 4.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flisikowska T, et al. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011;6:e21045. doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mashimo T, et al. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS One. 2010;5:e8870. doi: 10.1371/journal.pone.0008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geurts AM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 11.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 12.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 13.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 15.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbeel L, Freson K. Rab proteins and Rab-associated proteins: Major actors in the mechanism of protein-trafficking disorders. Eur J Pediatr. 2008;167:723–729. doi: 10.1007/s00431-008-0740-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osanai K, et al. Expression and characterization of Rab38, a new member of the Rab small G protein family. Biol Chem. 2005;386:143–153. doi: 10.1515/BC.2005.018. [DOI] [PubMed] [Google Scholar]
- 18.Brooks BP, et al. Analysis of ocular hypopigmentation in Rab38cht/cht mice. Invest Ophthalmol Vis Sci. 2007;48:3905–3913. doi: 10.1167/iovs.06-l464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninkovic I, White JG, Rangel-Filho A, Datta YH. The role of Rab38 in platelet dense granule defects. J Thromb Haemost. 2008;6:2143–2151. doi: 10.1111/j.1538-7836.2008.03178.x. [DOI] [PubMed] [Google Scholar]
- 20.Osanai K, et al. Altered lung surfactant system in a Rab38-deficient rat model of Hermansky-Pudlak syndrome. Am J Physiol Lung Cell Mol Physiol. 2010;298:L243–251. doi: 10.1152/ajplung.00242.2009. [DOI] [PubMed] [Google Scholar]
- 21.Loftus SK, et al. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci USA. 2002;99:4471–4476. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes VS, Wasmeier C, Seabra MC, Futter CE. Melanosome maturation defect in Rab38-deficient retinal pigment epithelium results in instability of immature melanosomes during transient melanogenesis. Mol Biol Cell. 2007;18:3914–3927. doi: 10.1091/mbc.E07-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasmeier C, et al. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osanai K, et al. A mutation in Rab38 small GTPase causes abnormal lung surfactant homeostasis and aberrant alveolar structure in mice. Am J Pathol. 2008;173:1265–1274. doi: 10.2353/ajpath.2008.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oiso N, Riddle SR, Serikawa T, Kuramoto T, Spritz RA. The rat Ruby (R) locus is Rab38: Identical mutations in Fawn-hooded and Tester-Moriyama rats derived from an ancestral Long Evans rat sub-strain. Mamm Genome. 2004;15:307–314. doi: 10.1007/s00335-004-2337-9. [DOI] [PubMed] [Google Scholar]
- 26.Di Pietro SM, Dell’Angelica EC. The cell biology of Hermansky-Pudlak syndrome: Recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 27.Wei ML. Hermansky-Pudlak syndrome: A disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- 28.Ma J, Plesken H, Treisman JE, Edelman-Novemsky I, Ren M. Lightoid and Claret: A rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc Natl Acad Sci USA. 2004;101:11652–11657. doi: 10.1073/pnas.0401926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Fernandez IA, Dell’Angelica EC. A data-mining approach to rank candidate protein-binding partners-The case of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Inherit Metab Dis. 2009;32:190–203. doi: 10.1007/s10545-008-1014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasty P, Abuin A, Bradley A. Gene targeting, principles, and practice in mammalian cells. In: Joyner AL, editor. Gene Targeting: A Practical Approach. 2nd Ed. Oxford: Oxford University Press; 2000. pp. 1–35. (The Practical Approach Series). [Google Scholar]
- 31.Kwan KM. Conditional alleles in mice: Practical considerations for tissue-specific knockouts. Genesis. 2002;32:49–62. doi: 10.1002/gene.10068. [DOI] [PubMed] [Google Scholar]
- 32.Friedel RH, Wurst W, Wefers B, Kuhn R. Generating conditional knockout mice. Methods Mol Biol. 2011;693:205–231. doi: 10.1007/978-1-60761-974-1_12. [DOI] [PubMed] [Google Scholar]
- 33.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sander JD, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 36.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 37.Moore M, Choo Y, Klug A. Design of polyzinc finger peptides with structured linkers. Proc Natl Acad Sci USA. 2001;98:1432–1436. doi: 10.1073/pnas.98.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Team RDC. R:A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.