Abstract
Nuclear export of mRNA is a key transport process in eukaryotic cells. To investigate it, we labeled native mRNP particles in living Chironomus tentans salivary gland cells with fluorescent hrp36, the hnRNP A1 homolog, and the nuclear envelope by fluorescent NTF2. Using light sheet microscopy, we traced single native mRNA particles across the nuclear envelope. The particles were observed to often probe nuclear pore complexes (NPC) at their nuclear face, and in only 25% of the cases yielded actual export. The complete export process took between 65 ms up to several seconds. A rate-limiting step was observed, which could be assigned to the nuclear basket of the pore and might correspond to a repositioning and unfolding of mRNPs before the actual translocation. Analysis of single fluorescent Dbp5 molecules, the RNA helicase essential for mRNA export, revealed that Dbp5 most often approached the cytoplasmic face of the NPC, and exhibited a binding duration of approximately 55 ms. Our results have allowed a refinement of the current models for mRNA export.
Keywords: Dbp5 ATPase cycle, nucleocytoplasmic transport, single molecule microscopy, translocation time
Eukaryotic cells store most of their genetic information on nuclear chromosomes and translate it into proteins in the cytoplasm. Thus, scores of RNA-encoded blueprints must be transported from the nucleus into cytoplasm through the supramolecular nuclear pore complexes (NPCs), which are embedded in the nuclear envelope (NE). Messenger RNA (mRNA) export is considerably more complex than protein transport through NPCs due to its tight connection to nuclear mRNA processing (1, 2). Nascent mRNA is cotranscriptionally loaded with mRNA-binding proteins, which are either transient components of the resulting ribonucleoprotein particle (mRNP) or, like the Chironomus tentans hrp36 (hnRNP A1 in mammals), escort the mRNA to its cytoplasmic destination (3). The key task in mRNA export is the remodeling of the mRNP by the ATP-dependent RNA helicase Dbp5. This process takes place at the cytoplasmic interface of the NPC, and presumably goes along with unloading of transport receptors from the mRNA. After remodeling, the mRNA cannot reenter the NPC and is released into the cytoplasm (2, 4). The most detailed analyses of mRNA export were accomplished by electron microscopy (EM) using C. tentans salivary gland cells (5, 6). By following the large 32–40 kb Balbiani ring 1 and 2 (BR) mRNAs, which encode salivary polypeptides, Daneholt and coworkers visualized native mRNPs during their NPC translocation and obtained a detailed view on this highly ordered process. The pivotal advantage of this system is the direct observation of a completely unmodified endogenous mRNA. However, EM provided only a static view to this highly dynamic cellular process. In vivo analysis of mRNA export requires fluorescence microscopy, which implies the generation of fluorescent mRNPs in situ. In recent studies on mRNA trafficking, this was accomplished by expressing modified mRNAs carrying multiple binding sites for loading with numerous copies of GFP-fused MS2, a bacteriophage coat protein (refs. 7–10) or for molecular beacons (11). To avoid introducing any artificial transcript or modifying an endogenous mRNA, we adopted the C. tentans system for light microscopy. We succeeded in visualizing single native mRNPs labeled by fluorescent hrp36. Light sheet fluorescence microscopy provided the required image contrast for tracing single mRNPs during nuclear export in the large nuclei. In addition, we resolved the reaction kinetics of single Dbp5 molecules at the NPC.
Results
Labeling Native mRNPs In Vivo.
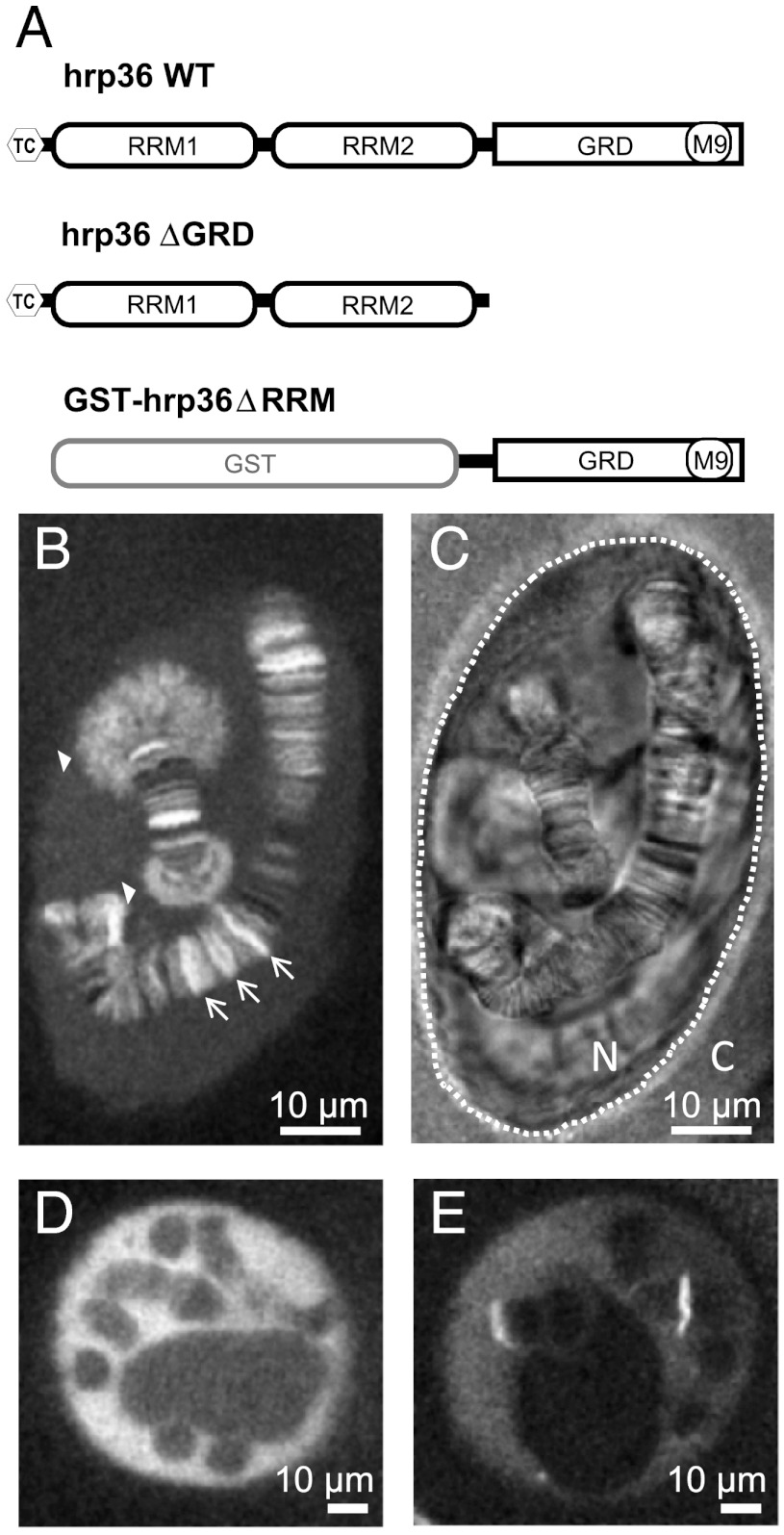
During transcription, the growing pre-mRNA associates with heterogeneous nuclear ribonucleoproteins (hnRNPs) to form mRNPs. It is known for the BR mRNPs that some hnRNPs leave the particle during intranuclear trafficking (6), while others escort the mRNA through the NPC. Hrp84 and hrp36 even remain bound to the mRNA when the polysomes start translation (3, 6). This stable association was also shown for the mammalian hrp36 homolog, hnRNP A1. Therefore, hrp36 constitutes a perfect probe for in vivo labeling of native endogenous mRNAs. C. tentans hrp36 (Mr ∼ 32,000) comprises two RNA recognition motifs (RRM) and a C-terminal glycine-rich domain (GRD; Fig. 1A). Previously, we demonstrated that microinjected fluorescently labeled recombinant hrp36 can be used to label and track single mRNPs in living salivary gland cell nuclei (12). After cytoplasmic microinjection the protein was rapidly translocated to the nucleus due to its C-terminal M9 transport signal, and became enriched at BR1 and 2 on chromosome IV (Fig. 1B, filled arrow heads) and other active transcription sites (arrows). To test whether the recombinant protein was stably incorporated in and not only transiently attached to mRNPs, we blocked transcription by either coinjection of actinomycin D or preincubation of the glands in hemolymph containing α-amanitin. Afterwards, the microinjected hrp36 was no longer seen at transcription sites (Fig. 1 D and E), and we concluded that the recombinant labeled protein, just like the endogenous hrp36, was cotranscriptionally integrated into mRNPs. This fact was further verified by microinjection of a C-terminal deletion mutant, hrp36ΔGRD, lacking the GRD. The GRD is not directly involved in mRNA binding, but its deletion abrogates the hrp36-mRNA interaction of the Drosophila homolog in vivo (13). As expected, hrp36ΔGRD did not label any transcription sites (SI Appendix, Fig. S1 A–C). Moreover, the transportin-dependent import of the protein was abolished because the M9 signal was also located in the GRD. Vice versa, a GST-GRD fusion protein without RRM domains was transported to the nucleus after cytoplasmic microinjection displaying the characteristic shuttling between nucleus and cytoplasm, but could not bind RNA any more (SI Appendix, Fig. S1 D–F).
Fig. 1.
hrp36-labeling of mRNPs. (A) Sketch of the used hrp36 expression constructs. Wild-type (WT) and hrp36ΔGRD were used as N-terminally tetra-cystein (tc) tagged proteins to improve fluorescence labelling. hrp36ΔRRM consists of the GRD and was microinjected as GST fusion protein to increase its molecular weight and prevent passive transport across the NE. (B) Fluorescence of WT hrp36-AlexaFluor647 in a cell nucleus 15 min after microinjection into the cytoplasm. (C) Bright field image of the image shown in (B). The polytene chromosomes show their characteristic banding pattern. Dotted line, NE; N, nucleus; C, cytoplasm. (D) hrp36 fluorescence in a nucleus after coinjection with actinomycin D imaged after 15 min incubation in hemolymph. (E) Hrp36 fluorescence in a nucleus preincubated for 1 h in hemolymph with α-amanitin to selectively block RNA polymerase II.
Characterization of Wild-Type and Mutant Dbp5 In Vivo.
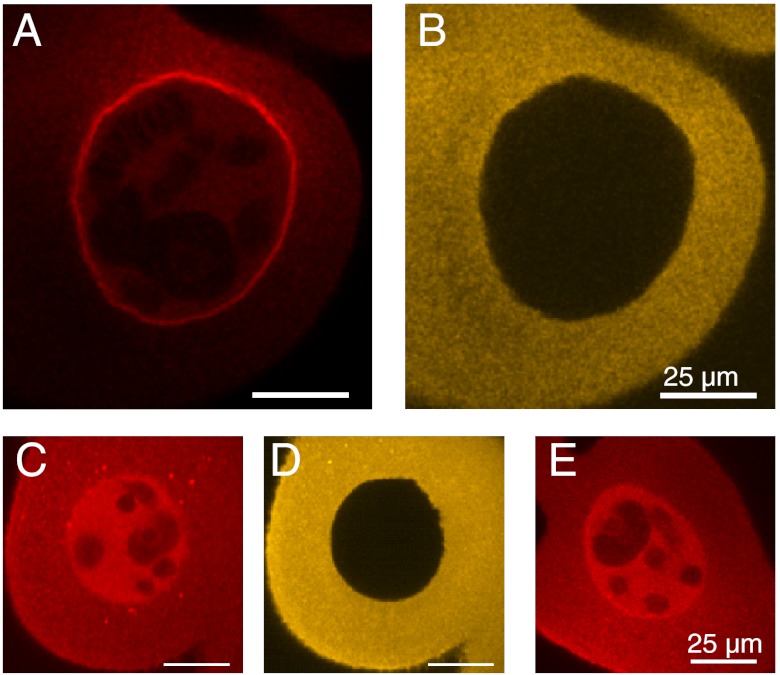
To resolve the binding kinetics of Dbp5 in comparison to mRNP export, we prepared fluorescence-labeled recombinant Dbp5. Microinjected Dbp5 was evenly distributed between cytoplasm and nucleus after microinjection, and strongly accumulated at the NE (Fig. 2 A and B). This characteristic localization was seen for endogenous Dbp5 in C. tentans (14), human (15), and yeast cells (16). Blocking transcription by actinomycin D in salivary gland cells abolished this localization, as was shown previously for endogenous Dbp5 (14) (Fig. 2 C and D). Finally, we analyzed the subcellular distribution of Dbp5 point mutants defective in NUP214 binding (R252A, Fig. 2E) or both NUP214 and mRNA interaction (D216R; SI Appendix, Figs. S2 and S3). We found that the interaction of wild-type C. tentans Dbp5 with NUP214 was sufficient to achieve the characteristic NPC localization. We concluded that the recombinant Dbp5 bound specifically and could functionally replace endogenous Dbp5 at the NPC.
Fig. 2.
Dbp5 within the cell. (A) Fluorescence of C.tentans Dbp5-AlexaFluor647 after microinjection into the cytoplasm of salivary gland cells imaged after 20 min incubation in hemolymph. (B) Fluorescence of coinjected BSA-AlexaFluor546. BSA cannot pass the NE, thus marking the injection site and demonstrating NE integrity. (C) Distribution of Dbp5 coinjected with actinomycin D, (D) distribution of coinjected BSA. Images shown in (C) and (D) were taken after approximately 20 min incubation of the gland in hemolymph. (E) Distribution of the Dbp5(R252A) point mutant after cytoplasmic injection. The highly conserved arginine at position 252 of C.tentans Dbp5 (corresponding to R259 in the human Dbp5 ortholog DDX19) mediates a critical salt bridge to NUP214. After replacing the arginine by alanine, the Dbp5-NPC interaction is almost completely lost. This point mutant showed the same subcellular distribution as WT Dbp5 after transcription blocking.
FISH Analysis of Transcription and mRNP Export.
C. tentans salivary gland cells represent an elegant but intricate biological system. In FISH experiments using various hybridization probes, we identified the conditions for successful mRNP export experiments before we began with the actual light microscopic analysis of mRNP export (see SI Appendix). Salivary gland dissection had to be performed in PBS. The FISH experiments revealed that transcription and export of BR1, BR2 or total poly(A) mRNA were only maintained if the glands were incubated and microscopically examined in hemolymph after dissection (SI Appendix, Fig. S4). In hemolymph transcription and export were functional for at least 60 min, but not in PBS. Thus, salivary glands were dissected and microinjected in PBS, but for microscopic transport measurements the sample chamber was filled with hemolymph to ensure that data were collected under physiological conditions.
Single Molecule Analysis of mRNP Export.
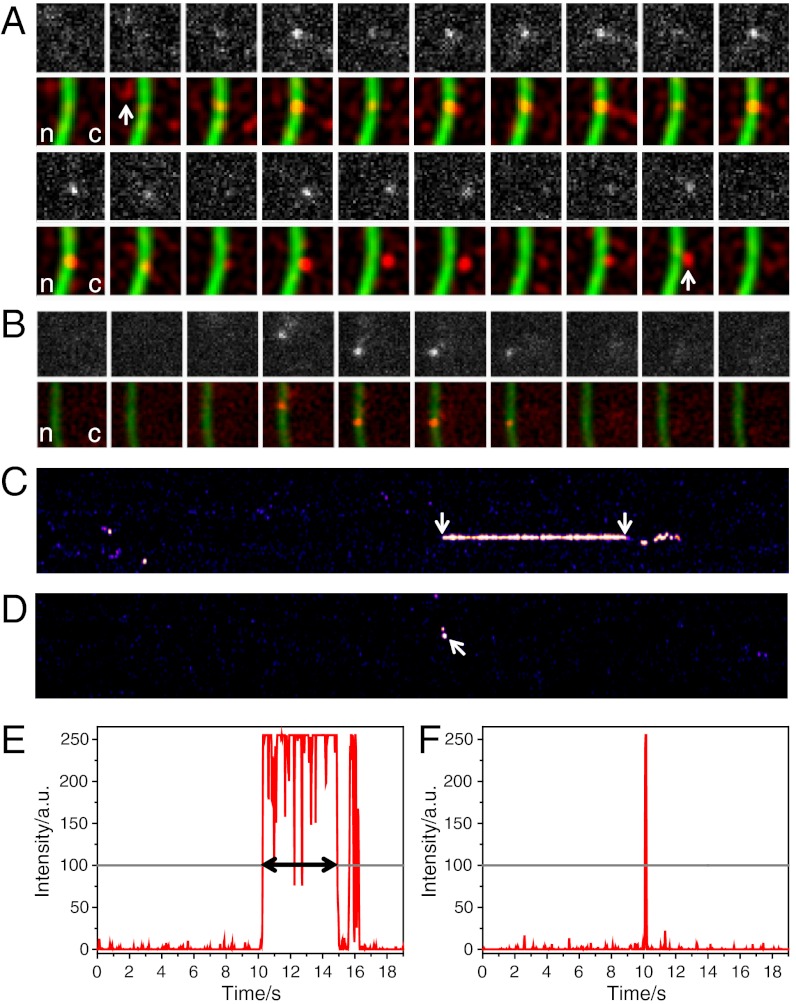
Fluorescence imaging of single molecules with high speed and the precise determination of single molecule positions is critically dependent on high signal-to-noise ratios. To achieve optimal signal-to-noise ratios, we used light-sheet fluorescence microscopy (17, 18). A thin light sheet was generated by a cylinder-lens optics, which illuminated the sample from the side perpendicular to the detection pathway of an inverted microscope (SI Appendix, Fig. S5). Thus, fluorophores near the focal region of the observation objective were selectively excited, which minimized fluorescence background, photobleaching, and phototoxicity (19). Very low amounts of AlexaFluor647-labeled hrp36 molecules were injected into the cytoplasm, and after nuclear import and their integration into mRNPs we could follow single hrp36-AlexaFluor647 labeled mRNPs in the gland cell nuclei about 120 μm deep in the sample. The concentration of injected hrp36 was so low that single mRNPs contained maximally a single labeled protein. We achieved high signal-to-noise ratios that yielded a localization precision of up to 10 nm for molecules bound to the NE (calculated according to ref. 20). In order to visualize the NE as the locus of mRNP export, we coinjected Alexa546-labeled NTF2 molecules. NTF2 is the transport receptor for RanGDP import and interacts with FG repeats of nucleoporins with high selectivity (21). NTF2- and hrp36-Alexa647 signals were sequentially acquired using exactly the same imaging path to minimize alignment problems in the dual color measurements. In movies that were acquired with an image integration time of 20 ms at 50 Hz frame rate, single hrp36-labeled mRNPs could well be discerned in the salivary gland cell nucleoplasm, cytoplasm, and near or at the NE. Unbound hrp36 molecules could not be resolved with this time resolution because they diffused too fast. Numerous complete mRNP export events were observed in which hrp36-labeled mRNPs approached the NE from the nucleoplasm, bound to it, and left it on the cytoplasmic face (e.g., Fig. 3A and Movie S1). In experiments aimed at the analysis of the interaction of single Dbp5 molecules with the NE, we proceeded in a similar manner using Alexa647-labeled Dbp5 molecules instead of hrp36. It was immediately obvious that Dbp5 showed exclusively short interactions with the NE (Fig. 3B and Movie S2).
Fig. 3.
Binding time analysis of mRNPs and Dbp5 at the NE. (A) Time series showing a single hrp36-labeled mRNP during its export from nucleus (n) to cytoplasm (c). The 1st and 3rd rows show raw images of hrp36 fluorescence; the 2nd and 4th rows show filtered data (red) and the NTF2-stained NPCs (green). Because the mRNP was observed at the NE for 230 frames (4.6 s), only the first and last 10 frames were shown. The nuclear approach to and cytoplasmic departure from the NE were marked. Single image size, 4.5 μm2. (B) A single Dbp5 molecule binds to the NE. The upper row shows raw data, and the lower row the corresponding filtered frames (red) and the NTF2-stained NPCs (green). The Dbp5 molecule hit the NE at two different spots dwelling for 1 and 3 frames, respectively. Image size 5.25 μm2. (C) Kymograph of the mRNP export shown in (A). Hrp36-fluorescence along the NE was plotted vs. time (total 19 s). When the mRNP left the NPC at the cytoplasmic face the signal showed a characteristic wobbling (see Movie S1). Only the membrane-bound phase (arrows) was considered for dwell time determination. (D) Kymograph of the membrane binding of Dbp5 from (B) (arrow). Fluorescence intensity along one row of the respective kymographs: (E) mRNP export shown in (A), (C), and (F). Dbp5 binding shown in (B) and (D). Fluorescence intensity was plotted in red, and the threshold in gray. The dwell time was determined as the time until the signal intensity fell below the threshold for three subsequent frames.
Kymograph-Based Dwell Time Analysis.
To efficiently screen the data for export events and to quantify the duration of mRNA export and Dbp5-NPC interactions, we evaluated the movies by kymographs which displayed the Alexa647-fluorescence signal of the linearized curve along the NE as a function of time in x-direction. The fluorescence along the NE was plotted in y-direction (21). Example kymographs are shown for hrp36-labeled mRNPs (Fig. 3C) and for wild-type Dbp5 (Fig. 3D). The kymographs revealed the recording of an interaction with the NE in a whole movie at a single glance. Also, the duration of such interactions could directly be determined in intensity plots of respective image lines (Fig. 3 E and F). We recorded several thousand movies, and their evaluation in terms of kymographs revealed numerous observations of hrp36 and Dbp5 interactions with the NE.
Analysis of hrp36 and Dbp5 Interaction with the NE.
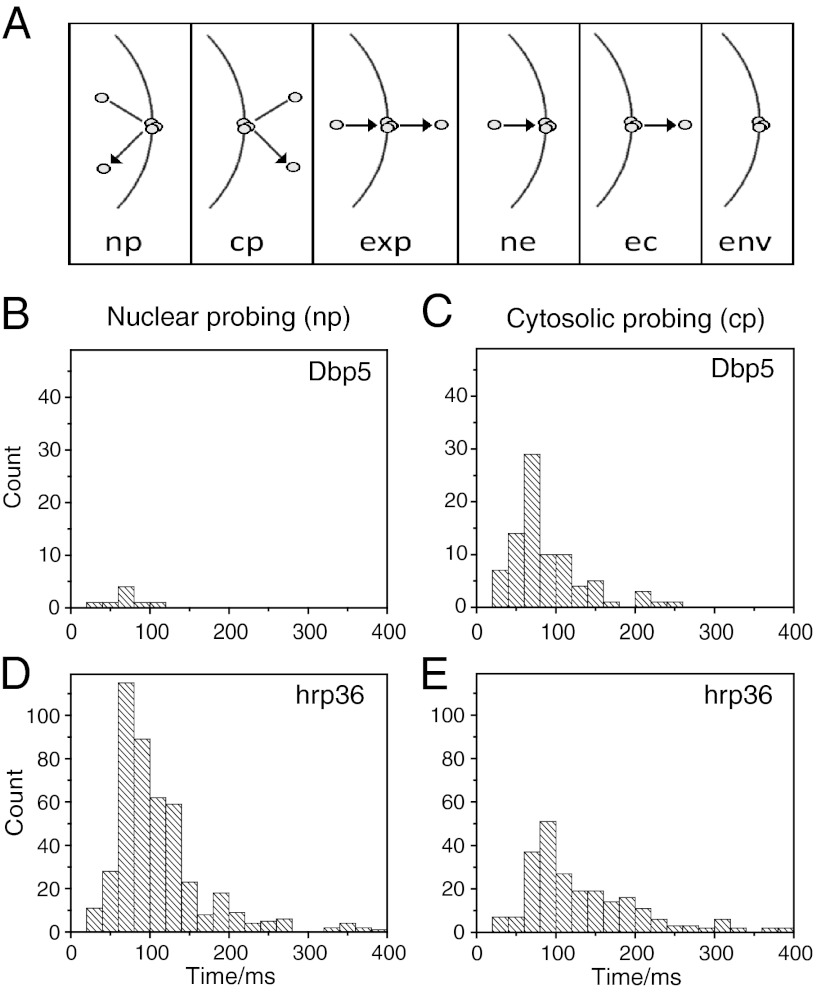
For all events detected in the kymographs, we carefully inspected the corresponding raw images. In these we determined the respective time-resolved single molecule movement in relation to the NE position that showed the approach, binding, and release of hrp36-labeled mRNPs and Dbp5 molecules to and from the NE. All observations were sorted into categories by discriminating among all possible interaction types for particles moving near the NE (Fig. 4A). Exemplary trajectories for all these types from the mRNP dataset are shown in SI Appendix, Fig. S6. We restricted the further analysis to the categories that comprised explicit NE approach, binding, and release. Trajectories of molecules approaching from nucleoplasm and hitting the NE but returning to the nucleus were designated as nuclear probing (np), and trajectories beginning and ending in the cytoplasm correspondingly as cytoplasmic probing (cp). The duration of the immediate membrane interaction was determined for all events in a category and plotted in histogram form. This procedure revealed that for Dbp5 cytoplasmic probing was prevailing (Fig. 4 B and C), whereas for hrp36-labeled mRNPs nuclear probing was observed most frequently (Nnp = 313 and Ncp = 184) (Fig. 4 D and E).
Fig. 4.
mRNP and Dbp5 binding time distributions. (A) Classification of NE binding events (np: nuclear probing; cp: cytoplasmic probing; exp: export; ne: nucleus to envelope; ec: envelope to cytoplasm; env: only envelope). Frequency histograms of (B) Dbp5 nuclear probing, (C) Dbp5 cytoplasmic probing, (D) nuclear probing of hrp36-labeled mRNPs, and (E) cytoplasmic probing of hrp36-labeled mRNPs.
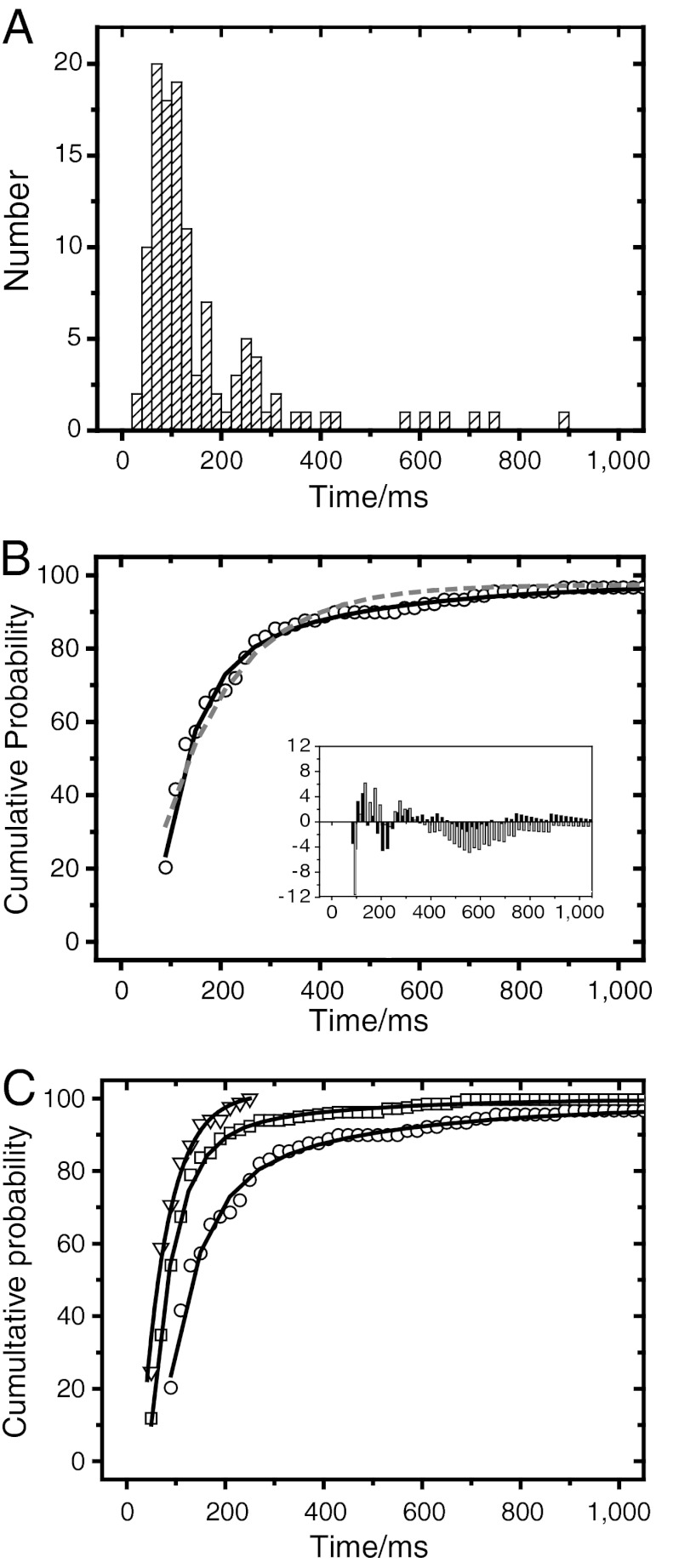
Obviously, only mRNP trajectories starting in nucleoplasm, hitting the NE, and ending in cytoplasm were unambiguous nuclear export processes (exp). The interaction times were compiled in a histogram (Fig. 5A). In comparison to nuclear probing, the capture of complete export events succeeded less often. Nevertheless, the kymograph analysis made it possible to extract quite a number of export events (Nexp = 121). In order to determine the mean duration of mRNP translocation across the NPC, and also the mean interaction times of cytoplasmic and nuclear probing by mRNPs and Dbp5, respectively, we translated the above histograms into normed cumulative probability distributions. These distributions allowed us to directly quantify the interaction time constants of the examined molecules with the NE (for a similar analysis, see ref. 9) by fitting exponential decay functions to the data. The resulting cumulative probability distribution for mRNP export events required a double-exponential decay function to yield a satisfactory description of the data (Fig. 5B). The fit yielded two time constants, τ1 = 65 ± 5 ms (87%) and τ2 = 350 ± 25 ms (13%). The occurrence of more than a single time constant was expected, because we observed export of the complete mRNA pool comprising genes of differing size. Time constants τ1 and τ2 presumably corresponded to the export of mRNA with a size of up to a few kilobases. However, some export events lasted conspicuously long. We observed eight bona fide export events with durations longer than 0.5 s, up to 6 s. We assume that these rare long events referred to the export of extremely large mRNA like the BR transcripts (5, 22). To support this assumption, we estimated the expected export duration for BR mRNA, and our chances to observe such an export (see SI Appendix). We found that we could expect to observe the export of a fluorescently labeled BR mRNP only rarely. Considering the transcription rate of BR mRNPs and the number of simultaneous export processes that can be deduced from electron microscopic data, we estimated the mean export event duration of BR mRNPs to be about 20 s. Hence, the long observed export events could well correspond to the observation of BR mRNPs.
Fig. 5.
mRNP and Dbp5 kinetics binding to the NE. (A) Analysis of mRNP export kinetics. Membrane events that represented export events were collected and their duration plotted as histogram. (B) The normed cumulative counts of the histogram in (A) were determined and fitted by a mono- (dashed gray curve) and biexponential (solid black curve) function. The residuum plot (insert) demonstrated that the export data were better described by a biexponential (black columns) than a monoexponential function (gray columns). (C) Comparison of the kinetics of cytoplasmic probing of Dbp5 (triangles), nuclear probing (squares) and export (circles) of hrp36-labeled mRNPs. Solid curves indicate the mono- (nuclear/cytoplasmic probing) and biexponential (for export only) fits.
Like the complete export, the duration of nuclear probing of mRNPs required a biexponential kinetics for a satisfactory fit, and yielded NE interaction times of τ1 = 50 ± 6 ms (94%) and τ2 = 275 ± 15 ms (6%) (Fig. 5C). We suspect that the short time corresponded to mRNP collisions with the NE, whereas the long one corresponded to interrupted or unsuccessful exports, when the particles entered the nuclear basket but did not proceed to the cytoplasm. In contrast, hrp36-labeled particles that approached the NE from the cytoplasm resided at the NE with a single binding time of τ = 95 ± 6 ms. This longer duration compared to the short nuclear probing time probably just reflected the lower mobility of mRNPs in cytoplasm compared to nucleoplasm. Cytoplasmic NE probing of Dbp5 had a time constant of τ = 54 ± 5 ms (Fig. 5C). Due to the very low number of events, we did not further analyze nuclear probing of Dbp5. All time constants are summarized in Table 1.
Table 1.
Summary of mRNP and Dbp5 Kinetics
| Probe | Mode | N | τ1 (ms) | τ2 (ms) |
| Dbp5 | Cytoplasmic probing (cp) | 83 | 54 ± 5 | - |
| hrp36 | Nuclear probing (np) | 313 | 50 ± 6 | 275 ± 15 |
| hrp36 | Complete export (exp) | 121 | 65 ± 5 | 350 ± 25 |
| hrp36 | Cytoplasmic probing (cp) | 184 | 95 ± 6 | - |
Trajectories of Single mRNPs within the NPC.
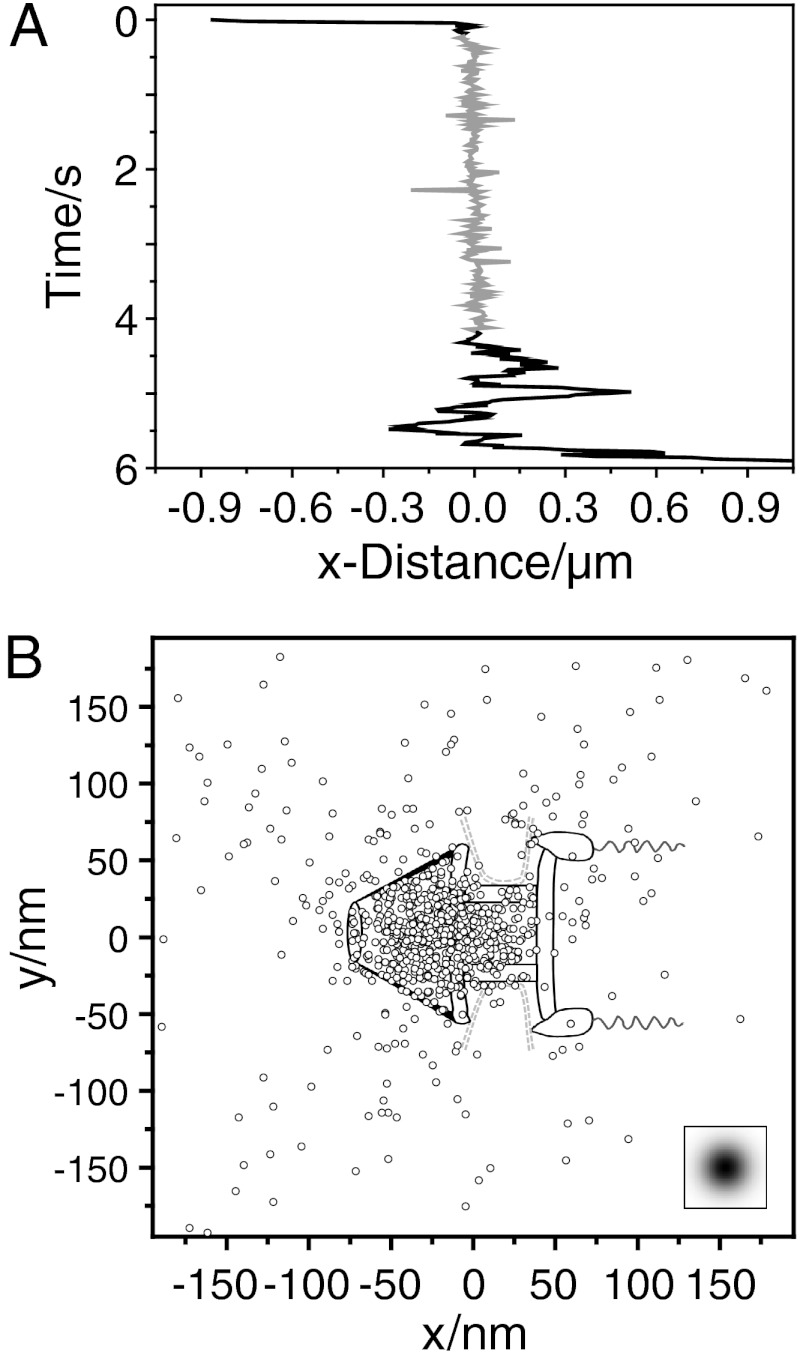
Finally, we focused on the actual transit of single mRNA molecules through the NPC. For this purpose, we traced single mRNPs labeled by hrp36 with nanometer precision during their pore transit (see Materials and Methods). Remarkably, such trajectories did not show reproducible signs of a progressive movement through the NPC (Fig. 6A). However, in long trajectories a pause in the movement was seen. Obviously the respective particles halted for a certain time period. In the shown example, the stop lasted for 4 s. We used a trajectory analysis introduced by Lowe et al. (23), who examined the nuclear import of quantum dots. Seventeen trajectories with binding durations between 0.3 s and 5.6 s were rotated onto a common axis corresponding to the export direction to account for the various positions of the events on the curved NE. Then we searched for the best overlap of these trajectories with a virtual NPC scheme that was designed according to the NPC geometry of salivary gland cell nuclei of C. tentans (24) (Fig. 6B). The excellent correspondence of the coordinate distribution and the NPC dimensions suggested that the mRNA lingered predominantly in the nuclear basket and the nuclear entrance of the NPC, probably before the actual translocation took place.
Fig. 6.
Binding site distribution at the NPC of mRNPs during export. (A) X-coordinates of a long mRNP trajectory crossing the NE. The trajectory contained a characteristic section where the movement paused (shown in gray). (B) Superposition of all export trajectories with durations > 300 ms. The trajectories were rotated onto each other, and aligned for optimal overlay with the NPC structure. The sketch shows the single positions superimposed onto an NPC scheme drawn to scale. The insert indicates the experimental single molecule localization precision.
Discussion
The first observation of a single mRNA traversing the nuclear pore succeeded 20 y ago by electron microscopy of BR mRNPs in salivary glands of C. tentans (25). The BR transcripts are embedded in doughnut-shaped granules of 50 nm diameter, which dock at the nuclear basket, rearrange therein, and enable the mRNA to enter the pore with the 5′ end first. Recently several researchers used the MS2 system to produce GFP-labeled mRNPs to study mRNA export. In order to create particles sufficiently bright for single molecule detection, multiple stem loops were introduced (7–9), which bound up to 48 MS2 coat proteins (MCPs) fused to GFP. The total molecular mass of the MCP/RNA tag was roughly 3,000,000 (9). Considering this molecular mass as a compact protein the MCP load alone corresponded to a particle of 16 nm diameter. Native mRNPs on the average have elongated shapes of 5–10 nm thickness and 15–30 nm length (26). Thus, the MS2-derived mRNA tags were as large as entire untagged mRNPs (11). Mor et al. (8) introduced different gene constructs based on human dystrophin cDNA, followed the MCP-GFP tagged mRNAs, and estimated the maximum export duration for a 10 kb mRNA construct with 0.5 s, but could not resolve the actual translocation process with their frame rate of 1 Hz. Grünwald and Singer (9) employed a MCP-tagged β-actin mRNA and sampled its interaction with the NE at 20 ms temporal resolution yielding a total translocation time of their mRNA construct of 180 ms. However, these authors excluded peculiarly slow events from the analysis, and their mRNA probe was preferentially seen at both the nuclear and cytoplasmic faces of the NPC.
To avoid any significant mRNP modification we established a labeling approach based on the endogenous mRNA binding protein hrp36, which is an integral part of C. tentans mRNPs. After microinjection, the fluorescence labeled protein was stably incorporated into mRNPs during transcription and allowed visualization of single native mRNPs in the salivary gland cell nuclei during nuclear export. The mRNPs were often perceived interacting with nuclear interface of the NE, but then returning to the nucleoplasm. We observed transient and short-lived (τ1 ∼ 50 ms) and long (τ2 ∼ 275 ms) interactions. Probably the shorter ones can be related to collisions with the NE without an entry into the nuclear basket. The longer ones could reflect unsuccessful docking, where mRNPs dissociate from the NPC before a suitable final position or conformation for translocation could be achieved.
About 25% of the encounters with the NE resulted in a successful mRNP export. These actual export events showed widely differing durations spanning two orders of magnitude in time. We derived two time constants (65 ms and 350 ms) describing the dissociation kinetics, but detected also events lasting up to almost 6 s. This time was not limited by bleaching of the fluorescent probe, because the actual approach to the NPC, binding, and dissociation into the cytoplasm were observed. Considering the fact that we labeled the complete mRNP pool comprising small and large particles, we suspect that a continuous distribution of binding times ranging from about 50 ms up to several seconds would appropriately describe the data, but it is not possible to prove the existence of such a distribution from real data.
Several times we observed mRNPs, possibly large BR transcripts, that arrested for several seconds at the NPC. The spatial region of this arrest was very well defined, and corresponded to a region with an extension of ± 25 nm only. While the colocalization precision was not good enough to directly relate this region to a specific NPC domain, we speculate that it might correspond to the nuclear basket, and that the pause might reflect the rearrangment and unpacking of the RNP fibril that had previously been made out by electron microscopy (22, 25). A best-fitting overlay of especially long export trajectories and the NPC structure of C. tentans supported this notion (Fig. 6B). Interestingly, the overlay indicated an accumulation of positions in the basket region and adjacent entry of the central framework of the NPC. Thus, the rate-limiting step of the translocation process is probably located in that domain and not at the cytoplasmic NPC face, where the mRNA is remodeled by the RNA helicase Dbp5 (4, 27, 28). An important cofactor of Dbp5 is Gle1, which is also tethered to the NPC via a specific NUP interaction. Gle1 binds Dbp5 together with soluble Inositol-6-phosphate, IP6, which might trigger ATP hydrolysis (4, 29). It was postulated that this reaction is the rate-limiting step in the Dbp5 ATPase cycle (29). Recently Hodge et al. (16) analyzed the interaction of Dbp5-GFP with the NE of yeast cells by photobleaching and determined a recovery time of < 1 s, suggesting a rapid exchange kinetics of the molecule at the NPC. These authors proposed a model wherein single Dbp5 go through multiple ATP-dependent remodeling cycles before being replaced. We found that single Dbp5 approached the NPC predominantly from the cytoplasm, and dissociated into this compartment after a transient binding with a mean duration of 55 ms. This duration was significantly longer than the binding times of protein transport receptors (10), and it is tempting to speculate that the 55 ms correspond to a full mechanochemical cycle of Dbp5 that was characterized by Montpetit, et al. (29). Future experiments using respective Dbp5 mutants are required to clarify this issue.
Our results suggest different scenarios for mRNA export. The mRNP trajectory data suggest that there is a rate-limiting step at the nuclear basket, which could refer to finding a suitable mRNP configuration for entering the pore interior. The duration of this step could vary largely for mRNPs of different sizes. The actual translocation could be achieved by the action of Dbp5. An alternative hypothesis is suggested by comparison of the Dbp5 binding time (55 ms) with the export duration of the larger mRNPs (350 ms up to several seconds). Several Dbp5 molecules might act in sequence on a single mRNP to achieve the required export factor release and mRNP translocation. In this case the observed translocation pause is just valid for the specific incorporation site of the labeling hrp36 that remains in the basket until it is finally moving through the pore. Furthermore, several Dpb5 molecules could act at different locations on a single mRNP simultaneously. Thus, we speculate that the action of several dozens of Dbp5 molecules might be required to achieve translocation. This hypothesis is compatible with the characteristic and strong labeling of the NE by fluorescent Dbp5, indicating the presence of a large number of Dbp5 molecules at the NE.
In summary, we made the well known C. tentans system usable for light microscopic study of mRNA export at the single molecule level. Notably, this system allows the analysis of nuclear export of fully native mRNA. Additional single molecule experiments using Förster resonance energy transfer between the translocating mRNA and Dbp5 will probably allow further insight into the mRNA translocation process.
Materials and Methods
Preparation and Microinjection of C. tentans Salivary Glands.
C. tentans midges were cultivated as described before (12). Salivary glands were transferred either to cover slides or custom made cuvettes designed for light sheet microscopy. For injection, the glands were kept in PBS, which was replaced afterwards with 20–40 μl hemolymph. Hemolymph was prepared freshly or used as premade stock flash frozen in liquid nitrogen and stored at -80 °C.
Preparation of Fluorescent Proteins.
Dbp5 cDNA was amplified from an epithelial C. tentans cell line. All proteins were expressed as GST fusion proteins in E.coli BL21[DE3],(pLysS), and purified using Glutathion agarose (Sigma). The N-terminal deletion mutant hrp36ΔRRM (aa 201–297) was used as GST fusion protein. From all other proteins the GST moiety was removed by thrombin digestion. For labeling of hrp36, an N-terminal tetra-cystein tag [Cys-Cys-Pro-Gly-Cys-Cys, tc (30)] was introduced. Tc-hrp36 proteins were labeled with AlexaFluor647 maleimide, and Dbp5 and NUP214 were labeled with AlexaFluor647 and AlexaFluor546 NHS-ester, respectively.
Light Microscopy.
Confocal images were made with a Zeiss LSM-510-META using a 63x (LCI Plan Neofluar, NA 1.2, Carl Zeiss MicroImaging) water immersion objective. For light sheet fluorescence microscopy of single fluorescent mRNPs, a modified Axiovert 200 (Carl Zeiss MicroImaging) was employed. For details of the instrument, see ref. 18 and SI Appendix, Fig. S5.
Data Evaluation and Analysis.
For single molecule imaging, movies were collected (integration time 20 ms per frame) and split into substacks of 1,000 frames for evaluation. The first fifty frames showed the NTF2-stained NPCs. Movies S1 and S2 were contrast-enhanced and background-subtracted before evaluation. For mRNP export, we examined 140 nuclei from 70 salivary glands. We evaluated 7,180 movies of 20 s duration. For Dbp5, we analyzed 1,300 movies of 20 s duration from 24 nuclei (12 salivary glands). For dwell time analysis of single mRNPs or Dbp5 molecules kymographs were created, which allowed direct read-out of the respective times (21). Kymographs were created automatically using ImageJ (31) and custom-made plugins as described in the SI Appendix. Routines for manual tracking of single molecule signals with subpixel precision were written in Matlab (MathWorks). Further data analysis and handling was done with Origin 8.0 (OriginLab).
Supplementary Material
Acknowledgments.
We thank C. Nietzel for excellent technical assistance, and B. Daneholt, R. Veith, and N. Visa for constructive discussions. U.K. and T.K. acknowledge financial support by the German Research Foundation Grant Ku 2474/7-1 and German National Academic Foundation, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9228.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201781109/-/DCSupplemental.
References
- 1.Bjork P, Wieslander L. Nucleocytoplasmic mRNP export is an integral part of mRNP biogenesis. Chromosoma. 2011;120:23–38. doi: 10.1007/s00412-010-0298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stewart M. Nuclear export of mRNA. Trends Biochem Sci. 2010;35:609–617. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Visa N, et al. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- 4.Ledoux S, Guthrie C. Regulation of the Dbp5 ATPase cycle in mRNP remodeling at the nuclear pore: a lively new paradigm for DEAD-box proteins. Genes Dev. 2011;25:1109–1114. doi: 10.1101/gad.2062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 6.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci USA. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mor A, et al. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]
- 9.Grunwald D, Singer RH. In vivo imaging of labeled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 12.Siebrasse JP, et al. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Natl Acad Sci USA. 2008;105:20291–20296. doi: 10.1073/pnas.0810692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zu K, Sikes ML, Beyer AL. Separable roles in vivo for the two RNA binding domains of Drosophila A1-hnRNP homolog. RNA. 1998;4:1585–1598. doi: 10.1017/s135583829898102x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Jin SB, Bjorkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napetschnig J, et al. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci USA. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodge CA, et al. The Dbp5 cycle at the nuclear pore complex during mRNA export I: dbp5 mutants with defects in RNA binding and ATP hydrolysis define key steps for Nup159 and Gle1. Genes Dev. 2011;25:1052–1064. doi: 10.1101/gad.2041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 18.Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U. Light sheet microscopy for single molecule tracking in living tissue. PloS One. 2010;5:e11639. doi: 10.1371/journal.pone.0011639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynaud EG, Krzic U, Greger K, Stelzer EH. Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2008;2:266–275. doi: 10.2976/1.2974980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubitscheck U, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kylberg K, et al. Exclusion of mRNPs and ribosomal particles from a thin zone beneath the nuclear envelope revealed upon inhibition of transport. Exp Cell Res. 2010;316:1028–1038. doi: 10.1016/j.yexcr.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 23.Lowe AR, et al. Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking. Nature. 2010;467:600–603. doi: 10.1038/nature09285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 25.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 26.Batisse J, Batisse C, Budd A, Bottcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J Biol Chem. 2009;284:34911–34917. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt C, et al. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montpetit B, et al. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 31.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.