Abstract
Computer-aided lead optimization derives a unique, orally bioavailable inhibitor of the signal transducer and activator of transcription (Stat)3 Src homology 2 domain. BP-1-102 binds Stat3 with an affinity (KD) of 504 nM, blocks Stat3–phospho-tyrosine (pTyr) peptide interactions and Stat3 activation at 4–6.8 μM, and selectively inhibits growth, survival, migration, and invasion of Stat3-dependent tumor cells. BP-1-102–mediated inhibition of aberrantly active Stat3 in tumor cells suppresses the expression of c-Myc, Cyclin D1, Bcl-xL, Survivin, VEGF, and Krüppel-like factor 8, which is identified as a Stat3 target gene that promotes Stat3-mediated breast tumor cell migration and invasion. Treatment of breast cancer cells with BP-1-102 further blocks Stat3–NF-κB cross-talk, the release of granulocyte colony-stimulating factor, soluble intercellular adhesion molecule 1, macrophage migration-inhibitory factor/glycosylation-inhibiting factor, interleukin 1 receptor antagonist, and serine protease inhibitor protein 1, and the phosphorylation of focal adhesion kinase and paxillin, while enhancing E-cadherin expression. Intravenous or oral gavage delivery of BP-1-102 furnishes micromolar or microgram levels in tumor tissues and inhibits growth of human breast and lung tumor xenografts.
Signal transducer and activator of transcription (STAT) proteins mediate responses to cytokines and growth factors (1). Recruitment via the Src homology 2 (SH2) domain to receptor phospho-tyrosine (pTyr) peptide motifs facilitates STAT phosphorylation on a key tyrosyl residue by growth factor receptors and the Janus kinase (Jak) and Src kinase families. Phosphorylation induces STAT–STAT dimerization through a reciprocal pTyr–SH2 domain interaction. The active dimers in the nucleus induce gene transcription by binding to specific DNA-response elements in the promoters of target genes.
The aberrant activation of Stat3 occurs in many human cancers (2) and promotes tumor progression. The mechanisms of Stat3-mediated tumorigenesis include dysregulation of gene expression that leads to uncontrolled growth and survival of tumor cells, enhanced tumor angiogenesis, and metastasis (3, 4). Tumor cell-associated constitutively active Stat3 also regulates proinflammatory cytokine expression, including RANTES (regulated upon activation normal T cell expressed and secreted) and CXC motif chemokine 10 (CXCL10)/IFN-γ–induced protein 10 (IP-10), and the induction of VEGF, interleukin 10 (IL-10), and other soluble factors, which in turn activate Stat3 in dendritic cells and inhibit their maturation. These events suppress tumor-immune surveillance (5). Reports also show Stat3–NF-κB cross-talk that supports the malignant phenotype (6, 7).
Given its importance to cancer, Stat3 is the focus of drug discovery efforts, and the SH2 domain–pTyr interaction has gained much attention (2, 8–14). Although several dimerization-disrupting small-molecule Stat3 inhibitors have been reported (8, 9, 13–16), thus far none has reached the clinic for several reasons, including the suitability of the scaffolds and pharmacokinetic issues. The leading dimerization-disrupting agent, S3I-201.1066 (17), was subjected to computer-aided lead optimization. We describe the derivation and characterization of the analog, BP-1-102, an orally bioavailable Stat3 SH2 domain ligand that inhibits Stat3 activation and functions in vitro and in vivo, and thereby inhibits growth of mouse xenografts of human breast and non–small-cell lung cancers.
Results
Computer-Aided Design of BP-1-102 as an Analog of S3I-201.1066.
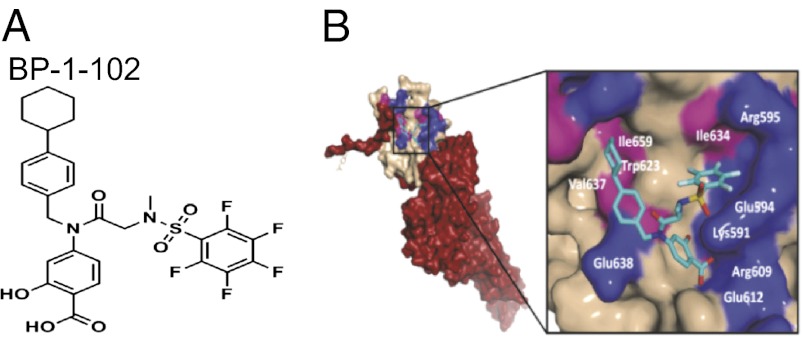
Analysis of the structural composition and topology of the Stat3 SH2 domain-binding “hotspot” shows three solvent-accessible subpockets on the protein surface, including the key pTyr705-binding region, which are accessed by S3I-201.1066 and most of the reported Stat3 inhibitors. BP-1-102 (Fig. 1A) has appendages that promote interactions with all three subpockets (Fig. 1B). Synthesis, chemical characterization, and detailed structural information and further discussion are presented in SI Results, SI Materials and Methods, and Scheme S1.
Fig. 1.
(A) Structure of BP-1-102. (B) Computational modeling of BP-1-102 binding to the Stat3 SH2 domain. (Left) Monomer Stat3 with the solvent-accessible surface of the SH2 domain (off-white) color-coded with hydrophilic (blue) and hydrophobic residues (pink) and overlaid with BP-1-102 (cyan). (Right) The three solvent-accessible subpockets of the SH2 domain surface accessed by BP-1-102, with the pentafluorobenzene sulfonamide component projecting into the third subpocket composed of Lys591, Glu594, Ile634, and Arg595.
Inhibition of Stat3 Signaling and Function.
BP-1-102 binds Stat3 with a KD of 504 nM, determined by surface plasmon resonance (SPR) analysis (Fig. S1A). It preferentially disrupts Stat3 binding to phosphorylated, native high-affinity, IL-6R/gp130 peptide (pTyr, pY904), with an IC50 of 4.1 μM (Fig. S1B), as determined by fluorescence polarization (FP) assay, compared with its weaker activity against Stat1 binding to the IFN-γ receptor peptide (GpYDKPHVL-NH2) or Stat5 binding to the erythropoietin receptor peptide (GpYLVLDKW-NH2), with an IC50 of 25–30 μM (Fig. S1C). Isothermal titration calorimetry (ITC) was further used to examine BP-1-102 effects on the interaction between pY904 and Stat3 (Fig. S1D). ITC experiments in the absence and presence of BP-1-102 were conducted using identical sample concentrations and conditions (SI Materials and Methods). Precomplexing Stat3 with BP-1-102 dramatically alters the pY904 profile, with significantly reduced enthalpic contribution by ∼6 kcal/mol (Fig. S1D, Left vs. Right, respective y-axis intercepts). This result indicates a direct interference of pY904 binding to the SH2 domain. Similarly, the reverse ITC titration of BP-1-102 into free or pY904-bound Stat3 (Fig. S1E, Left and Right, respectively) confirms that these two ligands interfere with each other’s binding at the SH2 domain. In fact, titration of BP-1-102 to pY904-bound Stat3 shows an increasing endothermic profile (heat absorption), consistent with BP-1-102 displacing pY904 from Stat3. Taken together, these results suggest direct competition of the ligands to the same binding site on the SH2 domain.
The R2 relaxation filter NMR approach (18) was used to study pY904 binding to Stat3 and verify that BP-1-102 competes with pY904. The pY904 signal (Fig. S1F, black tracing) was attenuated upon addition of Stat3 (Fig. S1F, red tracing), indicating binding. Addition of BP-1-102 resulted in further attenuation (Fig. S1F, blue tracing), suggesting direct interference with pY904 binding to the SH2 domain. The interference could be more significant if BP-1-102 did not form soluble aggregates in the aqueous environment used for NMR studies (Fig. S1G and SI Materials and Methods). This result further supports the findings by SPR, FP, and ITC. BP-1-102 inhibits Stat3 DNA-binding activity in vitro, with an IC50 value of 6.8 ± 0.8 μM (Fig. 2A), and preferentially inhibits Stat3–Stat3, over Stat1–Stat3, Stat1–Stat1, or Stat5–Stat5 DNA-binding activity (Fig. 2B), as measured by EMSA analysis (9, 11, 17). BP-1-102 is substantially improved over the lead, S3I-201.1066 (SPR, KD of 2.7 μM; FP, IC50 of 23 μM; EMSA, IC50 of 36 μM) (17).
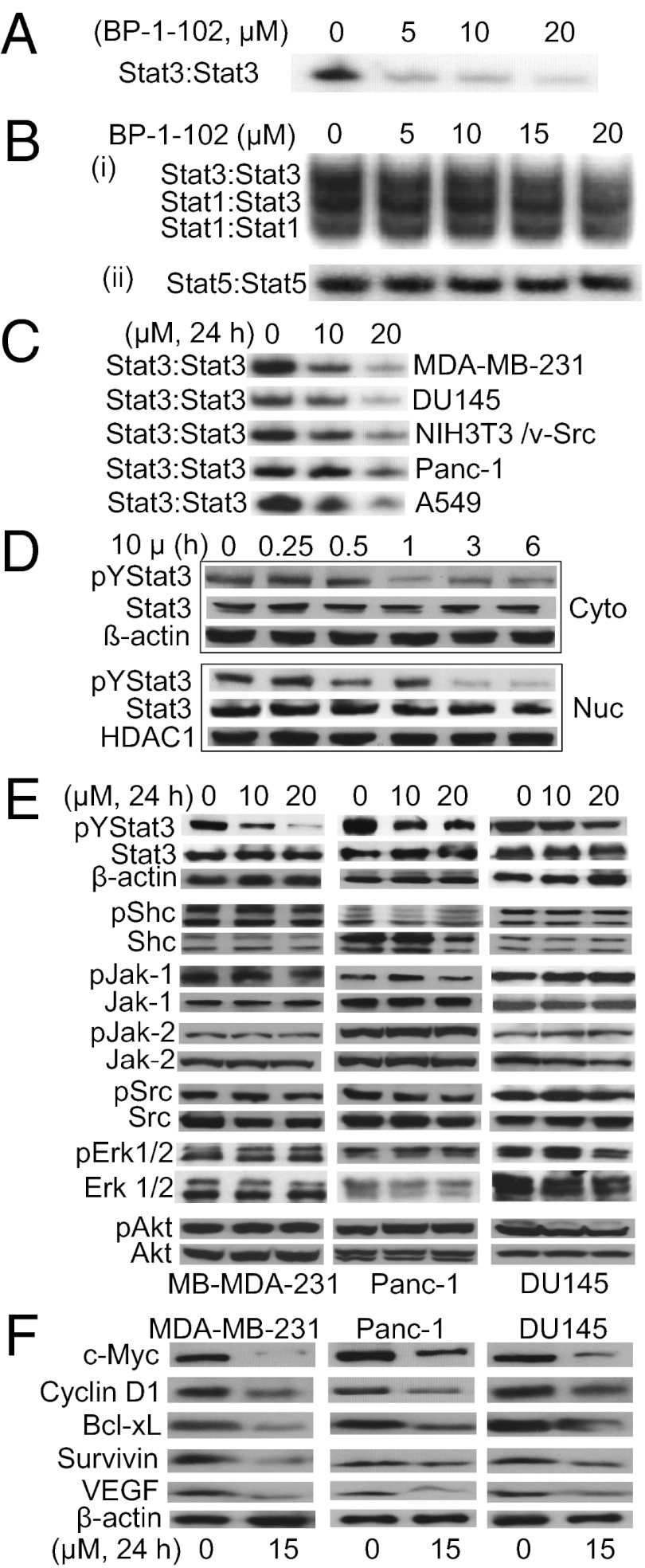
Fig. 2.
BP-1-102 inhibits Stat3 activation. (A and B) EMSA analysis of nuclear extracts of equal total protein containing activated STATs, preincubated with 0–20 μM BP-1-102 for 30 min before incubation with a radiolabeled (A and B, i) hSIE probe that binds Stat1 and Stat3 or (B, ii) mammary gland factor element probe that binds Stat5. (C) EMSA analysis using an hSIE probe for Stat3 DNA-binding activity in nuclear extracts of equal total protein prepared from the designated tumor cells treated with 0–20 μM BP-1-102. (D–F) Immunoblots of pY705Stat3, Stat3, histone deacetylase 1 (HDAC1), pY239/240Shc, Shc, pY1022/1023Jak1, Jak, pY1007/1008Jak2, Jak2, p416Src, Src, pT202/Y204Erk1/2, Erk1/2, pY473Akt, Akt, c-Myc, Cyclin D1, Bcl-xL, Survivin, VEGF, or β-actin in cytosolic (Cyto) or nuclear (Nuc) fractions or whole-cell lysates of equal total protein prepared from (D) MDA-MB-231 cells treated with 10 μM BP-1-102 for 0–6 h, or (E and F) the indicated cells treated with 0–20 μM BP-1-102 for 24 h. The positions of STAT–DNA complexes or proteins in the gels are labeled; control lanes (0) represent treatment with 0.05% DMSO. Data are representative of three or four independent determinations.
BP-1-102 inhibited constitutive Stat3 DNA-binding activity (Fig. 2C and Fig. S2A), Tyr705 phosphorylation (Fig. 2 D and E, and Fig. S2B, pYStat3), and Stat3-dependent luciferase reporter (pLucTKS3) (19, 20) induction (Fig. S2C, TKS3). The inhibition occurs in a dose- and time-dependent manner and as early as 30 min (Fig. 2 C and D and Fig. S2B). Stat3 is distributed in the cytoplasm, nucleus, and mitochondria (Fig. 2D and Fig. S2 D and E). Levels of pY705Stat3 are higher in the nucleus than in the cytoplasm, except in DU145 cells (Fig. S2 D and E), and are undetectable in mitochondria (Fig. S2E). BP-1-102 treatment attenuated both nuclear and cytoplasmic pY705Stat3 (Fig. 2D and Fig. S2E). By contrast, BP-1-102 had little or no effect on phospho-Shc, Src, Jak-1/2, Erk1/2, or Akt levels (Fig. 2E), induction of the Stat3-independent luciferase reporter, pLucSRE, which is driven by the serum-response element (SRE)/c-fos promoter (19, 20) (Fig. S2C, SRE), or the phosphorylation of many cellular kinases (Table S1). BP-1-102 treatment further suppressed c-Myc, Cyclin D1, Bcl-xL, Survivin, and VEGF expression (Fig. 2F), which occurred subsequent to Stat3 inhibition (Fig. 2D). Thus, inhibition of aberrantly active Stat3 suppresses Stat3-dependent gene regulation.
BP-1-102 Selectively Suppresses Growth, Survival, Malignant Transformation, Migration, and Invasion of Malignant Cells Harboring Constitutively Active Stat3.
Consistent with the dependency on abnormal Stat3 signaling of NIH 3T3/v-Src fibroblasts and malignant cells harboring aberrantly active Stat3 for the transformed phenotype (2, 4, 19, 20), BP-1-102 treatment suppressed cell proliferation, anchorage-dependent and -independent growth, and colony numbers of NIH 3T3/v-Src (v-Src), MDA-MB-231 (231), Panc-1, DU145, and A549 cells harboring aberrantly active Stat3 (Fig. 3A and Fig. S3 A and B). Overexpression of the artificially engineered, constitutively active Stat3C mutant (21) rendered MDA-MB-231 cells refractory to BP-1-102 (Fig. 3A, 231/St3C). Stat3C expressed in cells is insensitive to BP-1-102 (Fig. S2F). Moreover, BP-1-102 induced apoptosis in MDA-MB-231 cells (Fig. 3B, Lower, bars 3 and 4), which was attenuated (Fig. 3B, Lower, bars 6 and 7 vs. bars 3 and 4) by overexpressing Flag-tagged Stat3 SH2 domain (Fig. 3B, Upper, immunoblot), the target for BP-1-102. The data together show the specificity of BP-1-102 effects against Stat3-dependent tumor cells, which are dependent on disrupting Stat3 SH2 domain function. Treatment with BP-1-102 for 16 h, before observing the effect on proliferation (Fig. S3A, Insets), inhibited migration (Fig. 3C and Fig. S3C) and invasiveness (Fig. 3D and Fig. S3D) of Stat3-dependent tumor cells. By contrast, BP-1-102 had little or no similar antitumor cell effects on a range of nontarget cells, including normal NIH 3T3 (3T3), mouse thymus stromal epithelial cells, TE-71, Stat3-null mouse embryonic fibroblasts (−/−MEFs), NIH 3T3/v-Ras (v-Ras), or A2780S cells that do not harbor aberrantly active Stat3 (Fig. 3 A and C and Fig. S3 A–C). These effects are consistent with the down-regulation of known Stat3-inducible genes (Fig. 2F) (1, 3, 4).
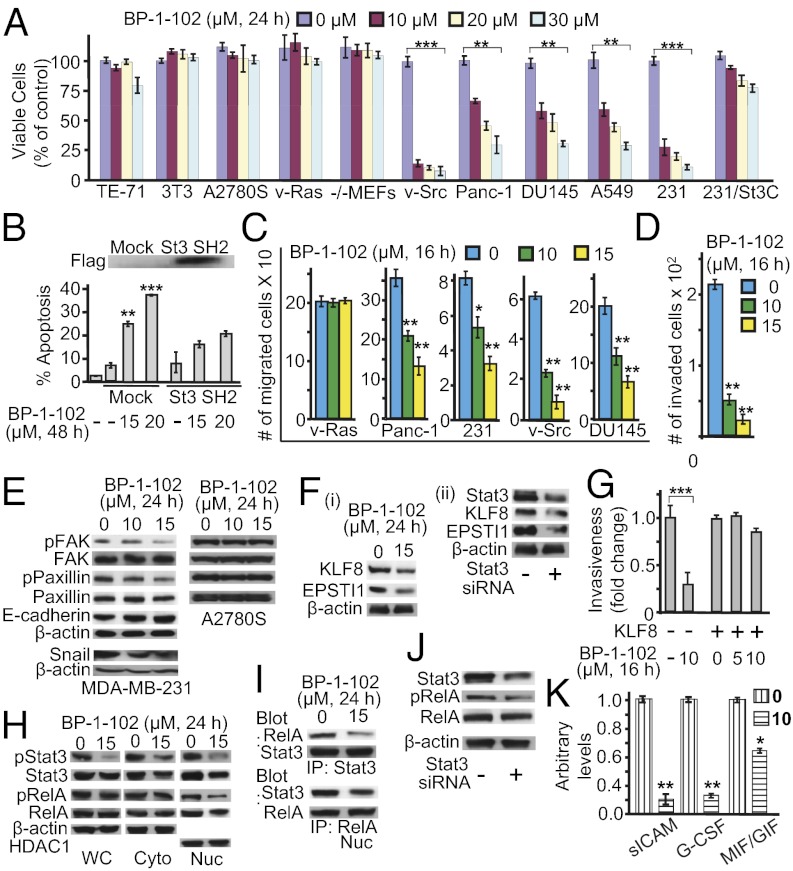
Fig. 3.
BP-1-102 induces antitumor cell effects in vitro and suppresses tumor-supporting factors. (A) Cultured MDA-MB-231, DU145, Panc-1, and NIH 3T3/v-Src cells harboring aberrantly active Stat3 and NIH 3T3, NIH 3T3/v-Ras, mouse thymus stromal epithelial cells, TE-71, Stat3-null mouse embryonic fibroblasts (Stat3−/− MEFs), cisplatin-sensitive ovarian cancer cells, A2780S cells that do not, were treated once with 0–30 μM BP-1-102 and subjected to CyQUANT cell proliferation assay. (B) Annexin V/flow cytometry analysis of MDA-MB-231 cells transfected with pcDNA-3 (mock) or Flag-tagged Stat3 (St3) SH2 domain and treated with 0–15 μM BP-1-102 (Lower); Flag immunoblot (Upper). (C) Cultured malignant cells were treated with BP-1-102, wounded, and allowed to migrate into a denuded area. (D) Number of invaded MDA-MB-231 cells in a BioCoat invasion chamber assay and the effects of BP-1-102. (E and F) Immunoblotting analysis of whole-cell lysates prepared from MDA-MB-231 cells (E and F, i) treated with 0–15 μM BP-1-102 or (F, ii) transfected with control (−) or Stat3 siRNA (+) and probing for FAK, pY576/577FAK, paxillin, pY118paxillin, E-cadherin, Snail, KLF8, EPSTI1, or β-actin. (G) Number of invaded MDA-MB-231 cells in a BioCoat invasion chamber assay and the impact of KLF8 overexpression on BP-1-102 effects. (H–J) Immunoblotting analysis of (H) whole-cell (WC), nuclear (Nuc), or cytosolic (Cyto) lysates, (I) immunecomplexes of Stat3 (Upper) or RelA (Lower) prepared from MDA-MB-231 cells treated with 0–15 μM BP-1-102, or (J) whole-cell lysates of MDA-MB-231 cells transfected with control (−) or Stat3 siRNA (+) and probing for pY705Stat3, Stat3, pS536RelA, RelA, β-actin, or HDAC1. (K) G-CSF, sICAM, and MIF/GIF levels assayed in conditioned medium from cultures of MDA-MB-231 cells treated for 48 h with BP-1-102. Positions of proteins in the gel are shown. Data are representative of three or four independent determinations. Values, mean ± SD, n = 4 or 9. *P < 0.05, **P < 0.01, ***P < 0.005.
BP-1-102 Modulates Factors That Regulate Cell Adhesion, Cell–Cell Interactions, Motility, Migration, and Invasion.
Except for Stat3 association with phospho-paxillin, focal adhesion kinase (FAK), and Src (22), little is known about the Stat3-dependent molecular events that promote tumor progression. BP-1-102–treated breast cancer MDA-MB-231 cells showed decreased phosphorylation of paxillin and FAK and increased E-cadherin expression (Fig. 3E). To exclude nonspecific effects, ovarian cancer A2780S cells that do not harbor aberrantly active Stat3 were treated with BP-1-102. No changes in p-FAK or p-paxillin levels were observed (Fig. 3E). The data that Stat3 inhibition occurs as early as 30 min (Fig. 2D), when FAK and paxillin are little affected (Fig. S4), suggest that the decreased p-FAK and p-paxillin levels at 24 h (Fig. 3E) are secondary events to Stat3 inhibition. BP-1-102 further suppressed Snail expression (Fig. 3E), a Stat3-regulated gene that controls E-cadherin expression.
FAK promotes Krüppel-like factor (KLF)8 induction (23). KLF8 and the tumor–stroma interaction factor, epithelial–stromal interaction 1 (EPSTI1) protein, promote tumor cell spread and invasiveness (24, 25). BP-1-102–treated breast cancer cells showed reduced KLF8 and EPSTI1 levels (Fig. 3F, i), which was validated by Stat3 knockdown by siRNA (Fig. 3F, ii). Furthermore, in normal NIH 3T3 fibroblasts, transiently cotransfected with the KLF8 promoter-driven luciferase reporter (pLucKLF8) and v-Src vector, the activation of Stat3 by v-Src (20) induced plucKLF8 expression by over twofold, which was repressed by BP-1-102 (Fig. S5A). The KLF8 promoter has three putative Stat3 binding sites (Fig. S5B, i). Site-specific mutation in the nucleotide sequence −253/−245 (site 1) severely impaired the mutant pLucKLF8/−253T/G reporter induction (Fig. S5A, compare bars 2 and 5). In vitro DNA-binding/EMSA analysis further showed a strong Stat3 binding to an oligonucleotide probe incorporating site 1 (−253/−245), compared with the standard high-affinity sis-inducible element (hSIE) probe, which was diminished by blocking anti-Stat3 antibody in a supershift assay (Fig. S5B, ii). No Stat3 binding was observed to site 2 or 3 (Fig. S5B, ii), although we do not exclude the possibility they contribute to Stat3 responsiveness. Thus, Stat3 directly induces the KLF8 gene.
To further study KLF8 importance, we evaluated BP-1-102 effects on cell motility and invasiveness in a KLF8 overexpression or knockdown background. By contrast to inhibition of invasiveness of the wild-type cells (Fig. 3 D and G, bar 2), KLF8 overexpression (24) in MDA-MB-231 cells abolished BP-1-102 effects (Fig. 3G, bars 3–5). Further, studies using tetracycline-inducible KLF8 shRNA (24) in MDA-MB-231-K8ikd cells show that KLF8 knockdown, as expected, suppressed cell migration (Fig. S5C, compare bar 2 vs. 5) and invasiveness (Fig. S5D, compare bar 2 vs. 5) and in turn minimized the BP-1-102–induced effect that is otherwise observed in the wild-type, uninduced cells (Fig. S5 C and D, compare the relative change between bars 2 and 3 to that of bars 5 and 6). Thus, KLF8 expression is one of the underlying mechanisms of Stat3-mediated tumor cell migration and invasiveness.
BP-1-102 Represses Stat3–NF-κB Cross-Talk and the Extracellular Production of Soluble Factors.
Stat3 cross-talks with factors such as NF-κB in the tumor microenvironment (6, 7) that redirect inflammation signal for oncogenic functions. BP-1-102–mediated attenuation of nuclear and cytoplasmic pY705Stat3 and of nuclear total Stat3 led to decreased nuclear pRelA and total RelA levels (Fig. 3H), whereas cytoplasmic RelA and pRelA levels appeared unchanged. To investigate the concurrent decline in nuclear RelA, we focused on the pStat3–pRelA complex (26), which is detected in the nucleus by coimmunoprecipitation analysis (Fig. 3I, lane 1) and as colocalization in immunofluorescence/confocal microscopy (Fig. S6A, control, merged). BP-1-102 treatment diminished the pStat3–pRelA interaction (Fig. 3I, lane 2 and Fig. S6A, compare 25 μM, 16 h to control), which was validated by Stat3 knockdown by siRNA (Fig. 3J). By contrast, BP-1-102 treatment had no effect on IκB–RelA interactions (Fig. S6B). Per the published report (26) that nuclear Stat3–NF-κB complex promotes nuclear NF-κB retention, BP-1-102–mediated inhibition of activated Stat3 diminishes nuclear Stat3 that in turn down-regulates nuclear pNF-κB.
To explore further the BP-1-102 effect on Stat3 cross-talks, we examined the production of soluble factors by tumor cells. Culture medium from BP-1-102–treated MDA-MB-231 cells showed lower granulocyte colony-stimulating factor (G-CSF), soluble intercellular adhesion molecule (sICAM) 1, and macrophage migration-inhibitory factor (MIF)/glycosylation-inhibiting factor (GIF) levels (Fig. 3K). Moreover, treatment of cells with exogenous G-CSF further induced Stat3 and RelA phosphorylation above constitutive levels (Fig. S6C, compare lanes 1 and 3) and consequently blocked the BP-1-102–repressive effect (Fig. S6C, compare lanes 2 and 4). Thus, BP-1-102 inhibits the production of soluble factors by tumor cells.
BP-1-102 Inhibits Growth of Human Breast and Non–Small-Cell Lung Tumor Xenografts and Modulates Stat3 Activity, Stat3 Target Genes, and Soluble Factors in Vivo.
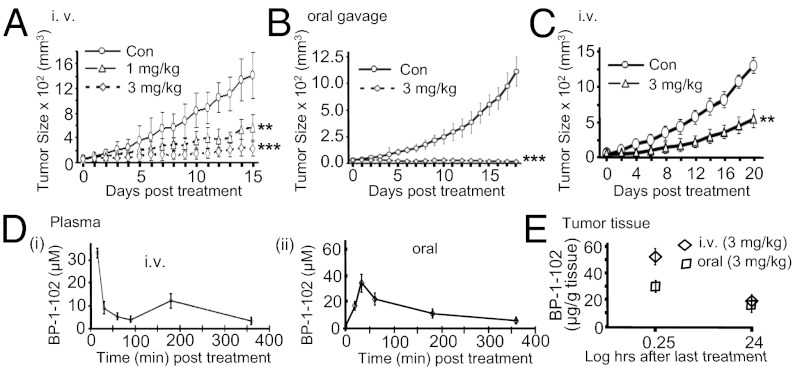
BP-1-102 inhibited growth of mouse xenografts of human breast (MDA-MB-231) and non–small-cell lung (A549) tumors that harbor aberrantly active Stat3 when administered via i.v. (tail vein injection, 1 or 3 mg/kg, every 2 or 3 d for 15 d) (Fig. 4 A and C) or oral gavage (3 mg/kg, 100 μL, every day) (Fig. 4B). No significant changes in body weights (Fig. S7 A and B) or obvious signs of toxicity, such as loss of appetite, decreased activity, or lethargy, were observed during the efficacy study or in a separate toxicity study where animals were dosed 1 or 3 mg/kg i.v. every 2 or 3 d for 21 d and monitored over 42 d, as shown by body weight and gross anatomical examination of organs (Fig. S7 C and D). The apparent stronger antitumor response to oral gavage is likely due to the daily dosing.
Fig. 4.
Human breast and non–small-cell lung tumor xenografts and the antitumor effects and in vivo pharmacokinetic properties of BP-1-102. (A–C) Mice bearing MDA-MB-231 (A and B) or A549 (C) tumors were administered BP-1-102 via i.v., 1 or 3 mg/kg or vehicle (0.05% DMSO in PBS) every 2 or 3 d or oral gavage, 3 mg/kg or vehicle (0.05% DMSO) every day. Tumor sizes, measured every 2 or 3 d, were converted to tumor volumes and plotted against days of treatment. (D and E) Graphical representations of BP-1-102 levels in (D) plasma samples collected from mice 15–360 min post-single dosing of 3 mg/kg via i.v. (i) or oral gavage (ii), or (E) tumor tissues extracted 15 min or 24 h after the last dosing with 3 mg/kg, i.v. or oral gavage. Values, mean ± SD, n = 7–10. **P < 0.01, and ***P < 0.005.
Analysis of tumor tissue lysates shows decreased Stat3 DNA-binding activity in treated tumors (T1 and T3) compared with nontreated control (Fig. S8A, Upper). Immunoblotting analysis of lysates from residual tumor tissues also showed suppression of pY705Stat3, c-Myc, Cyclin D1, Bcl-xL, Survivin, and VEGF expression that occurred in a dose-dependent manner (Fig. S8A, Lower), decreased pFAK, phospho-paxillin, KLF8, and EPSTI1 levels, enhanced E-cadherin expression (Fig. S8B), and diminished pRelA (Fig. S8C), compared with control tumors. Analysis of residual tumor tissue lysates (T1 and T3) also showed suppression of sICAM-1, MIF/GIF, serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 (Serpine 1), and interleukin 1 receptor antagonist (IL-1RA) production (Fig. S8D), whereas G-CSF was undetectable.
BP-1-102 Is Detectable at Micromolar Concentrations in Plasma and in Micrograms in Tumor Tissues.
In vivo pharmacokinetic profiling of plasma samples from a cohort of three mice collected at 15, 30, 60, 90, 180, and 360 min post-i.v. treatment (3 mg/kg) with a single dose showed BP-1-102 levels upward of 35 μM at 15 min postdosing, which declined by 30 min to a steady 5–10 μM level for up to 6 h (Fig. 4D, i), whereas plasma samples post-oral dosing at 3 mg/kg showed peak BP-1-102 levels of about 30 μM at 30 min, which steadily declined to 5–10 μM over a 6-h period (Fig. 4D, ii). Thus, blood levels over a prolonged period can exceed the IC50 values against Stat3. Moreover, BP-1-102 was detectable at 55 or 32 μg/g tumor tissue, respectively, for i.v. or oral delivery of 3 mg/kg, 15 min after the last dosing, and at 25 or 15 μg/g tumor tissue, respectively, for i.v. or oral delivery of 3 mg/kg, 24 h after the last dosing (Fig. 4E). Data together confirm that BP-1-102 is orally bioavailable and that the agent accumulates in tumor tissues at levels sufficient to inhibit aberrantly active Stat3 functions and inhibit tumor growth.
Discussion
BP-1-102 is designed as a Stat3–Stat3 dimerization disruptor with optimized structural features to enhance inhibitory activity. Stat3 binding is supported by SPR, FP, ITC, and NMR data that also show disruption of Stat3 SH2 domain–pTyr peptide interaction. Modeling predicts it binds to the three solvent-accessible subpockets of the Stat3–Stat3 dimer interface (27) (Fig. 1B), making hydrogen bonds in the third subpocket, and additional interactions with the charged Lys side chain via the unique pentafluorobenzene, which contribute to increased activity. Binding at the pTyr peptide binding site of the SH2 domain would disrupt preexisting Stat3–Stat3 dimers and block de novo Stat3 phosphorylation at the receptor and dimer formation. The reduced nuclear pYStat3 is the combination of diminished nuclear translocation, due to activation inhibition, and Stat3 nuclear exit upon dimer disruption (28).
The antitumor cell effects and the in vivo efficacy of BP-1-102 are consistent with thwarting Stat3’s key role in tumorigenesis, including dysregulation of gene expression leading to uncontrolled growth, survival, and angiogenesis (2, 4). The present study identifies KLKF8 (24) as a Stat3 target gene that promotes motility, migration, and invasion. Further, the induction of FAK and paxillin phosphorylation, EPSTI1 (25) expression, and the down-regulation of E-cadherin likely contribute to Stat3-mediated malignant progression (Fig. S9). Both KLF8 and EPSTI1 promote epithelial–mesenchymal transition and tumor invasiveness and are up-regulated in invasive and metastatic tumors (24, 25), as is aberrantly active Stat3. The induction of these events would repress epithelial cell assembly and cadherin-based cell–cell adhesions and promote dynamic regulation of cell–matrix adhesions that would drive tumor migration and invasiveness.
In the strong interplay of tumor cells with the microenvironment, Stat3 modulates inflammatory cytokines/chemokines that in turn suppress immune and inflammatory cells’ functions and tumor-immune surveillance (6). Of the factors regulated by Stat3 (5), IL-6, VEGF, and NF-κB also in turn promote Stat3 activation or are in cross-talks that perpetuate protumorigenic processes (6, 7, 26). The present study raises the possibility that sICAM (29), G-CSF, MIF/GIF, Serpine 1, and IL-1RA induction support Stat3-dependent tumor processes, including angiogenesis (30) (Fig. S9); MIF is overexpressed in breast cancer (31) and promotes disease progression (32), Serpine 1 expression correlated with advanced clear cell renal cell carcinoma and promoted tumor angiogenesis and aggressiveness (33). Further, IL-1RA blocked IL-1–induced antitumor cell effects in prostate cancer cells (34) and enhanced the proliferation (35) and growth of hepatic and glioblastoma cells (36), and sIL-1RA mRNA expression correlated with lymph node and hepatic metastases in gastric carcinoma patients (37). BP-1-102–mediated repression of these tumor-supporting factors likely contributes to its antitumor effects.
We present a unique Stat3 inhibitor, BP-1-102, that sufficiently accumulates in tumor tissues and induces antitumor responses in human tumor xenografts that harbor aberrantly active Stat3. The oral bioavailability of BP-1-102 represents a substantial advancement in the discovery of small-molecule Stat3 inhibitors as unique anticancer agents.
Materials and Methods
Cells and Reagents.
NIH 3T3, NIH 3T3/v-Src, and NIH 3T3/v-Ras cells, the human breast cancer line MDA-MB-231 and counterpart expressing inducible KLF8 shRNA (231-K8ikd), and human prostate (DU145), non–small-cell lung (A549), and pancreatic (Panc-1) cancer cells have all been previously reported (11, 17, 20, 24). Stat3-dependent (pLucTKS3), Stat3-independent (pLucSRE), and pLucKLF8 luciferase reporters, and the v-Src, β-galactosidase, Stat3 SH2 domain, Stat3C, and pLVUT-tTR-KRAB-KLF8 vectors have all been reported (9, 19–21, 23, 24). Other reagents are the Human Cytokine Array Kit (R&D Systems), G-CSF (Sigma-Aldrich) used at 100 ng/mL, and Stat1 and Stat5 proteins (SignalChem). Primary antibodies used were anti-Stat3, pStat3, pSrc, Src, pErk1/2, Erk1/2, pJak1, Jak1, Jak2, pJak2, pShc, Shc, Cyclin D1, c-Myc, Bcl-xL, Survivin, Akt, pAkt, pRelA, RelA, pFAK, FAK, phospho-paxillin, paxillin, E-cadherin, HDAC1, Snail, and β-actin (Cell Signaling Technology), VEGF (Santa Cruz Biotechnology), Flag and EPSTI1 (Sigma-Aldrich), and KLF8 (24). Cells were grown in DMEM containing 10% (vol/vol) heat-inactivated FBS.
Cloning and Protein Expression.
Molecular cloning, expression, and purification of His-tagged Stat3 have previously been reported (17). The cloning of the pXJ-FLAG-Stat3 SH2 domain and mutant KLF8 promoter is described in detail in SI Materials and Methods.
Nuclear Extract Preparation, Gel Shift Assay, and Densitometric Analysis.
Studies and analysis were carried out as previously described (9, 20, 28). Details are provided in SI Materials and Methods.
Immunoprecipitation and Western Blotting Analyses.
Studies were performed as previously described (9, 17, 28). Details are provided in SI Materials and Methods.
Cell Viability, Proliferation, Colony Survival, and Wound-Healing Assays.
Studies were performed as previously reported (17). Details are provided in SI Materials and Methods.
Transient Transfection of Cells, Luciferase, and Apoptosis Assays.
Studies were performed as previously reported (9, 20). After seeding for 12–24 h, cells were transfected with designated plasmid or siRNA, treated or not with BP-1-102, and processed for luciferase, immunoblotting, or Annexin V/flow cytometry analysis. Details of the Stat3 siRNA and studies are provided in SI Materials and Methods.
Immunostaining with Laser-Scanning Confocal Imaging.
Studies are described in detail in SI Materials and Methods.
Fluorescence Polarization Assay.
Assays were conducted as previously reported (17).
Surface Plasmon Resonance Analysis.
Studies were performed as previously reported (17).
Cell Migration/Invasion Assays.
Experiments were carried out and quantified as previously reported (9, 24, 28). Details are provided in SI Materials and Methods.
Cytokine Analysis.
The assay is described in detail in SI Materials and Methods.
Tumor Xenografts, Efficacy, and Pharmacokinetic Studies.
Studies were performed as previously reported (9, 17, 28). BP-1-102 concentrations in mouse plasma and tumor tissue lysates were assayed using a validated analytical procedure via HPLCy. Details are provided in SI Materials and Methods.
Statistical Analysis.
Statistical analysis was performed on mean values using Prism (GraphPad Software). The significance of differences between groups was determined by the paired t test at *P < 0.05, **P < 0.01, and ***P < 0.005.
Supplementary Material
Acknowledgments
We thank all colleagues and members of our laboratories for stimulating discussions, and the Sanford-Burnham Medical Research Institute Pharmacology Core for conducting the pharmacokinetic studies. This work was supported by National Cancer Institute Grants CA106439 (to J.T.), CA128865 (to J.T.), and CA132977 (to J.Z.); the University of Hawaii (J.T.); and the University of Toronto (P.T.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121606109/-/DCSupplemental.
References
- 1.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Yue P, Turkson J. Targeting STAT3 in cancer: How successful are we? Expert Opin Investig Drugs. 2009;18(1):45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 4.Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin Ther Targets. 2004;8:409–422. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10(1):48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21(1):11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiquee K, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunning PT, et al. Isoform selective inhibition of STAT1 or STAT3 homo-dimerization via peptidomimetic probes: Structural recognition of STAT SH2 domains. Bioorg Med Chem Lett. 2007;17:1875–1878. doi: 10.1016/j.bmcl.2007.01.077. [DOI] [PubMed] [Google Scholar]
- 11.Turkson J, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 12.Turkson J, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–269. [PubMed] [Google Scholar]
- 13.Mandal PK, et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J Med Chem. 2011;54:3549–3563. doi: 10.1021/jm2000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandal PK, Ren Z, Chen X, Xiong C, McMurray JS. Structure-affinity relationships of glutamine mimics incorporated into phosphopeptides targeted to the SH2 domain of signal transducer and activator of transcription 3. J Med Chem. 2009;52:6126–6141. doi: 10.1021/jm901105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, et al. Structure-based design of conformationally constrained, cell-permeable STAT3 inhibitors. ACS Med Chem Lett. 2010;1(2):85–89. doi: 10.1021/ml100010j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin L, et al. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res. 2010;70:2445–2454. doi: 10.1158/0008-5472.CAN-09-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, et al. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol. 2010;79:1398–1409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalvit C, et al. High-throughput NMR-based screening with competition binding experiments. J Am Chem Soc. 2002;124:7702–7709. doi: 10.1021/ja020174b. [DOI] [PubMed] [Google Scholar]
- 19.Turkson J, et al. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol. 1999;19:7519–7528. doi: 10.1128/mcb.19.11.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turkson J, et al. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 22.Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: Localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–3558. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Urvalek AM, Liu J, Zhao J. Activation of KLF8 transcription by focal adhesion kinase in human ovarian epithelial and cancer cells. J Biol Chem. 2008;283:13934–13942. doi: 10.1074/jbc.M709300200. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, et al. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30:1901–1911. doi: 10.1038/onc.2010.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen HL, Rønnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002;79:703–710. doi: 10.1006/geno.2002.6755. [DOI] [PubMed] [Google Scholar]
- 26.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fletcher S, Turkson J, Gunning PT. Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. ChemMedChem. 2008;3:1159–1168. doi: 10.1002/cmdc.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siddiquee KAZ, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol. 2007;2:787–798. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. Anti-inflammatory effects of 8-hydroxydeoxyguanosine in LPS-induced microglia activation: Suppression of STAT3-mediated intercellular adhesion molecule-1 expression. Exp Mol Med. 2006;38:417–427. doi: 10.1038/emm.2006.49. [DOI] [PubMed] [Google Scholar]
- 30.Gho YS, Kim PN, Li HC, Elkin M, Kleinman HK. Stimulation of tumor growth by human soluble intercellular adhesion molecule-1. Cancer Res. 2001;61:4253–4257. [PubMed] [Google Scholar]
- 31.Jesneck JL, et al. Do serum biomarkers really measure breast cancer? BMC Cancer. 2009;9:164. doi: 10.1186/1471-2407-9-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe R, Peng T, Sailors J, Bucala R, Metz CN. Regulation of the CTL response by macrophage migration inhibitory factor. J Immunol. 2001;166:747–753. doi: 10.4049/jimmunol.166.2.747. [DOI] [PubMed] [Google Scholar]
- 33.Zubac DP, Wentzel-Larsen T, Seidal T, Bostad L. Type 1 plasminogen activator inhibitor (PAI-1) in clear cell renal cell carcinoma (CCRCC) and its impact on angiogenesis, progression and patient survival after radical nephrectomy. BMC Urol. 2010;10:20. doi: 10.1186/1471-2490-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsieh TC, Chiao JW. Growth modulation of human prostatic cancer cells by interleukin-1 and interleukin-1 receptor antagonist. Cancer Lett. 1995;95(1-2):119–123. doi: 10.1016/0304-3835(95)03876-x. [DOI] [PubMed] [Google Scholar]
- 35.Yamada Y, Karasaki H, Matsushima K, Lee GH, Ogawa K. Expression of an IL-1 receptor antagonist during mouse hepatocarcinogenesis demonstrated by differential display analysis. Lab Invest. 1999;79:1059–1067. [PubMed] [Google Scholar]
- 36.Oelmann E, et al. Autocrine interleukin-1 receptor antagonist can support malignant growth of glioblastoma by blocking growth-inhibiting autocrine loop of interleukin-1. Int J Cancer. 1997;71:1066–1076. doi: 10.1002/(sici)1097-0215(19970611)71:6<1066::aid-ijc25>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 37.Iizuka N, et al. Interleukin-1 receptor antagonist mRNA expression and the progression of gastric carcinoma. Cancer Lett. 1999;142(2):179–184. doi: 10.1016/s0304-3835(99)00162-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.