Abstract
The replication/transcription complex of severe acute respiratory syndrome coronavirus is composed of at least 16 nonstructural proteins (nsp1–16) encoded by the ORF-1a/1b. This complex includes replication enzymes commonly found in positive-strand RNA viruses, but also a set of RNA-processing activities unique to some nidoviruses. The nsp14 protein carries both exoribonuclease (ExoN) and (guanine-N7)-methyltransferase (N7-MTase) activities. The nsp14 ExoN activity ensures a yet-uncharacterized function in the virus life cycle and must be regulated to avoid nonspecific RNA degradation. In this work, we show that the association of nsp10 with nsp14 stimulates >35-fold the ExoN activity of the latter while playing no effect on N7-MTase activity. Nsp10 mutants unable to interact with nsp14 are not proficient for ExoN activation. The nsp10/nsp14 complex hydrolyzes double-stranded RNA in a 3′ to 5′ direction as well as a single mismatched nucleotide at the 3′-end mimicking an erroneous replication product. In contrast, di-, tri-, and longer unpaired ribonucleotide stretches, as well as 3′-modified RNAs, resist nsp10/nsp14-mediated excision. In addition to the activation of nsp16-mediated 2′-O-MTase activity, nsp10 also activates nsp14 in an RNA processing function potentially connected to a replicative mismatch repair mechanism.
Keywords: RNA proofreading, viral evolution, protein-protein interaction, capping
In 2003, an outbreak of an unusually pathogenic agent spread from China to the whole world. More than 8,400 people were infected with ∼800 case-fatalities by this novel coronavirus now known as severe acute respiratory syndrome coronavirus (SARS-CoV). Viruses from the Coronavirus and Torovirus genera constitute the Coronaviridae virus family, which together with Arteriviridae and Roniviridae belong to the Nidovirales order (1). The coronavirus genome consists of a single-stranded, positive-sense RNA of 27–32 kb, the largest size for an RNA virus genome. The former is directly translated into two polyprotein precursors corresponding to ORF-1a (called pp1a) and ORF-1a elongated by ORF-1b (called pp1ab) following a ribosomal frameshift. The ribosomal frameshift frequency allows a three- to fivefold excess of pp1a over pp1ab. The pp1a and pp1ab polyproteins are intracellularly processed by viral proteases to yield 11 and 16 (11 + 5) nonstructural proteins (nsps), respectively. These nsps assemble together with cellular factors to form a huge replication/transcription complex (RTC) associated with membrane structures derived from the endoplasmic reticulum (ER) (2, 3). Apart from enzyme activities usually essential for RNA genome replication/transcription, coronaviruses also encode for a set of RNA-processing activities that are either unique to genera inside Nidovirales or are found only in a few other groups of RNA viruses (4). Among these activities are two RNA nucleases, an exoribonuclease (nsp14, named ExoN) (5), and a uridylate-specific endoribonuclease (nsp15, named NendoU) (6). Additionally, it has recently been demonstrated that SARS-CoV nsp1 in association with the 40S ribosome subunit induces an endonucleolytic degradation of host mRNAs, though the enzyme responsible of the cleavage has not been identified (7).
The presence of RNA endo- and exonucleases is puzzling, and the roles of nsp14 and nsp15 along the virus life cycle are unknown. Nsp14 is bifunctional, with a 3′ to 5′ ExoN activity residing in its N-terminal part (5), whereas a (guanine-N7)-methyltransferase (N7-MTase) activity is embedded in the C-terminal part (8, 9). The N7-MTase domain is not functionally separable from the ExoN domain because N-terminal deletions of the protein impair nsp14 N7-MTase activity (8). Nsp14 contains essential ExoN [exo(ribo)nucleases] containing a conserved Asp-Glu-Asp-Asp motif (DEDD) ExoN superfamily motifs and has been shown to hydrolyze single- and double-stranded RNAs (ssRNA and dsRNA) to final products of 8–12 nt and 5–7 nt, respectively. This enzyme has been proposed to be involved in replication and recombination during minus-strand discontinuous transcription (5, 10, 11).
However, murine hepatitis virus (MHV) and SARS-CoV viruses ExoN mutants exhibit growth defects but are competent for replication in cell culture (12, 13). Notably, nsp14 mutant viruses exhibit a mutator phenotype, with an overall 12- to 20-fold increase in mutation frequency and up to 14-fold increase in mutation rate compared with WT. Nsp14 was therefore proposed to be involved in proofreading, repair, and/or recombination mechanisms essential to maintain the integrity of the astonishingly long CoV’s RNA genome (for a review, see ref. 14). However, the mechanisms by which nsp14 ExoN safeguards replication fidelity remain to be discovered.
Nsp14 is known to interact with nsp10 (15, 16), whose crystal structure is known (17, 18). Nsp10 also interacts with nsp16 in vitro, forms a complex whose structure has been recently solved, and switches on an RNA-cap 2′-O-MTase activity carried by nsp16 (9, 19–21).
In this work, we demonstrate a second regulatory role for SARS-CoV nsp10. Nsp14 is converted by nsp10 into a >35-fold more potent exoribonuclease. Because substrate requirements of the nsp10/nsp14 complex include dsRNAs having a mismatched or Watson–Crick base-paired 3′-end, our results suggest that nsp14 is a proofreading enzyme in agreement with the mutator phenotype observed for coronavirus nsp14 mutants (12–14).
Results
SARS-CoV nsp10 Protein Interacts with the nsp14 Protein.
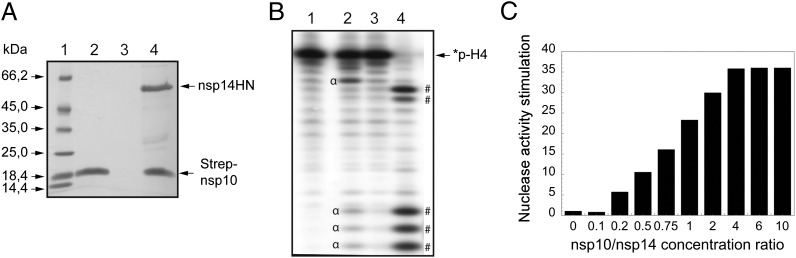
The nsp10 and nsp14 proteins were previously shown to interact using yeast and mammalian two-hybrid systems (15, 16). We sought to demonstrate this interaction using an in vitro system. For this purpose, SARS-CoV Strep-nsp10 and nsp14HN proteins were coexpressed in Escherichia coli and the bacterial cell lysate was incubated with Strep-Tactin beads to bind the Strep-tagged nsp10 protein. As shown in Fig. 1A, nsp14 remains associated with nsp10 when both proteins are coexpressed, whereas nsp14 alone is unable to bind to the beads. We thus confirm that nsp14 stably interacts with nsp10.
Fig. 1.
Nsp10 interacts with nsp14, and nsp10/nsp14 shows enhanced ExoN activity. (A) SARS-CoV nsp14HN and Strep-nsp10 proteins coexpressed or expressed alone were incubated with Strep-Tactin Sepharose. Strep-Tactin–bound proteins were eluted with d-desthiobiotin and analyzed by SDS/PAGE and Coomassie blue staining. Lane 1 corresponds to the molecular size markers; lane 2 to Strep-nsp10 expressed alone; lane 3 to nsp14HN expressed alone; and lane 4 to Strep-nsp10 coexpressed with nsp14HN. (B) Autoradiogram of RNA cleavage products. Synthetic *p-H4 RNA was radiolabeled at its 5′-end using PNK in the presence of [γ32P]ATP (the asterisk indicates the 32P-labeling position). *p-H4 RNA was incubated at 37 °C in Tris⋅HCl buffer 40 mM (pH 8), DTT 5 mM with no protein (lane 1), 0.7 μM of nsp14 (lane 2), nsp10 (lane 3), or both proteins (lane 4) during a 90-min period. The reaction products were then separated on a 20% (wt/vol) denaturing Urea-PAGE and revealed using photostimulated plates and a FujiImager (Fuji). (C) *p-H4 RNA was hydrolyzed with fixed nsp14 concentration (50 nM) in the presence of increasing concentration of nsp10 ranging from 0 to 1,600 nM (nsp10/nsp14 is indicated below the bar graph). ExoN activity was quantified using denaturing Urea-PAGE followed by measuring the hydrolysis of *p-H4 corresponding band using a FujiImager and Image Gauge software analysis.
Nsp10 Protein Activates nsp14 ExoN Activity.
To discover a potential function for this interaction, we produced both proteins individually in E. coli and purified them separately using affinity followed by size-exclusion chromatography. Nsp10 was recovered with an N-terminal hexahistidine tag [(His)6], and nsp14 was recovered as an untagged protein (Materials and Methods).
We first incubated a 5′-end 32P-labeled ssRNA substrate named *p-H4 (Table S1) with nsp14 in the presence or absence of nsp10. Reaction products were separated by denaturing Urea-PAGE and revealed using autoradiography. Nsp14 alone exhibits nuclease activity, albeit weak with this RNA substrate (Fig. 1B, lane 2, major degradation products are indicated by α), in agreement with others (5, 11). The nsp10 protein does not carry any nuclease activity under these conditions (Fig. 1B, lane 3), but incubation of both proteins results in a strong nuclease activity (lane 4, major degradation products are indicated by #). As the RNA was labeled at its 5′-end, the laddering degradation pattern is suggestive of a 3′ to 5′ directionality.
We determined optimal reaction conditions as well as metallic ion requirements of the nsp10/nsp14 ExoN activity. Interestingly, we found that the latter critically depends both on metallic ions such as Mg2+, and on the presence of the Zn2+ ions of nsp10 (Fig. S1 and SI Text). We also determined nsp10/nsp14 concentration ratio yielding the maximal ExoN activity. As shown in Fig. 1C, the ExoN activity is stimulated by nsp10 in a dose-dependent manner until a >35-fold stimulation, reached with a fourfold excess of nsp10 over nsp14. At equimolar ratio, as used in Fig. 1B, the ExoN activity is stimulated around 20-fold compared with the ExoN activity exhibited by nsp14 alone. We infer that besides the recently identified stimulation effect of nsp10 onto nsp16 MTase activity (9), nsp10 also stimulates the ExoN activity of nsp14. In contrast, the presence of nsp10 has no effect on nsp14-mediated N7-MTase activity, as reported before (9).
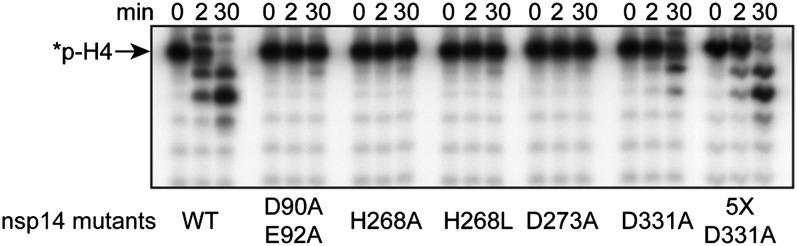
ExoN Activity Exhibited by nsp10/nsp14 Involves the DEDDh Catalytic Residues of nsp14.
To formally demonstrate that nsp10 does not have any nuclease activity switched on by the presence of nsp14, we mutated the nsp14 conserved catalytic residues D90XE92, D243, H268, and D273 of the DEDDh ExoN motifs to alanine residues. We also substituted residue D331, from the DxG motif implicated in S-adenosyl-methionine (SAM) binding (8) and used it as a negative control. As shown in Fig. 2A, mutations of residues belonging to motif Exo I, II, or III completely abrogate ExoN activity. Nsp14 mutants migrate at their expected molecular weight after purification and are known to keep their native folding because they exhibit N7-MTase activity in vitro (9). We also observe that the SAM binding-site mutation within the N7-MTase domain (D331A) dampens ExoN activity. However, a fivefold excess of nsp14 D331A allows one to observe an ExoN activity comparable to that of WT. These results demonstrate that nsp10/nsp14 ExoN activity involves the nsp14 DEDDh catalytic residues and that nsp10 acts as an activation cofactor devoid of nuclease activity per se. Moreover, the SAM-binding mutant (D331A) reveals a cross-talk between nsp14 N7-MTase and ExoN catalytic sites, a result in agreement with the observation made by others that deletions in the ExoN domain of nsp14 lead to inactivation of the N7-MTase activity (8).
Fig. 2.
Mutagenesis analysis of nsp14 ExoN activity. Residues from the nsp14 ExoN catalytic site and from the SAM-binding site of the nsp14 MTase domain were mutated into alanine. Equal amounts of each nsp14 mutant were incubated with nsp10 and *p-H4 RNA for 0, 2, and 30 min. In lane 5XD331A, the concentration of the mutant was fivefold higher. The panel shows the time-dependent hydrolysis of *p-H4 RNA after Urea-PAGE separation and autoradiography.
Activation of SARS-CoV nsp14 ExoN Activity Requires Direct Interaction with nsp10.
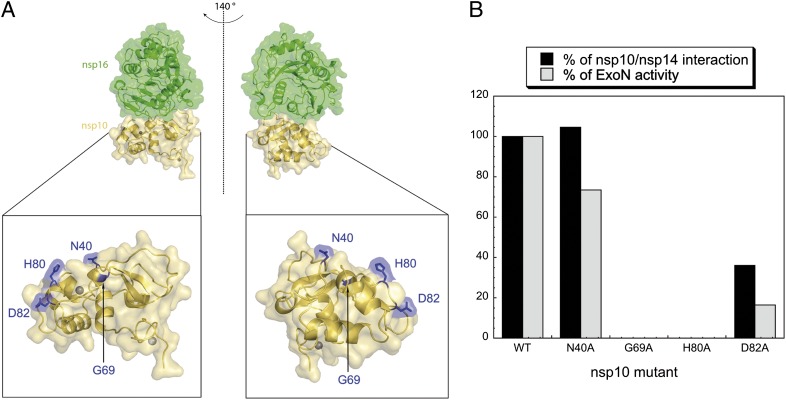
We selected residues (N40, G69, H80, D82) localized on the surface of the nsp10 protein to perform alanine substitutions (Fig. 3A). Strep-tagged nsp10 mutants were coexpressed with nsp14 and purified using Strep-Tactin resin. Proteins eluted from Strep-Tactin were separated by LabChip (Caliper), and peaks corresponding to nsp10 and nsp14 were quantified. Fig. 3B presents the percentage of interaction between nsp14 and nsp10 mutants relative to nsp10 WT (black bars). G69A and H80A mutants completely lose their ability to interact with nsp14, whereas N40A and, to a lesser extent, D82A keep their interaction properties. We next analyzed the consequence of these nsp10 mutations on the nsp10/nsp14 ExoN activity. *p-H4 RNA was incubated with nsp14 in the presence of nsp10 WT or nsp10 mutants. RNA hydrolysis was quantified upon denaturing Urea-PAGE. Fig. 3B presents the percentage of nsp14 ExoN activity obtained with each nsp10 mutant relative to nsp10 WT (gray bars). The results show that interaction of nsp10 and nsp14 is required for the stimulation of ExoN activity. Moreover, we note that residues annihilating both nsp10/nsp14 interaction and ExoN activity (e.g., G69 and H80) are localized within a surface area involved in the nsp10/nsp16 interaction (Fig. 3A) (19, 20). Because these data suggested a common interaction surface for both nsp14 and nsp16 with nsp10, we performed competition experiments to evaluate complex stabilities. As shown in Fig. S2A, a 16-fold molar excess of nsp16 is unable to alter nsp10/nsp14-mediated ExoN activity. We then tried to destabilize nsp10/nsp16 interaction using nsp14. As shown in Fig. S2B, the presence of nsp14 increases the 2′-O-MTase activity of nsp10/nsp16, and the effect was even higher in the presence of the nsp10/nsp14 complex. We conclude that once formed, both complexes are stable. Nsp10/nsp14 might stimulate further nsp10/nsp16 2′-O-MTase activity, suggesting a cross-talk between nsp14 and nsp16 MTase activities, both of which are involved in RNA capping.
Fig. 3.
Nsp10/nsp14 interaction is required for nsp14 ExoN stimulation. (A) Selection of nsp10 surface mutants and their position in the nsp10/nsp16 dimer. The nsp10/nsp16 complex image (Upper) was represented using PyMOL, and an enlargement of nsp10 is shown (Lower) with the position of mutated amino acids (highlighted in blue). nsp16 is represented in green, nsp10 in gold, and nsp10 zinc structural ions as gray spheres. (B) Quantification of nsp10/nsp14 interaction and corresponding ExoN activity. Nsp10 WT or mutants proteins carrying a Strep-TagII were coexpressed with nsp14HN. After purification on Strep-Tactin beads, eluted proteins were separated using LabChip (Caliper), and the intensities of peaks corresponding to nsp10 and nsp14 proteins were quantified. Molecular ratio obtained for nsp10 WT/nsp14 complex was taken as 100%. Results are presented in percentage of interaction compared with the nsp10 WT/nsp14 complex (black bars). For ExoN quantification, equal amounts of nsp10 mutants were incubated with nsp14 and *p-H4 RNA. After 30 min of incubation, reaction products were separated using Urea-PAGE, revealed using a FujiImager, and quantified using Image Gauge software. Results are presented in percentage of ExoN activity (nsp10 WT/nsp14 was taken as 100% of activity; gray bars).
Substrate Requirements of the SARS-CoV nsp10/nsp14 ExoN.
To determine the nsp10/nsp14 ExoN activity substrate requirements, we used a set of 5′-end radiolabeled RNAs as substrates; their secondary structures were predicted using the Mfold RNA modeling server and are presented in Fig. S3. ExoN assays indicate that nsp10/nsp14 ExoN activity requires an RNA 3′-end engaged in a duplex structure (SI Text and Fig. S4). This finding is also illustrated in Fig. 4, where a ssRNA oligonucleotide (RNA11) incubated with the enzyme complex (nsp10/nsp14) is barely degraded (major degradation products are indicated by α). Upon annealing of its complementary strand carrying a biotin (RNA11revbiot) at its 3′-end, the resulting dsRNA heteroduplex is rapidly hydrolyzed by nsp10/nsp14 into shorter products of ∼3–4 nt (indicated by # in Fig. 4). Together, these results show that nsp10/nsp14 ExoN shows no obvious sequence preference, but requires the 3′-end to be engaged in a stable RNA duplex.
Fig. 4.
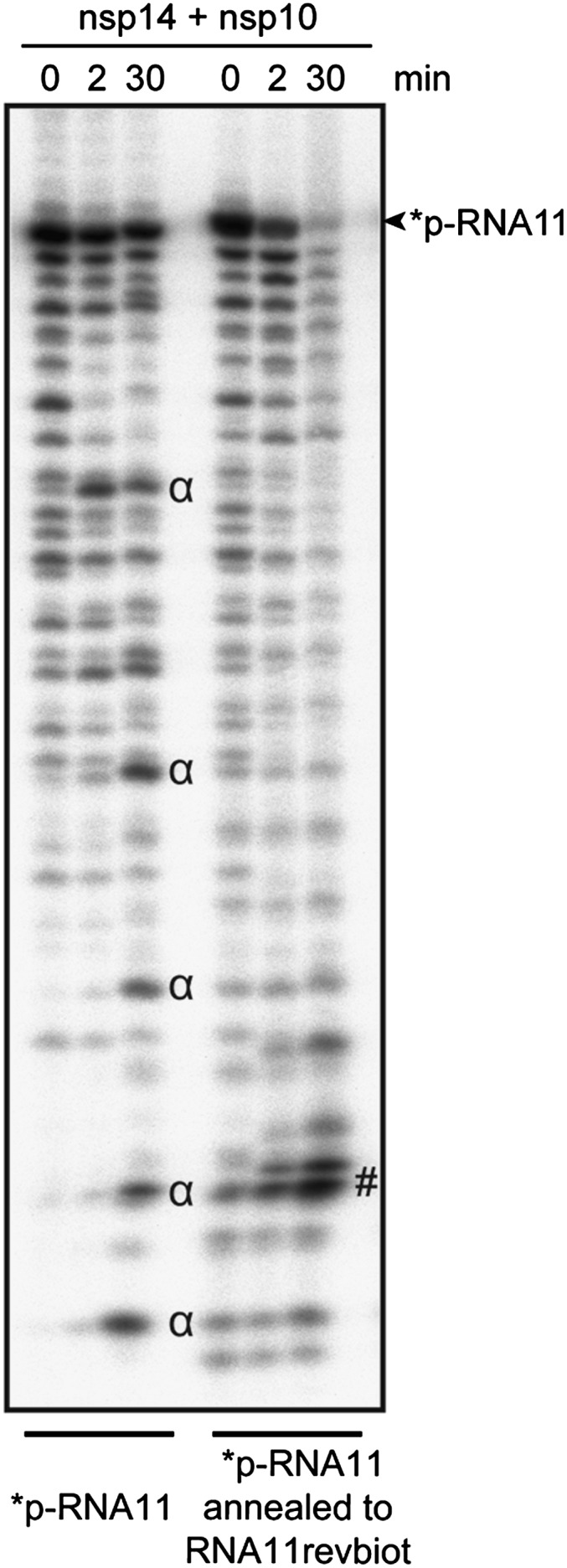
The nsp10/nsp14 ExoN activity is a dsRNA-dependent exonuclease. Autoradiogram of an ExoN assay. HPLC-purified ssRNA (RNA11) was labeled at its 5′-end using PNK and [γ32P]ATP, and hybridized to its complementary strand modified at its 3′-end with biotin (RNA11revbiot). Single- or double-stranded labeled-RNAs were subjected to nsp10/nsp14 ExoN digestion for 0, 2, and 30 min. Digestion products were separated on Urea-PAGE and revealed by autoradiography.
Nsp14 ExoN Activity Requires a Free 3′-Hydroxyl End and Proceeds 3′ to 5′.
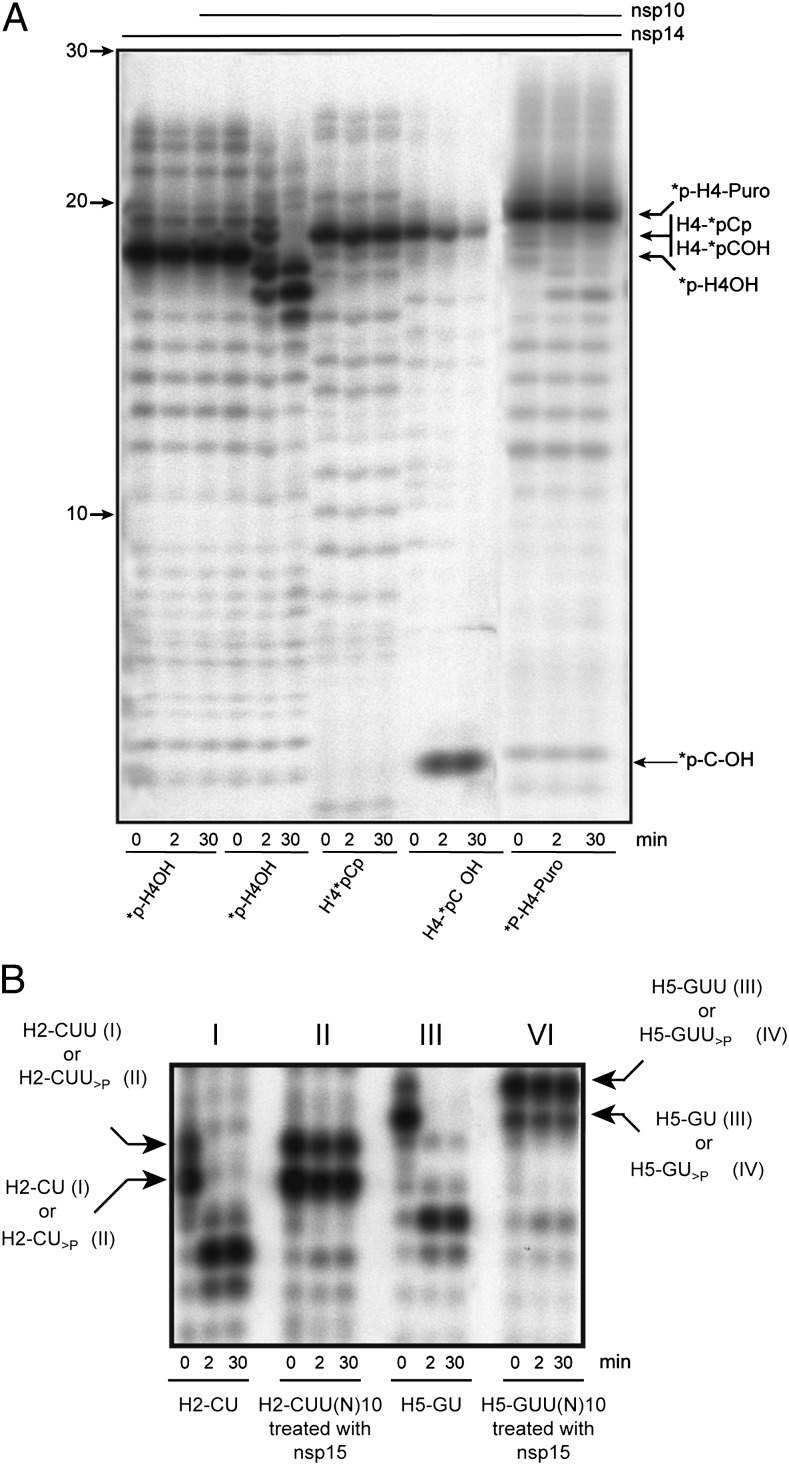
We next investigated the effect of a modification of the RNA 3′-OH end on nsp10/nsp14 ExoN activity. Fig. 5A shows that dsRNA oligonucleotides carrying either a 3′-terminal puromycin (*p-H4-puro) or phosphate (H4-*pCp vs. H4-*pCOH) resist nsp10/nsp14-mediated hydrolysis. We also analyzed the hydrolysis of 3′-end-labeled H4-*pCOH; its hydrolysis leads to the immediate recovery of a unique product comigrating with a *pCOH control, demonstrating that nsp10/nsp14 proceeds 3′ to 5′.
Fig. 5.
Nsp10/nsp14 requires a free 3′-hydroxyl end. (A) Autoradiogram of an ExoN assay performed with RNA carrying 3′-phosphate, 3′-puromycin, or 3′-hydroxyl end. The 3′ modified RNAs (*p-H4 as control, *p-H4-puromycin, H4-*pCp, and H4-*pCOH) were incubated with nsp14 or nsp10/nsp14 for 0, 2, and 30 min (asterisk indicates the 32P-labeling position). Digestion products were separated on 20% (wt/vol) Urea-PAGE and revealed by autoradiography. (B) Autoradiogram of nsp15-generated 2′-3′-cyclic phosphate RNAs cleavage assay. RNA substrates H2-CUU(N)10 and H5-GUU(N)10 were synthesized using T7 RNA polymerase and subsequently dephosphorylated and radiolabeled. These RNAs were then incubated with nsp15 to generate *p-H2-CU>P and *p-H2-CUU>P and, to a lesser extent, *p-H5-GUU>P and *p-H5-GU>P. Control RNAs *p-H2-CUOH and *p-H5-GUOH were synthesized using T7 RNA polymerase and led to the production of the side products *p-H2-CUUOH and *p-H5-GUUOH, respectively. After purification, these substrates were incubated with nsp10/nsp14 for 0, 2, and 30 min. Digestion products were separated on 20% (wt/vol) Urea-PAGE and revealed by autoradiography.
Because the nsp10/nsp14 ExoN activity was proposed to be functionally linked to the nsp15 NendoU activity, e.g., in a proofreading mechanism (14), we assayed nsp15 NendoU products as substrates for the ExoN activity of nsp10/nsp14. For this purpose, RNAs containing an internal CUU or GUU sequences [H2-CUU(N)10 and H5-GUU(N)10] were incubated with nsp15 to generate a terminal 2′,3′-cyclic phosphate RNAs [H2-CU(U)>P and H5-GU(U)>P] (22, 23). As shown in Fig. 5B, in contrast to unmodified RNAs used as controls, RNAs carrying a 2′,3′-cyclic phosphate end resist nsp10/nsp14-mediated hydrolysis. We conclude that, irrespective of a putative role of nsp14 in mismatch repair, nsp15-mediated RNA endonucleolytic cleavage is unlikely to serve as an entry point for nsp14 ExoN.
SARS-CoV nsp14 ExoN Activity Is Able to Excise a 3′ Mismatched Nucleotide.
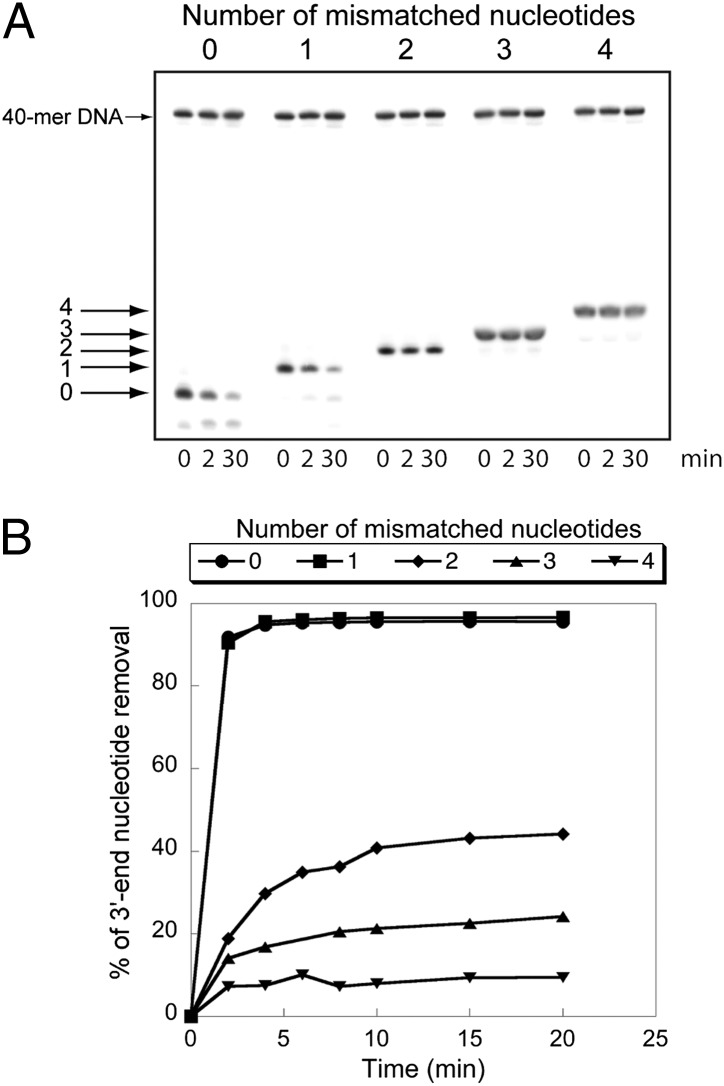
We assayed the ability of nsp14 to excise 3′-end mismatched nucleotides on a dsRNA mimicking an erroneous replication product (Fig. S5). For this purpose, a 40-mer RNA (LS1) carrying a 3′-biotin group was annealed to a radiolabeled reverse-complement RNA carrying no mismatch (LS2) or adding one, two, three, or four mismatched nucleotides to its 3′-end (LS3–LS6). Fig. 6A shows that nsp10/nsp14 is able to excise a single 3′-terminal mismatch, but excision capability strongly decreases when a longer mismatch is introduced at the 3′-end. The time course of RNA degradation shown in Fig. 6B indicates that a mismatched nucleotide is excised at the same rate as that of a regular Watson–Crick base pair, whereas the RNA excision rate decreases as a function of the extension of unpaired sequences. To determine the substrate preference between a Watson–Crick and a mismatched 3′-end base pair, we performed competition experiments using a radiolabeled substrate carrying either a U:A or a U:C 3′-end base pair. Cold substrates were used as challengers competing for nsp10/nsp14 binding and degradation. As shown in Fig. S6 A and B, a substrate carrying a 3′-end mismatch competes out more efficiently the perfectly matched 3′-end substrate than the other way around. This result indicates that a mismatched 3′-end base pair might actually be the preferred substrate for nsp10/nsp14 ExoN activity. Finally, we compared the excision efficiency using different mismatched base pairs. Single mismatches of any type (A:G, A:A, A:C, U:G, U:C, U:U) were removed efficiently; initial velocity measurements of nsp14 mismatch excision indicate that the ability of the ExoN to degrade these substrates does not depend on the nature of the nucleotide misincorporated at the 3′-end (Fig. S6C).
Fig. 6.
Comparison of nsp10/nsp14 ExoN activity on paired and mismatched 3′-end nucleotide base pairs. (A) A 40-nt RNA (LS1) was annealed with 5′-radiolabeled oligoribonucleotides carrying zero, one, two, three, or four noncomplementary bases at its 3′-end (respectively, LS2, LS3, LS4, LS5, or LS6 (Table S1 and Fig. S5). To avoid nsp10/nsp14-mediated LS1 degradation, this RNA carries a biotin group at its 3′-end. Duplex RNAs were then incubated (0, 2, and 30 min) with purified nsp10/nsp14 (200 nM/50 nM). Reaction products were separated on a 20% (wt/vol) denaturing Urea-PAGE and revealed by autoradiography. A radiolabeled 40-mer DNA was introduced in the reaction mixture as a quantification reference. (B) Kinetics of mismatch excision. The assay was performed as in A, using 100 nM of nsp14 and 400 nM of nsp10. RNA cleavage was quantified at 0, 2, 4, 6, 8, 10, 15, and 20 min. Data are presented as percent of 3′-nucleotide removal.
Because the main RNA-dependent RNA polymerase (RdRp) nsp12 is known to interact with nsp14 (15), we conclude that nsp10/nsp14 is able to efficiently remove a 3′-terminal mismatch present on an RNA supposedly being synthesized by nsp12. Longer mismatched structures, unlikely to be synthesized by the viral polymerase, do not act as nsp10/nsp14 ExoN substrates.
Discussion
SARS-CoV encodes several RNA processing activities, including RNA nucleases such as the nsp15 NendoU and the nsp14 ExoN. Those RNases activities are expected to be highly specific and regulated in vivo to avoid any unwanted cleavage of viral and/or cellular RNAs in infected cells. In this work, we present a unique regulation mechanism directed by the nsp10 protein on the nsp14 ExoN activity. We demonstrate that nsp14 possesses weak 3′–5′ ExoN activity that is strongly enhanced by the addition of the nsp10 protein. The stimulation occurs through direct interaction between nsp10 and nsp14 without affecting the N7-MTase activity of nsp14 (9). Nsp10 mutants losing the capacity to interact with nsp14 do not activate ExoN activity any longer. Nsp10 is already known to act as an allosteric regulator of nsp16, a 2′-O-MTase involved in RNA capping (9). The role of nsp10 described here is especially remarkable because it depends on residues already identified as essential for nsp16 2′-O-MTase activation, indicating that the same nsp10 surface might be involved in regulating both nsp16 and nsp14. Precise mapping of the nsp10/nsp14 interface will tell the degree of surface overlap and thus might inform us of a possible mutually exclusive activation of nsp14 and nsp16. However, ribosomal frame-shifting generating pp1ab is expected to produce a ∼three- to sixfold excess of nsp10 over nsp14 or nsp16, indicating that both complexes might coexist in the infected cell.
Nsp14 has been proposed to be involved in nucleotide misincorporation repair notably because of its affiliation to the DEDDh nuclease superfamily, which also includes DNA proofreading exonucleases. This finding was recently strengthened by studies of nsp14 mutants devoid of ExoN activity in MHV and SARS-CoV, which were shown to exhibit increased mutation rate of viral progeny genomes in infected cell cultures (12, 13). Our work indicates that nsp14 can efficiently hydrolyze dsRNA as well as dsRNA with one 3′ mismatched nucleotide mimicking a polymerase misincorporation product. Therefore, our work provides a biochemical basis of the involvement of nsp14 ExoN in a still-putative proofreading pathway. Nucleotide misincorporation repair has not yet been evidenced, reported, or demonstrated in the RNA virus world even if a study suggested a “proofreading” mechanism associated with primer processing by the influenza virus RNA polymerase (24). It is commonly believed that RNA viruses, having small-sized genomes, produce an abundant viral progeny, including a large number of subtypes, mainly due to the intrinsically high error rate of viral RdRps. Proofreading has been reported for DNA and cellular RNA synthesis (25). In the case of DNA proofreading, nucleotide misincorporation generates a kinetic slow-down leading to hydrolysis of the nascent DNA 3′-end by an exonuclease recruited by or present on the DNA polymerase. Known DNA proofreading exonucleases belong to the DEDD superfamily (26). In the presence of dNTPs, DNA synthesis resumes once the kinetic block is relieved. Cellular RNA proofreading mechanisms differ by several points. The nuclease catalytic site and polymerization site reside in the same “tunable” active center as shown for RNA polymerase II and bacterial RNA polymerase (27, 28). Nucleotide misincorporation leads to a transcriptional slow-down (29) and allows backtracking and subsequent cleavage of the 3′ terminal dinucleotide, containing the mismatched nucleotide, by the polymerase intrinsic ribonuclease activity (30–32). Dinucleotide cleavage is template assisted and generates a 3′-OH end used by the polymerase to resume transcription.
In the case of the nsp14-mediated increase of base-pairing fidelity revealed by others for MHV and SARS-CoV (12, 13), the ExoN activity of nsp14 is clearly involved. Therefore, because nsp12 RdRp and nsp14 are distinct but interacting proteins, the mechanism used by coronaviruses to increase their replication fidelity may be similar to DNA proofreading activities observed in both eukaryotic and prokaryotic DNA synthesis machineries of cellular and viral origins. The implication of the nsp15 NendoU activity in an RNA misincorporation repair mechanism is yet unclear, but our data weakens the previous hypothesis of a direct cooperation between nsp14 and nsp15.
Viral RNA nucleases may potentially serve other roles apart from mismatch correction in the infected cell. Recent studies showed that the nucleoprotein (NP) of Lassa virus contains a domain structurally related to the DEDDh nuclease subfamily (33, 34). It is likely that this 3′–5′ ExoN domain is involved in the degradation of viral dsRNA replication intermediates, leading to type I IFN signaling inhibition (35). A similar mechanism of IFN production inhibition might be at work for nsp14 ExoN, because it was reported that MHV and SARS-CoV did not induce IFN production while pattern recognition receptors are functional in the infected cell (36, 37). The formal involvement of nsp14 ExoN in this process is indeed conceivable, and remains to be investigated.
In conclusion, we have demonstrated that SARS-CoV nsp14 is strongly (>35-fold) activated by nsp10 into a potent 3′–5′ dsRNA ExoN able to remove a 3′-terminal mismatch. The nsp10 protein, despite its small size and unknown enzymatic function, appears to be central in the coronavirus RTC through regulation of at least two RNA-processing enzymes. Because the 2′-O-MTase activity of nsp16, also regulated by nsp10, has recently been implicated in host cell immune evasion processes (38–40), the nsp10 protein might be a viral modulator of the coronavirus immune response suppression through its effect on nsp16 and perhaps nsp14, implying that nsp14 ExoN may well be involved in both mismatch repair and escape of host innate immunity. Because of its central role and atomic level structural definition, nsp10 is a promising target for the discovery of pleiotropic anticoronavirus drugs.
Materials and Methods
Reagents.
All radioactive reagents were purchased from Perkin-Elmer.
Cloning of the SARS-CoV nsp10 and nsp14 Genes.
Expression plasmids pDest14/(His)6-nsp10, pDest14/(His)6-nsp14, and pTXB1-nsp14 were obtained as described previously (9). Nsp10 and nsp14 genes were also cloned into a dual-promoter expression plasmid described previously (41). In this backbone, SARS-CoV nsp10 is expressed in fusion with an N-terminal Strep-TagII (named Strep-nsp10), whereas nsp14 is in fusion with a N-terminal hexahistidine tag (named nsp14HN).
Single-point mutant plasmids were generated by PCR using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions.
Expression and Purification of SARS-CoV nsp10 and nsp14 Proteins.
Expression and purification of nsp10 and nsp14 from pDest14/(His)6-nsp10, pDest14/(His)6-nsp14, and pTXB1-nsp14 vectors were performed as described previously (9). SARS-CoV nsp10/nsp14 coexpression was performed as described for nsp10/nsp16 in (41). SARS-CoV nsp15 protein expression and purification was done as described previously (22).
RNA Synthesis and Purification.
Synthetic RNAs used in this study are listed in Table S1, and their predicted structure is presented in Fig. S3. Synthetic RNAs were purchased from Jena Bioscience or Biomers (HPLC grade). H2-CUU(N)10, H2-CUOH, H5-GUU(N)10, and H5-GUOH RNAs were synthesized using MEGAshortscript kit (Ambion), subsequently dephosphorylated using shrimp alkaline phosphatase (Fermentas) according to the manufacturer’s instructions, and finally radiolabeled using polynucleotide kinase (PNK) and [γ32P]ATP. H4 RNA was also labeled with 32P-*pCp using T4 RNA ligase (New England Biolabs) according to the manufacturer’s instructions.
Exonuclease Assay.
Standard reactions contained 50 nM nsp14, 200 nM nsp10, and 750 nM radiolabeled RNA as substrate. Reactions were performed in 40 mM Tris (pH 8), 5 mM DTT, and 1 mM MgCl2. After incubation at 37 °C, reactions were stopped by the addition of an equal volume of loading buffer (formamide containing 10 mM EDTA). The digestion products were then analyzed in 7 M urea containing 20% (wt/vol) polyacrylamide gels (acrylamide/bisacrylamide ratio 19:1) buffered with 0.5× Tris-taurine-EDTA and visualized using a Fluorescent Image Analyzer FLA3000 (Fuji).
Endonuclease Assay Using SARS-CoV nsp15.
Digestion of 500 nmols of H2-CUU(N)10 and H5-GUU(N)10 RNAs were performed in a buffer containing 40 mM Tris (pH 7.5), 50 mM KCl, 1 mM DTT, 5 mM, and 1.5 μM of purified SARS-CoV nsp15.
Supplementary Material
Acknowledgments
We thank Barbara Selisko, Stéphane Priet, Hugues Tolou, and Bruno Coutard for helpful discussion, comments, and assistance. This work was supported by French National Research Agency ANR-08-MIEN-032, Fondation pour la Recherche Médicale (Programme Équipe FRM), European Union Seventh Framework Programme FP7/2007-2013 [Project SILVER (small inhibitor leads against emerging RNA viruses) Grant 260644], Infectiopole Sud, and Direction Générale pour l’Armement Contract 07co404. M.B. received a fellowship from the Direction Générale pour l’Armement.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201130109/-/DCSupplemental.
References
- 1.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoops K, et al. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Hemert MJ, et al. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder EJ, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minskaia E, et al. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov KA, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, et al. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7:e1002433. doi: 10.1371/journal.ppat.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci USA. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouvet M, et al. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6:e1000863. doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almazán F, et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen P, et al. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J Biochem Mol Biol. 2007;40:649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- 12.Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imbert I, et al. The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan J, et al. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph JS, et al. Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol. 2006;80:7894–7901. doi: 10.1128/JVI.00467-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su D, et al. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol. 2006;80:7902–7908. doi: 10.1128/JVI.00483-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lugari A, et al. Molecular mapping of the RNA Cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome coronavirus nsp10 and nsp16. J Biol Chem. 2010;285:33230–33241. doi: 10.1074/jbc.M110.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Decroly E, et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricagno S, et al. Crystallization and preliminary X-ray diffraction analysis of Nsp15 from SARS coronavirus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:409–411. doi: 10.1107/S1744309106009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj K, et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem. 2008;283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishihama A, Mizumoto K, Kawakami K, Kato A, Honda A. Proofreading function associated with the RNA-dependent RNA polymerase from influenza virus. J Biol Chem. 1986;261:10417–10421. [PubMed] [Google Scholar]
- 25.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr Opin Struct Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Shevelev IV, Hübscher U. The 3′ 5′ exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 27.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 28.Opalka N, et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 29.Alic N, et al. Selectivity and proofreading both contribute significantly to the fidelity of RNA polymerase III transcription. Proc Natl Acad Sci USA. 2007;104:10400–10405. doi: 10.1073/pnas.0704116104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 31.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 32.Cramer P. Molecular biology. Self-correcting messages. Science. 2006;313:447–448. doi: 10.1126/science.1131205. [DOI] [PubMed] [Google Scholar]
- 33.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468:779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez-Sobrido L, Giannakas P, Cubitt B, García-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology. 2007;361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou H, Perlman S. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol. 2007;81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Sastre A. 2 methylate or not 2 methylate: Viral evasion of the type I interferon response. Nat Immunol. 2011;12:114–115. doi: 10.1038/ni0211-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Debarnot C, et al. Crystallization and diffraction analysis of the SARS coronavirus nsp10-nsp16 complex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:404–408. doi: 10.1107/S1744309111002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.