Abstract
Movement of the plant hormone abscisic acid (ABA) within plants has been documented; however, the molecular mechanisms that regulate ABA transport are not fully understood. By using a modified yeast two-hybrid system, we screened Arabidopsis cDNAs capable of inducing interactions between the ABA receptor PYR/PYL/RCAR and PP2C protein phosphatase under low ABA concentrations. By using this approach, we identified four members of the NRT1/PTR family as candidates for ABA importers. Transport assays in yeast and insect cells demonstrated that at least one of the candidates ABA-IMPORTING TRANSPORTER (AIT) 1, which had been characterized as the low-affinity nitrate transporter NRT1.2, mediates cellular ABA uptake. Compared with WT, the ait1/nrt1.2 mutants were less sensitive to exogenously applied ABA during seed germination and/or postgermination growth, whereas overexpression of AIT1/NRT1.2 resulted in ABA hypersensitivity in the same conditions. Interestingly, the inflorescence stems of ait1/nrt1.2 had a lower surface temperature than those of the WT because of excess water loss from open stomata. We detected promoter activities of AIT1/NRT1.2 around vascular tissues in inflorescence stems, leaves, and roots. These data suggest that the function of AIT1/NRT1.2 as an ABA importer at the site of ABA biosynthesis is important for the regulation of stomatal aperture in inflorescence stems.
Keywords: guard cells, liquid chromatography-tandem mass spectrometry, water stress
Plant hormones are a group of naturally occurring compounds that induce various physiological responses at low concentrations (less than 10−6 M). Endogenous hormone levels are determined principally by the balance between biosynthesis and catabolism; however, the rate of biosynthesis and catabolism in a cell cannot account for the hormone concentration at the site of action if the hormones are mobile. In animals, hormones are synthesized in particular cell types and are transported to target cells to induce physiological processes. In plants, however, although some plant hormones have been reported to be mobile, the significance of hormone transport in controlling physiological responses is largely unknown, except in the case of auxin (indole-3-acetic acid). Auxin transporters have been identified by characterizing mutants isolated based on defects in their organ development or their responses to exogenously applied auxins or environmental stimuli (1, 2). In contrast, forward genetics approaches based on physiological or hormone responses have not succeeded in isolating mutants defective in the transport of other hormones. This suggests that the transport systems are highly redundant, or that hormone transport is not required for physiological functions.
In plants, the hormone abscisic acid (ABA) plays crucial roles in the responses to abiotic and biotic stresses, as well as in the induction and maintenance of seed dormancy (3–6). Long-distance transport of ABA from roots to leaves has been extensively discussed in relation to stomatal closure in response to water deficit (7–11). Stomata closed when a part of the root system was exposed to water stress, whereas the leaf water status was unchanged (11). The stomatal aperture was correlated with the ABA content in the xylem sap, suggesting that ABA synthesized in roots was transported to leaves via the xylem to induce stomatal closure. Nevertheless, it appears that ABA transport from roots to leaves is not always required for stomatal closure, because reciprocal grafting between WT and ABA-deficient mutants showed that leaf genotypes, but not root genotypes, were correlated with leaf stomatal conductance (12, 13).
Almost all the enzymes involved in the main ABA biosynthetic pathway have been identified (14, 15). Immunohistochemical studies using antibodies raised against Arabidopsis ABA biosynthetic enzymes AAO3, AtABA2, and AtANCED3 suggested that ABA is actively synthesized in vascular tissues (parenchyma cells) in response to water stress (16, 17). This, in turn, indicates that ABA synthesized in vascular tissues must be translocated to guard cells to induce stomatal closure. Recently, two ATP-binding cassette (ABC)-type transporters were identified as ABA transporters; one mediated ABA export from the inside to the outside of cells (AtABCG25) and the other mediated ABA uptake from the outside to the inside of cells (AtABCG40) (18, 19). The promoter activity of AtABCG25 was detected in vascular tissues whereas that of AtABCG40 was detected in guard cells, supporting the idea that ABA is transported from vascular tissues to guard cells. It should be noted, however, that the loss-of-function abcg25 and abcg40 mutants did not show the phenotype observed in typical ABA-deficient mutants such as aba1, aba2, and aba3, indicating that the ABA transport system might be highly complex and redundant. Here, we developed a modified yeast two-hybrid system to functionally screen ABA transporters by using the recently identified ABA receptor complex (20, 21) as a sensor to monitor ABA levels in the cells. By using this approach, we found that some members of the previously characterized transporter family NRT1/PTR (22) efficiently imported ABA into the cells. We discuss the physiological significance of ABA transport mediated by NRT1/PTR proteins, especially in relation to the regulation of stomatal aperture.
Results
Functional Screening of ABA Transporters.
It has been reported that some of the PYR/PYL/RCAR ABA receptors interact with a group of PP2C-type protein phosphatases in the presence of ABA, and that ABA-dependent interactions between the receptor and PP2C can be monitored by using a yeast two-hybrid system (21). We constructed yeast two-hybrid systems with ABI1 or HAB1 PP2C fused to the GAL4 activation domain (AD; AD-ABI1 and AD-HAB1, respectively) and the PYR1 receptor fused to the GAL4 DNA-binding domain (BD; BD-PYR1); the expression of downstream HIS3 served as a selection marker (Fig. S1A). To identify candidates for ABA importers, cDNA clones capable of inducing interactions between the receptor and PP2C in the presence of 0.1 μM ABA [racemic mixture, (±)-ABA], at which concentration there is almost no interaction between PYR1 and PP2C, were screened from Arabidopsis cDNA libraries (Fig. S1 A and B). Overrepresentation of At1g69850 and At1g27040 sequences in the screened population (SI Materials and Methods) suggested that they were good candidates for genes encoding ABA importers. The two candidate genes belong to the NRT1/PTR transporter family, which consists of 53 related sequences (22) (Fig. S1C). Some members of this family encode proteins capable of transporting nitrate or di/tripeptides (22) and At1g69850 was previously characterized as a low-affinity nitrate transporter NRT1.2 (23). As we identified two closely related sequences from the group I NRT1/PTR family in our screen, we examined whether other proteins in this group could also induce interactions between the receptor and PP2C under low ABA concentrations. We cloned 11 cDNAs encoding group I NRT1/PTRs and determined their effects on the AD-ABI1/BD-PYR1 interactions (Fig. 1). Of the 11 cDNAs, four of them (At1g69850, At1g27040, At3g25260, and At3g25280) were able to induce an interaction between AD-ABI1 and BD-PYR1 in the presence of 0.1 μM ABA. For convenience, we designated the four candidate genes, At1g69850 (NRT1.2), At1g27040, At3g25260, and At3g25280, as ABA-IMPORTING TRANSPORTER (AIT) 1, 2, 3, and 4, respectively.
Fig. 1.
Effects of group I NRT1/PTR family on ABA-dependent interactions between AD-ABI1 and BD-PYR1. cDNAs encoding group I NRT1/PTR or empty vector (Control) were introduced into yeast cells containing AD-ABI1 and BD-PYR1, and their interactions were detected by using HIS3 as a selection marker in the presence of 0.1 μM ABA in the medium. Four independent transformants were tested for their growth on selection medium (synthetic dextrose; -Leu, -Trp, -Ura, -His).
Although it was reported that AtABCG40 mediates ABA uptake into cells (18), AtABCG40 cDNA did not induce an interaction between AD-ABI1 and BD-PYR1 in the presence of 0.1 μM ABA (Fig. S1D). On the contrary, it was reported that AtABCG25 mediates ABA export from the inside to the outside of cells (19). If this is the case, AtABCG25 should inhibit the AD-ABI1/BD-PYR1 interaction in our yeast system. However, AtABCG25 cDNA did not affect the ABA-dependent AD-ABI1/BD-PYR1 interaction (Fig. S1D).
Biochemical Characterization of AITs.
We compared the effects of AITs on the ABA-dependent AD-ABI1/BD-PYR1 interaction by monitoring expression levels of a marker gene, LacZ, by β-gal assays with o-nitrophenyl β-D-galactopyranoside as the substrate (Fig. 2A). Cells expressing AIT1 showed the highest β-gal activity, and 10 nM (±)-ABA was sufficient to significantly induce β-gal activity. Among the four AITs, AIT4 was least effective in inducing β-gal activity; however, 10 μM ABA significantly induced β-gal activity compared with that in cells containing the control vector. The dose-dependent reporter expression levels suggested that AIT1 and AIT3 might have relatively high affinity for ABA compared with that of AIT2 and AIT4.
Fig. 2.
Hormone import activities of AITs. (A) AIT1–4 or empty vector (Control) was introduced into yeast cells containing AD-ABI1 and BD-PYR1. ABA-dependent AD-ABI1/BD-PYR1 interactions in the presence of 0, 0.01, 0.1, 1, or 10 μM (±)-ABA in the medium were detected as the expression of lacZ determined by β-gal assay with o-nitrophenyl β-D-galactopyranoside (ONPG). Units of β-gal activity represent 1,000× OD420/min/OD600 cells. Values are means ± SD of three biological replicates. (B) ABA import activities (fmoles ABA/1 × 107 cells/min) of AIT1 in the presence of 0.5, 1, 2, 5, or 10 μM (+)-ABA were determined by analyzing ABA incorporated into yeast cells by LC-MS/MS. Amounts of ABA incorporated into the cells with empty vector are also shown (Control). Mean values of two biological replicates are shown in Michaelis–Menten plot. (C) ABA import activities (fmoles ABA/1 × 107 cells/min) of AIT3 in the presence of 1, 10, or 20 μM (+)-ABA were determined by analyzing ABA incorporated into yeast cells by LC-MS/MS. Amounts of ABA incorporated into cells with empty vector are also shown (Control). Values are means of two biological replicates. (D) ABA-import activities of insect cells expressing AIT1. ABA-import activities (picomoles ABA/1 × 106 cells/min) of insect cells expressing AIT1 or a control gene (Control) in the presence of 0.1 or 1 μM (±)-ABA were determined by LC-MS/MS. Values are means of two biological replicates. (E) ABA import activities (fmoles ABA/1 × 107 cells/min) of AIT1 and AIT3 in the presence of 10 μM (+)- or (−)-ABA were determined by analyzing ABA incorporated into yeast cells by LC-MS/MS. Amounts of ABA incorporated into cells with empty vector are also shown (Control). Values are means ± SD of three biological replicates. (F) Activities of AIT1 and AIT3 to import GA3, IAA, or JA (fmoles hormone/1 × 107 cells/min) were determined by analyzing compounds incorporated into yeast cells by LC-MS/MS. Amounts of compounds incorporated into the cells with empty vector are also shown (Control). Values are means ± SD of three biological replicates. For A–F, similar results were obtained at least in two independent experiments using different cell cultures.
The ABA-importing activities of AIT1 and AIT3 in yeast were confirmed by directly analyzing ABA incorporated into the cells by LC-MS/MS (Fig. 2 B and C). We could not detect the activities of AIT2 and AIT4 because of the detection limit of this assay system. The ABA import activity of AIT1 was confirmed in insect cells (Fig. 2D). Michaelis–Menten plots indicated that the Km value of AIT1 for ABA [(S) enantiomer, (+)-ABA] was ∼5 μM (Fig. 2B). We were unable to determine the Km value of AIT3 because of the high and variable levels of background ABA incorporated into the control cells when higher ABA concentrations were used for assays. Interestingly, AIT1 showed a higher affinity for (+)-ABA than for the synthetic (R) enantiomer of ABA [(−)-ABA] as its substrate (Fig. 2E). Also, AIT1 did not import gibberellin (GA3), indole-3-acetic acid (IAA), or jasmonic acid (JA) (Fig. 2F). In contrast, AIT3 did not discriminate between (S) and (R) configurations of ABA (Fig. 2E), and was able to import GA3 (Fig. 2F).
Characterization of Loss-of-Function Mutants and Overexpressors.
To determine the physiological functions of AITs in plants, we obtained T-DNA insertion mutants in AIT genes and characterized their phenotypes. We obtained three T-DNA insertion alleles for both AIT1 and AIT2 and one for AIT3 (Fig. S2A). We were unable to obtain insertion mutants for AIT4. We also obtained two insertion alleles for At5g62730, which is in the same clade as the four AITs in the phylogenetic tree (Figs. S1C and S2A). First, we examined the effect of exogenously applied (±)-ABA on seed germination and/or postgermination growth. ait1 was less sensitive than WT to ABA at relatively low concentrations (0.5–1 μM), although the phenotype was milder than that of a dominant abi1-1 mutant (24) (Fig. 3A and Fig. S2B). Next, we tested the effect of an ABA agonist, pyrabactin. Uptake of d6-ABA into yeast cells mediated by AIT1 was inhibited by an excess amount of nonlabeled ABA, whereas it was not inhibited by pyrabactin (Fig. S2C). This indicated that pyrabactin is not a substrate of AIT1. Thus, if ait1 is defective in ABA signaling rather than ABA import, the mutant should show altered responses to pyrabactin as well as to ABA, like abi1-1. However, inhibition of seed germination by pyrabactin was equivalent in ait1 and WT (Fig. 3A). If AIT1 mediates the import of ABA into cells, overexpression of AIT1 should enhance ABA sensitivity. We selected two transgenic lines, a moderate overexpressor and an extreme overexpressor, for germination assays (Fig. 3B). As expected, AIT1 overexpressors were more sensitive to ABA than the control lines containing an empty vector, and their sensitivity was related to the expression levels of AIT1.
Fig. 3.
Phenotypes of ait1 and AIT1 overexpressors during germination. (A) Germination rates determined as cotyledon greening of WT, ait1, and abi1-1 in Col background (abi1-1C) on one-half Murashige and Skoog medium (Control) or one-half Murashige and Skoog medium containing ABA (0.5 or 1 μM) or pyrabactin (Pyr; 5 or 20 μM). Values are means ± SD of three biological replicates. Similar results were obtained in at least two independent experiments using different seed batches. (B) Expression of AIT1 in 2-wk-old control plants containing a empty vector (C-1 and 2) and AIT1 overexpressors (OX-1 and 2) (Top). Expression levels relative to 1/1,000 diluted 18S rRNA are shown. Values are means ± SD of three technical replicates. Similar results were obtained in two independent experiments using different plant cultures. Seeds were stratified and germination rates determined as cotyledon greening in the presence of 0, 0.1, 0.2, or 0.5 μM ABA after 10 d at 22 °C in light conditions (Bottom). Values are means ± SD of three biological replicates. Similar results were obtained in two independent experiments using different seed batches.
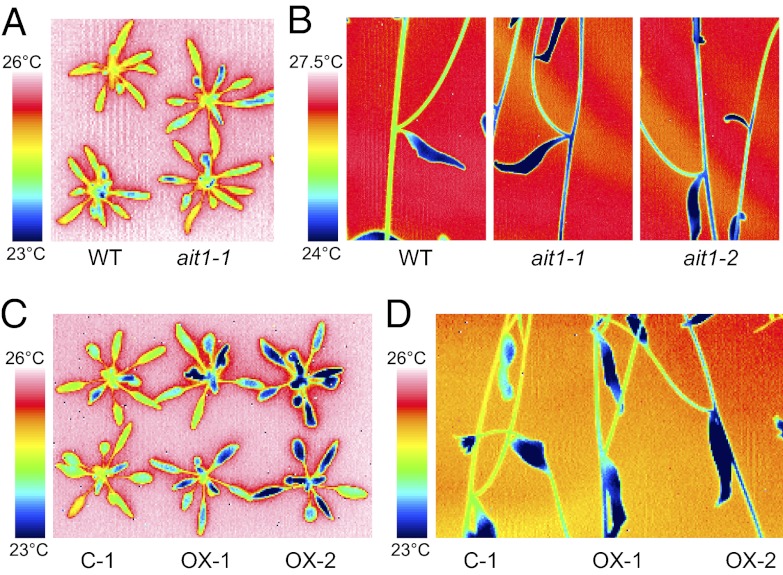
It has been suggested that ABA synthesized in vascular tissues is transported to guard cells (16, 17). Therefore, we investigated whether AITs are involved in this process. The stomatal aperture can be monitored indirectly by measuring surface temperature with an infrared thermal camera (25). Thus, we expected that, if some of the AITs are involved in ABA transport, the mutants would show altered surface temperature. The leaf surface temperatures of ait1, ait2, and ait3, as well as mutants defective in At5g62730, were comparable to that of WT (Fig. 4A and Fig. S3A). However, the surface temperature of the inflorescence stems was clearly lower in ait1 than in WT (Fig. 4B). Measurements of stomatal aperture showed that ait1 had more open stomata than WT (Fig. S3B). On the contrary, mutants defective in AtABCG25 or AtABCG40 were comparable to WT in terms of surface temperature of leaves and inflorescence stems (Fig. S3 C–E). These data suggest that ABA transport mediated by AIT1 is important for stomatal closure in inflorescence stems. Next, we investigated the effects of AIT1 overexpression on surface temperature. Loss-of-function mutants and overexpressors of a certain gene often show opposite phenotypes. In fact, ait1 and AIT1 overexpressors showed opposite phenotypes during seed germination in terms of sensitivity to exogenously applied ABA (Fig. 3). However, AIT1 overexpressors had reduced surface temperature in inflorescence stems and also in leaves (Fig. 4 C and D).
Fig. 4.
Surface temperature of leaves and inflorescence stems as determined by using a thermal imaging camera. (A) Leaf surface temperature of WT and ait1. (B) Surface temperature of inflorescence stems of WT and ait1. (C) Leaf surface temperature of AIT1 overexpressors (OX-1 and OX2) and control line carrying an empty vector (C-1). (D) Surface temperature of inflorescence stems of AIT1 overexpressors (OX-1 and OX2) and a control line (C-1).
Expression of AIT1.
Because heterologous expression of AIT1 in yeast and insect cells enhanced ABA uptake into the cells (Fig. 2), we hypothesized that this protein mediates ABA import at the plasma membrane. To test this idea, the AIT1 protein fused to GFP was expressed under the control of the 35S promoter (p35S:AIT1-GFP) in the WT background. As expected, GFP fluorescence was preferentially detected at the plasma membrane (Fig. 5A). To gain more insight into the physiological role of AIT1, we analyzed the spatial expression pattern of AIT1 by using a promoter-reporter system. We generated transgenic plants expressing β-glucuronidase (GUS) under the control of the AIT1 promoter (pAIT1:GUS), and the promoter activity was visualized by GUS staining (Fig. 5 B–F). Consistent with the observation that ait1 was insensitive to exogenously applied ABA during germination, AIT1 promoter activity was detected in imbibed seeds (Fig. 5B). After germination, GUS activity was detected in vascular tissues in cotyledons, true leaves, hypocotyls, roots, and inflorescence stems (Fig. 5 C–F).
Fig. 5.
Subcellular localization of AIT1 and spatial expression patterns of AIT1. (A) GFP fluorescence (a and d), bright-field (b and e), and merged (c and f) images of root tissues of 2-wk-old 35S:AIT1-GFP transgenic plants before (a–c) and after (d–f) osmolysis with 20% sucrose. (Scale bars: a, 20 μm; d, 10 μm.) Similar localization pattern was observed in three independent transgenic lines. (B–F) GUS activities in pAIT1:GUS transgenic plants: imbibed seeds (B), 2-wk-old seedling (C), main root of 2-wk-old seedling (D), lateral root of 2-wk-old seedling (E), and transverse section of inflorescence stem (F). (Scale bars: 1 mm.) Expression pattern was similar in six independent transgenic lines.
Discussion
Biochemical Characteristics of AITs.
Here, we identified four closely related proteins (AIT1–4) belonging to the NRT1/PTR family as candidates for ABA importers (Fig. 1 and Fig. S1C). NRT1 proteins are low-affinity nitrate transporters (22, 26–30), except for the dual-affinity nitrate transporter CHL1/NRT1.1, which catalyzes both low- and high-affinity nitrate uptake in a phosphorylation-dependent manner (31). AIT1 was previously characterized as NRT1.2, which mediates low-affinity nitrate uptake into cells with a Km value of ∼5.9 mM (23). However, our results showed that the Km value of AIT1 for ABA was approximately 5 μM (Fig. 2B) and that AIT1 could significantly induce the AD-ABI1/BD-PYR1 interaction even at the ABA concentration of 10 nM (Fig. 2A). Also, it appeared that AIT1 specifically recognized the chemical structure of ABA because it did not import GA3, IAA, JA, or pyrabactin (Fig. 2F and Fig. S2C), and it showed greater affinity for (+)-ABA rather than for (−)-ABA as its substrate (Fig. 2E). These data suggest that ABA is a better substrate than nitrate for AIT1/NRT1.2. The ABA import activity of AIT3 was relatively high (Fig. 2 A and C). Interestingly, AIT3 imported both GA3 and ABA (Fig. 2F), and it did not discriminate between (+)- and (−)-ABA (Fig. 2E), indicating that AIT1 and AIT3 have different biochemical characteristics.
Physiological Function of AIT1.
Heterologous expression of AIT1 in yeast or insect cells promoted the uptake of ABA into the cells (Fig. 2). AIT1 was preferentially localized to the plasma membrane of plant cells (Fig. 5A). These observations indicate that AIT1 functions as an ABA transporter mediating cellular ABA uptake. Interestingly, the surface temperature of inflorescence stems was lower in ait1 than in WT (Fig. 4B). Consistent with this, the stomata in inflorescence stems of ait1 had a wider aperture than those of WT (Fig. S3B), indicating that the defect in AIT1 function resulted in increased transpiration through stomata. Based on the biochemical function of AIT1, we expected that AIT1 would mediate ABA uptake into the guard cells in inflorescence stems. However, AIT1 was expressed in the vascular tissues in inflorescence stems (Fig. 5F). The expression pattern of AIT1 in vascular tissues resembles the localization of ABA biosynthetic enzymes such as AAO3, ABA2, and NCED3 (16, 17). Accordingly, we propose a model in which AIT1 functions to maintain the ABA pool size at the site of biosynthesis (vascular parenchyma cells), and this might be required for efficient translocation of ABA to guard cells (Fig. S4). In this model, the loss of AIT1 function results in decreased ABA levels at the site of biosynthesis because of diffusion into the apoplastic space within stem tissues. It could be also possible that AIT1 is expressed very low levels in guard cells. This would help to explain the phenotype observed in ait1. Whereas the inflorescence stem of ait1 had a lower surface temperature than that of WT (Fig. 4B), the leaf surface temperature was comparable between WT and ait1. It is possible that AIT1 is not required for stomatal closure or that ABA transport is highly complex and redundant in leaves. As ABA can be transported on a long-distance basis through the xylem or phloem (9), it is interesting to speculate that the expression of AIT1 in vascular tissues might be related to loading/unloading of ABA into/out of xylem/phloem vessels. However, ABA measurements showed that bulk ABA contents in the inflorescence stems were comparable between WT and ait1 (Fig. S5A). This suggested that the defective stomatal closure in the inflorescence stems of ait1 results from altered local distribution of ABA.
Overexpression of AIT1 resulted in lower surface temperature of leaves and inflorescence stems (Fig. 4 C and D). One might expect opposite phenotypes between loss-of-function mutants and constitutive overexpressors, but that was not the case. In germination assays in which the ability of the seeds to germinate depends on the amount of exogenously applied ABA, ait1 and AIT1 overexpressors showed opposite phenotypes (Fig. 3). On the contrary, the reduced surface temperature phenotypes of ait1 and AIT1 overexpressors were observed in the absence of exogenous ABA, indicating that the movement and localization of endogenous ABA were altered in these plants. Thus, it is possible that the apparently similar effects of the loss of function and overexpression of AIT1 on stomatal aperture were caused by different mechanisms. We speculate that ABA remains at the site of its biosynthesis (parenchyma cells) and thus it cannot reach the guard cells in the AIT1 overexpressors (Fig. S4B). This idea is supported by the observation that the movement of exogenously applied ABA was inhibited in the inflorescence stems of AIT1 overexpressors, but not in those of ait1 (Fig. S5B).
Roles of Different Types of ABA Transporters.
Although it has been reported that AtABCG25 mediates ABA import and AtABCG40 mediates ABA export (18, 19), we were unable to detect these predicted activities in our assay system (Fig. S1D). It is possible that these transporters are not functionally expressed in our yeast system. It appears that the two possible ABA importers, AIT1 and AtABCG40, have distinct physiological roles, because the reduced stem surface temperature was observed in ait1, but not in abcg40 (Fig. 4B and Fig. S3E). The inability of the abcg40 mutation to enhance the ait1 phenotype supports this idea (Fig. S3E). In addition, germination of abcg40 and WT were similarly inhibited by ABA in our experimental conditions, in which we clearly observed ABA-resistant germination of ait1 (Fig. 3A and Fig. S6). It also appears that AtABCG25 is not involved in the physiological processes mediated by AIT1 in inflorescence stems (Fig. S3E). Although it appears that AIT1, ABCG40, and ABCG25 have distinct physiological roles, these three transporters cannot fully explain all the physiological processes mediated by ABA. There may be other ABA transporters that are yet to be identified. Alternatively, it is possible that some physiological responses do not require ABA transport.
Crosstalk with Nitrate Signaling.
Mutants affected in nitrate assimilation or nitrate transport show altered responses to ABA, and crosstalk between nitrate and ABA signals has been discussed (32, 33). Because AIT1/NRT1.2 imports nitrate in addition to ABA, it is possible that the ABA-related phenotypes observed in ait1 might be caused indirectly by disruption of nitrate signaling. If this is the case, ait1 should exhibit an ABA-hypersensitive phenotype, because nitrate negatively regulates ABA signaling. However, the phenotype observed in ait1 was opposite to that deduced from the function of AIT1/NRT1.2 as a nitrate transporter. Interestingly, the nitrate transporter NRT1.1 transports IAA in addition to nitrate (34). It was suggested that the induction of lateral root development by high nitrate concentrations was caused by the inhibition of endogenous IAA transport through NRT1.1 by competition with nitrate, leading to accumulation of IAA at the lateral root tips (34). Similarly, it is possible that the inhibition of ABA signaling by nitrate is caused by the inhibition of endogenous ABA transport through AITs. It was also reported that the mutant defective in NRT1.1 (chl1) had more closed stomata in the leaves and was more resistant than WT to water stress (35). It was suggested that nitrate transport mediated by NRT1.1 functions to regulate stomatal aperture. However, as chl1 and ait1 show opposite phenotypes in terms of stomatal aperture, the physiological and biochemical functions of NRT1.1 are distinct from those of AIT1/NRT1.2.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis WT (Col-0) and mutant plants (all in Col-0 background) were grown in soil containing vermiculite and Metro-mix 350 (Sun Gro Horticulture) at a 3:1 ratio. Nutrients were provided by a 1/1,000 dilution of Hyponex (Hyponex Japan). Plants were grown in growth chambers at 22 °C under continuous white light (∼10 W/m−2) with a relative humidity of 75%. T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center (Figs. S2 and S6). Homozygous mutants were selected by PCR by using primers designed according to the Salk Institute Genomic Analysis Laboratory Web site (http://signal.salk.edu/tdnaprimers.2.html).
Yeast Screening.
cDNA libraries constructed from RNA extracted from 2-wk-old Arabidopsis seedlings or a full-length cDNA library (36) were introduced into the yeast strain PJ69-4A containing AD-ABI1 and BD-PYR1, or AD-HAB1 and BD-PYR1, and colonies surviving on selection medium (synthetic dextrose; -Leu, -Trp, -Ura, -His) in the presence of 0.1 μM ABA were selected. Screening was performed according to standard protocols. Detailed procedures are described in SI Materials and Methods.
Cloning of NRT1/PTR Family.
cDNAs for 11 members of the group I NRT1/PTR were amplified with primer combinations listed in Table S1, and cloned into pENTR/D-TOPO (Invitrogen).
Transport Assays.
ABA import activities were determined indirectly by detecting the ABA-dependent interactions between AD-ABI1 and BD-PYR1 or by analyzing hormones incorporated into yeast cells or insect cells by using LC-MS/MS as described in SI Materials and Methods. Conditions for LC and parameters for MS/MS are shown in Tables S2 and S3.
ABA Measurements from Plant Tissues.
Extraction and purification of ABA from plant tissues were conducted essentially as described previously (37, 38) with some modifications. Details of the procedure are given in SI Materials and Methods. Conditions for LC and parameters for MS/MS are shown in Tables S2 and S3.
Transgenic Plants.
To produce AIT1-overexpressing Arabidopsis plants, AIT1 cDNA was cloned into pBE2113 (39). For promoter-GUS analysis, 2,000 bp of the AIT1 promoter region was cloned into pGWB3 (40). To express the C terminus GFP fusion of AIT1 under the control of the 35S promoter, AIT1 cDNA was cloned into pGWB5 (40). Each construct was introduced into Agrobacterium strain GV3101 and transformed into Arabidopsis Col-0 accession by the floral-dip method. GUS staining was performed as described previously (41) and observed by stereomicroscopy (MZ FLIII; Leica). GFP fluorescence was observed by confocal laser scanning microscopy (LSM5 Pascal; Carl Zeiss). Details of the procedure are given in SI Materials and Methods.
Measurements of Stomata Aperture.
Images of stomatal apertures were obtained by the Suzuki Universal Micro-Printing (SUMP) method using SUMP liquid and a SUMP plate (SUMP Laboratory). The copied SUMP images were observed by light microscopy (Axioplan 2; Carl Zeiss).
Thermal Imaging.
Thermal images were obtained using a thermal video system (TVS-8500; Nippon Avionics).
Germination Tests.
Seeds were surface-sterilized in a solution containing 5% (vol/vol) NaClO and 0.05% (vol/vol) Tween 20, rinsed with water, and sown on 0.8% (wt/vol) agar plates (pH 6.5) containing 0.5× Murashige and Skoog salts and Mes (0.5 g/L). Chemical stock solutions (1,000× concentration) were prepared in DMSO and added to the media after autoclaving. After sowing seeds, plates were incubated at 4 °C in the dark for 3 d and then transferred to light conditions at 22 °C.
Supplementary Material
Acknowledgments
We thank Dr. Tadao Asami (The University of Tokyo) for providing [13C2]abscisic acid, Dr. Takashi Hirayama (Okayama University) for providing abi1-1C, Dr. Tsuyoshi Nakagawa (Shimane University) for providing pGWB vectors, the Arabidopsis Biological Resource Center for providing transfer DNA insertion mutants, Dr. Jutaro Fukazawa (RIKEN Plant Science Center) for technical advice on yeast experiments and confocal microscopy, and Dr. Yusuke Jikumaru (RIKEN Plant Science Center) for technical advice on LC-MS/MS analysis and for providing jasmonic acid. We also thank Dr. Eiji Nambara (University of Toronto), Dr. Akira Endo (University of Toronto), Dr. Takeshi Nishimura (Tokyo Metropolitan University), Dr. Annie Marion-Poll (Institut National de la Recherche Agronomique Versailles), and Dr. Helen North (Institut National de la Recherche Agronomique Versailles) for their critical reading of this manuscript and for valuable discussions. Hormone analysis using the Nexera/TripleTOF 5600 system was supported by the Japan Advanced Plant Science Network.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203567109/-/DCSupplemental.
References
- 1.Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- 2.Petrásek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 3.Cao FY, Yoshioka K, Desveaux D. The roles of ABA in plant-pathogen interactions. J Plant Res. 2011;124:489–499. doi: 10.1007/s10265-011-0409-y. [DOI] [PubMed] [Google Scholar]
- 4.Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 5.Nambara E, et al. Abscisic acid and the control of seed dormancy and germination. Seed Sci Res. 2010;20:55–67. [Google Scholar]
- 6.Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:439–473. [Google Scholar]
- 7.Davies WJ, Zhang JH. Root signals and the regulation of growth and development of plants in drying soil. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:55–76. [Google Scholar]
- 8.Jiang F, Hartung W. Long-distance signalling of abscisic acid (ABA): The factors regulating the intensity of the ABA signal. J Exp Bot. 2008;59:37–43. doi: 10.1093/jxb/erm127. [DOI] [PubMed] [Google Scholar]
- 9.Sauter A, Davies WJ, Hartung W. The long-distance abscisic acid signal in the droughted plant: The fate of the hormone on its way from root to shoot. J Exp Bot. 2001;52:1991–1997. doi: 10.1093/jexbot/52.363.1991. [DOI] [PubMed] [Google Scholar]
- 10.Seo M, Koshiba T. Transport of ABA from the site of biosynthesis to the site of action. J Plant Res. 2011;124:501–507. doi: 10.1007/s10265-011-0411-4. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson S, Davies WJ. ABA-based chemical signalling: The co-ordination of responses to stress in plants. Plant Cell Environ. 2002;25:195–210. doi: 10.1046/j.0016-8025.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- 12.Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi: 10.1111/j.1365-313X.2007.03234.x. [DOI] [PubMed] [Google Scholar]
- 13.Holbrook NM, Shashidhar VR, James RA, Munns R. Stomatal control in tomato with ABA-deficient roots: response of grafted plants to soil drying. J Exp Bot. 2002;53:1503–1514. [PubMed] [Google Scholar]
- 14.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 15.North HM, et al. The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 2007;50:810–824. doi: 10.1111/j.1365-313X.2007.03094.x. [DOI] [PubMed] [Google Scholar]
- 16.Endo A, et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008;147:1984–1993. doi: 10.1104/pp.108.116632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koiwai H, et al. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004;134:1697–1707. doi: 10.1104/pp.103.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang J, et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA. 2010;107:2355–2360. doi: 10.1073/pnas.0909222107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuromori T, et al. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA. 2010;107:2361–2366. doi: 10.1073/pnas.0912516107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK. Nitrate transporters and peptide transporters. FEBS Lett. 2007;581:2290–2300. doi: 10.1016/j.febslet.2007.04.047. [DOI] [PubMed] [Google Scholar]
- 23.Huang NC, Liu KH, Lo HJ, Tsay YF. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell. 1999;11:1381–1392. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merlot S, et al. Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J. 2002;30:601–609. doi: 10.1046/j.1365-313x.2002.01322.x. [DOI] [PubMed] [Google Scholar]
- 26.Almagro A, Lin SH, Tsay YF. Characterization of the Arabidopsis nitrate transporter NRT1.6 reveals a role of nitrate in early embryo development. Plant Cell. 2008;20:3289–3299. doi: 10.1105/tpc.107.056788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan SC, Lin CS, Hsu PK, Lin SH, Tsay YF. The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell. 2009;21:2750–2761. doi: 10.1105/tpc.109.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JY, et al. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell. 2010;22:1633–1646. doi: 10.1105/tpc.110.075242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SH, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell. 2008;20:2514–2528. doi: 10.1105/tpc.108.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YY, Tsay YF. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell. 2011;23:1945–1957. doi: 10.1105/tpc.111.083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu KH, Tsay YF. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003;22:1005–1013. doi: 10.1093/emboj/cdg118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alboresi A, et al. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005;28:500–512. doi: 10.1111/j.1365-3040.2005.01292.x. [DOI] [PubMed] [Google Scholar]
- 33.Matakiadis T, et al. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009;149:949–960. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krouk G, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 35.Guo FQ, Young J, Crawford NM. The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell. 2003;15:107–117. doi: 10.1105/tpc.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichikawa T, et al. The FOX hunting system: An alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- 37.Kanno Y, et al. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of abscisic acid biosynthesis, abscisic acid transport and hormone interactions. Plant Cell Physiol. 2010;51:1988–2001. doi: 10.1093/pcp/pcq158. [DOI] [PubMed] [Google Scholar]
- 38.Saika H, et al. Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol. 2007;48:287–298. doi: 10.1093/pcp/pcm003. [DOI] [PubMed] [Google Scholar]
- 39.Mitsuhara I, et al. Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa T, et al. Development of series of gateway binary vector, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:33–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 41.Tan BC, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.