Abstract
Traditional approaches to protein–protein docking sample the binding modes with no regard to similar experimentally determined structures (templates) of protein–protein complexes. Emerging template-based docking approaches utilize such similar complexes to determine the docking predictions. The docking problem assumes the knowledge of the participating proteins’ structures. Thus, it provides the possibility of aligning the structures of the proteins and the template complexes. The progress in the development of template-based docking and the vast experience in template-based modeling of individual proteins show that, generally, such approaches are more reliable than the free modeling. The key aspect of this modeling paradigm is the availability of the templates. The current common perception is that due to the difficulties in experimental structure determination of protein–protein complexes, the pool of docking templates is insignificant, and thus a broad application of template-based docking is possible only at some future time. The results of our large scale, systematic study show that, surprisingly, in spite of the limited number of protein–protein complexes in the Protein Data Bank, docking templates can be found for complexes representing almost all the known protein–protein interactions, provided the components themselves have a known structure or can be homology-built. About one-third of the templates are of good quality when they are compared to experimental structures in test sets extracted from the Protein Data Bank and would be useful starting points in modeling the complexes. This finding dramatically expands our ability to model protein interactions, and has far-reaching implications for the protein docking field in general.
Keywords: protein modeling, protein recognition, structural bioinformatics, structure alignment
Protein–protein interactions (PPI) are a key component of life processes at the molecular level, and the number detected in genome-wide studies is fast growing. We want to understand their properties and be able to manipulate them for structure-based drug design. For this purpose, we must characterize PPI structurally, but their study by X-ray and NMR methods is demanding and slow, and computational methods appear to be a necessary complement.
The structural predictions of PPI generally rely on docking procedures that can be roughly divided into: (i) template-free docking, where many or all the possible binding modes of two proteins are explored with no a priori knowledge of the structure of the complex, and (ii) template-based docking, where the similarity with previously known complexes determines the prediction. Template-free docking methods rely on the geometric and chemical-physical complementarity of the protein surfaces (1), now often supplemented by statistical potentials (2, 3), and subject to a variety of constraints (4). The template-free modeling can also be applied to prediction of domain–domain structures (5, 6). The Critical Assessment of Predicted Interactions (CAPRI) blind prediction experiment (7) has shown that the template-free methods can yield good models of protein–protein complexes (8), but their ability to sample the conformational space is limited, and the multiple minima generate many false positives.
With single proteins, template-based modeling starts with the target sequence being aligned on the sequences of putative templates, or threaded on their structures. This has long been known to be more reliable and efficient than ab initio model building (9), and can be used for genome-wide modeling. In model organisms such as Escherichia coli or yeast, the Protein Data Bank (PDB) offers valid templates for a significant part of their soluble proteins (10), including those in known PPI (85% in E. coli and 39% in yeast, according to our results). To model a PPI, the procedure may be applied separately to each partner protein, but a template must also be found for their assembly (11–17). Cocrystallized protein–protein structures are still few, and the availability of such templates is a key issue. Nevertheless, we show here that good-quality structural templates can be found for almost all known PPI that involve proteins for which a structure is known or can be built by homology. This suggests a new paradigm in PPI modeling: When there is a template for the components, there is also a template for the complex. Its rationale is that model building uses sequence-based similarity in the case of a monomer, but structure similarity for a complex. Two complexes with sequence identities > 40% adopt similar binding modes in a majority of cases (18, 19). As protein structure is more conserved than sequence, similar binding modes should also occur in the absence of sequence similarity. Our results indicate that this is very frequent, which dramatically expands our ability to model PPI.
Results and Discussion
The study is based on 126,897 PPI involving pairs of proteins with both interacting partners from the same organism for 771 species spanning the entire universe of life from Archaea to human. Models of individual proteins were built by NEST (20) from either profile-to-profile or BLAST sequence alignments. The structure alignment-based models of complexes were generated by TM-align (21) (see Methods).
The structural similarity of two complexes was evaluated by the TMm scores (the smallest of the receptor and the ligand TM-scores). The similarity of the binding mode in two complexes was assessed by two root-mean-square distances (rmsd). The interface rmsd (IF), measured on the Cα atoms of interface residues, is widely used to compare binding modes of the same monomer [e.g., assessing the quality of CAPRI models (8)]. The interaction rmsd (IA) was introduced by Aloy et al. (18) to compare binding modes involving dissimilar monomers. It is measured on a standard set of 14 points, seven attached to the ligands after superposing the receptors, and seven to the receptors after superposing the ligands (18). IA correlates well with IF when it is used to compare binding modes of the same monomer.
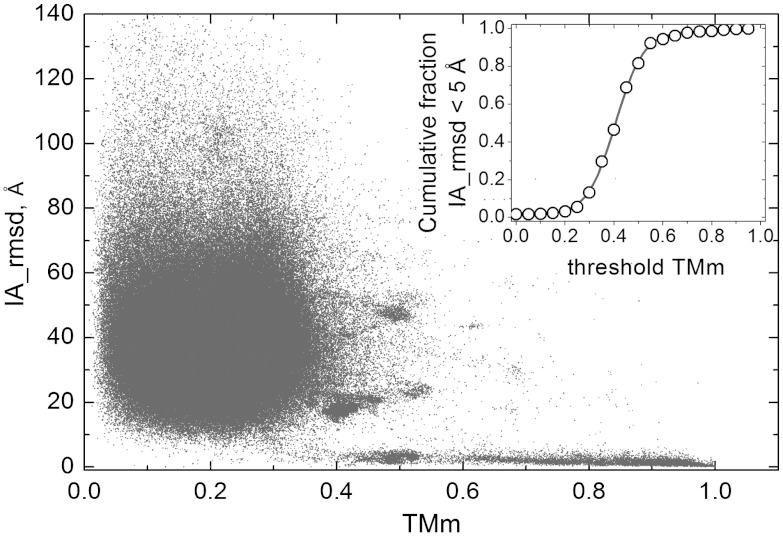
Fig. 1 shows how IA, a measure of the binding mode similarity, correlates with TMm, a measure of the structural similarity between the complexes, in an all-to-all pairwise comparison of 989 from the Dockground (22) public resource for protein recognition studies (http://dockground.bioinformatics.ku.edu) purged at 95% sequence identity level. TMm values above 0.5 imply that both components of the two complexes share the same fold (21). Fig. 1 shows that nearly all such complexes have IA < 5 Å, meaning that they also share the same binding mode (18). The inset represents the phase transition that occurs near TMm = 0.4. Binding modes are mostly similar above, and mostly different below, this threshold. Thus, TMm = 0.4 threshold is a good quantitative measure for distinguishing similar binding modes and as such will be used in this the paper, although some results will be presented for other threshold values as well.
Fig. 1.
Correlation of the structural difference in binding modes with the structure alignment score. The interaction rmsd (IA) is plotted against the lowest of the two components protein TM-scores (TMm), in all-to-all pairwise comparison of 989 complexes extracted from Dockground at 95% sequence identity. In the inset, the cumulative fraction of the complex pairs with IA < 5 Å and TMm > threshold TMm is plotted as a function of threshold TMm to show the transition that occurs near the threshold TMm = 0.4.
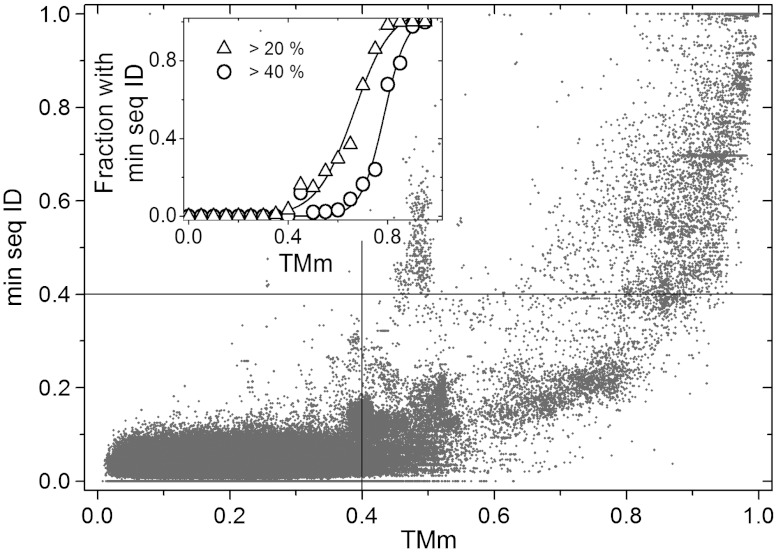
In Fig. 2, we plot TMm against the lowest of the two sequence identity values of the two components of the same complexes. The bottom left quadrant contains pairs of complexes that differ in both the sequences and the binding mode. They make up 96.3% of the data, reflecting the diversity of the complexes in Dockground. The top right quadrant, with 0.9% of all data and 24.9% of those with TMm > 0.4, contains pairs where conserved sequences correlate with similar folds and binding modes. The bottom right quadrant (75.1% of the data with TMm > 0.4) contains pairs where the folds and binding mode are conserved, but the sequences are not. Such pairs are three times as many as those where the conservation of the binding mode correlates with that of the sequences. Last, the top left quadrant indicates that a similarity is detected by the sequence, but not the structural alignment. It is nearly empty, meaning that the structural alignments find essentially all the templates detectable from the sequences.
Fig. 2.
Correlation of structure and sequence similarity. The lowest of the sequence identity fractions for two aligned components of a complex is plotted against the corresponding TMm, in all-to-all pairwise comparison of 989 complexes extracted from Dockground at 95% sequence identity. The lines separate quadrants below and above a sequence and a structure-based threshold (see Text). In the inset, the fraction of the complex pairs with lowest sequence identity > 20% and 40% is plotted in 0.05 bins of TMm values to show that many pairs with a similar binding mode (TMm > 0.4) have a low sequence identity, below 20 or 40%.
The data in Fig. 1 show less diversity of interaction modes at high degrees of structural similarity, compared to the interactions at high sequence identity [see figure 2 in ref. (18)]. As seen from Fig. 2 of this paper, correlation of high sequence identities and structure similarities is spread, indicating considerable variation in sequence/structure relationship. The observed difference between the distributions of sequence identity vs. interaction mode and structure similarity vs. interaction mode reflects a more direct relation of the structure to the binding.
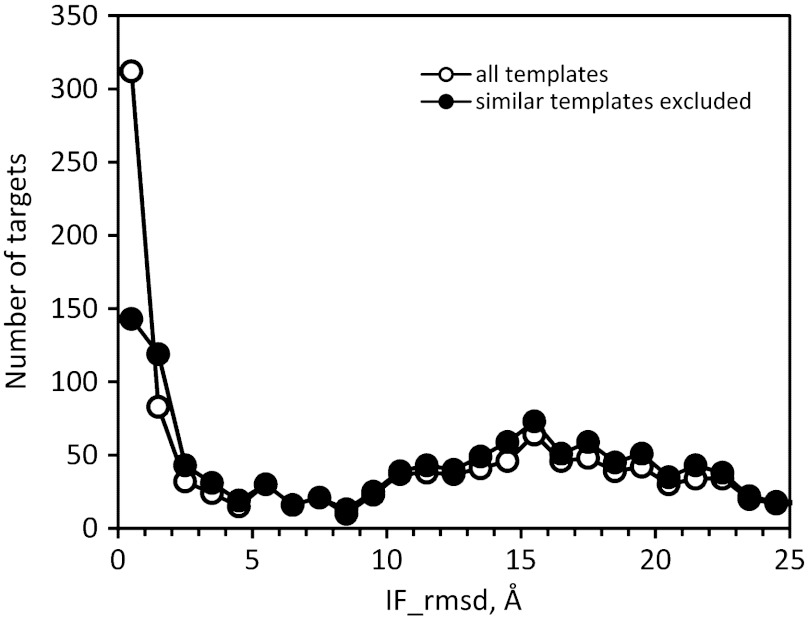
In order to assess its predictive value, we tested the approach on new protein–protein structures in PDB released in 2009–2011, utilizing older template structures released before 2009. For 1296 new complexes, templates with TMm > 0.4 were identified for all but nine complexes, and their quality was estimated by the IF criterion as in CAPRI predictions. A majority (55%) of templates were homodimers, which are much more numerous than heterodimers in PDB, whereas all targets were heterodimers. Fig. 3 shows the distribution of the IF values. It is bimodal, with 36% of the modeled complexes below IF = 5 Å, which should make them good starting points for further modeling. When high similarity templates (TM-score > 0.9) were excluded from the search, templates were found for 1279 complexes, and 28% had IF < 5 Å (Fig. 3), out of which only 4% account for homodimeric templates (Fig. S1). Moreover, 1,194 complexes had a sequence identity with at least one template monomer < 40%, below the threshold for correlation between sequence and binding mode conservation. Out of those, 23% had structure alignment-based models with IF < 5 Å. Higher TMm threshold values predictably decrease the number of detected templates, while increasing the overall quality of templates. However, this increase is relatively small (Fig. S2). For example, the threshold increase from 0.4 to 0.5 leads to 22% fewer detected templates, while the fraction of good templates increases by 7% only. This adds an argument for the use of TMm = 0.4 threshold as the optimal value that provides a broad spectrum of templates, while maintaining their quality.
Fig. 3.
Benchmarking of template-based docking. The distribution of targets is shown according to the interface rmsd (IF) from the cocrystallized structure. The benchmarking of PDB complexes released in 2009–2011 was based on template structures from 2008 and earlier.
We also tested the 31 complexes that have been submitted as targets to CAPRI in the years 2001–2009, and for which the X-ray structure has been published. Template-based docking yielded at least one model with IF < 10 Å for 11 targets, and one with IF < 5 Å for four (SI Text). Thus, the fraction of good quality templates was about the same as for the larger set of new complexes in the PDB, but the coverage was comparatively low. A possible reason is that CAPRI avoids “trivial” targets similar to complexes already in PDB, whereas such similarities are of obvious interest when modeling a complex of biological interest.
Benchmarking of the predictive approaches, based on the X-ray structures in PDB or on those available in blind experiments, like critical assessment of structure prediction (9) and CAPRI (7), has its limitations. Such benchmarking is performed on the pool of structures that reflects capabilities and priorities of structural biologists and under-represents significant areas of the structure universe, membrane proteins for instance. Thus, a “true” predictive quality of the modeling (e.g., the percentage of correct templates, the added value of structural alignment vs. sequence alignment) remains unknown. However, its approximation by benchmarking a set of known structures is an accepted practice in template-based modeling of individual proteins, as well as in the younger field of the template-based modeling of complexes.
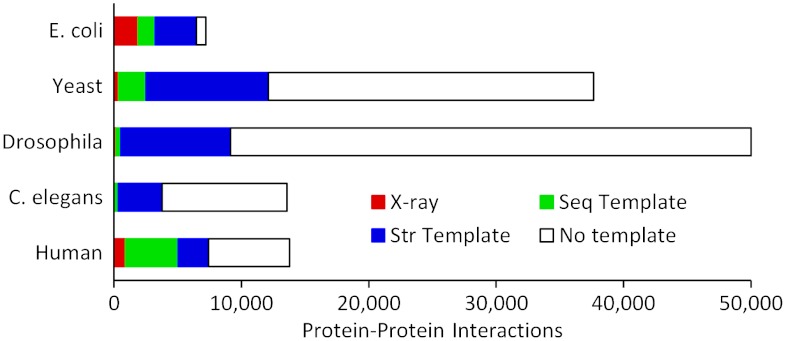
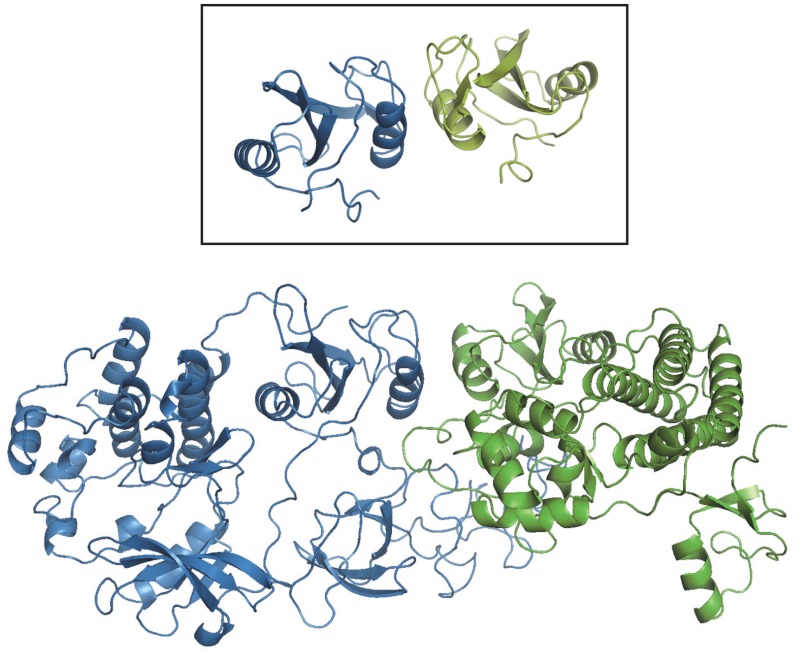
We next asked to what extent template-based docking can be used to model known PPI in whole genomes. Homology models of individual proteins were built, and templates of their complexes searched for in PDB as described in Methods. The results shown in Fig. 4 concern the five genomes with the largest number of known PPI. X-ray structures of the complexes represent 26% of the known PPI in E. coli, and 6.7% in human. Very few (1% or less) are available in yeast, Drosophila and Caenorhabditis elegans, and sequence-based templates can be found for only 0.8–6% of their known PPI. Structural alignments yield a dramatic increase of the coverage: from 7% to 32% of the PPI in yeast, 1% to 18% in Drosophila, and 2.5% to 28% in C. elegans. Fig. 5 shows an example of a complex built by structural alignment of monomers’ homology models and a cocrystallized template, with low target/template sequence identity.
Fig. 4.
Structural coverage of PPI. The data for five genomes with the largest number of known PPI shows different categories of complexes structures: (red) complexes with a X-ray structure, (green) complexes with a sequence template, (blue) complexes for which the structure of the monomers is known or can be built by homology; a structural template is found for 99% of the latter complexes.
Fig. 5.
A complex built by structural alignment of the homology models of its components and a non-homologous structural template. The modeled complex comprises the human Lyn A tyrosine kinase (sequence GI code 198941, blue ribbon) and the tyrosine kinase binding domain (TKB) of the mouse Cbl (Casitas B-lineage) lymphoma protein (sequence GI code 6680858, green ribbon). The inset shows the homo-dimeric SH2 domain of the human Grb10 protein (PDB entry 1NRV), that was used as a template to build the model (TM-scores 0.90 and 0.69). Sequence identities between the target and the template monomer are 26 and 2.8%. The homology model of Lyn A was based on the human Src tyrosine kinase (chain A of PDB entry 2H8H, sequence identity 52%), that of the mouse Cbl TKB, on the human homolog (chain A of PDB entry 1FBV, sequence identity 94%).
Remarkably, structural templates were found for nearly all (33,537 out of 33,840, or 99%) the complexes in which both components could be built. In Fig. 4, “no template” therefore indicates a failure to find a template for one or both components, not of the complex. The coverage should therefore improve as more individual proteins have their structure determined, but also as more sophisticated high-throughput methods allow their modeling at lower levels of target/template similarity. One can think of such ability to detect templates for virtually all complexes as a consequence of the monomers modeling by sequence similarity, followed by modeling of the complex by structure similarity, which is significantly broader in scope (structure is more conserved than sequence).
Our observation correlates with the conclusions of the study by Honig and co-workers (23) on the completeness of current structural databases of protein–protein interactions. A significant part of complexes with the structural templates in Fig. 4 (46%, Fig. S3) have a higher accuracy template with TMm > 0.6, further increasing the probability of the correct binding mode prediction. A fraction of the PPI for which we failed to find a template may be false positives, which are frequent in existing databases (24), and conversely, not all structures of the complexes built by the structure alignment will be correct. However, the significant success rates in the benchmarking (see above) give confidence to the results and suggest that a substantial part of the docked complexes is of sufficient quality to inspire and guide experiments. In any case, our finding that the existing pool of cocrystallized protein–protein complexes provides docking templates for nearly all the known PPI that involve structurally characterized proteins, has far-reaching implications for the docking field, placing template-free docking in a narrow niche, similar to the template-free prediction of individual proteins.
Methods
The PPI data were imported from the BIND (25) and DIP (26) databases. Redundancy between BIND and DIP, defined as a difference in one or more residues in at least one interacting partner, was excluded. Putative templates for modeling individual proteins included all non-redundant protein structures in the PDB, with the same definition of redundancy. The Needleman-Wunsch algorithm (27) with affine gap penalty (28) was used to search for sequence homology templates in PDB. The scoring matrices were based on sequence profiles from PSI-BLAST runs (29), as implemented in BLASTPGP, on the non-redundant sequence database from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), with substitution matrix BLOSUM62 (30), five iterations, and all other parameters set to default values (for details of the procedure, see ref. 31).
An alignment was considered as significant if its normalized raw score was > 1. If no significant profile-to-profile alignment was found, a single iteration was performed with BLASTPGP. Alignments were retained for modeling if the sequence identity was > 20% and it covered ≥40% of the target sequence. If template complexes were found in PDB, the pair of the alignments with the highest normalized raw score (for profile alignments) or lowest e-value (for PSI-BLAST alignments) was retained to build a model of the complex. In case of more than one alignment with the same score, the one with the highest sequence identity between the target and the template was used. In case of more than one alignment with the same sequence identity, we used the ones with the highest coverage of the target to the template (for the PSI-BLAST alignments) and/or with the template of highest PDB quality (best resolution, least amount of missing atoms, etc.). If no template complex was found, models of the components were built using the best alignments (with the above criteria) on single protein templates. The protein models were built by the NEST program from the JACKAL package (20) with the default parameters. More elaborate, multitemplate methods could not be considered in view of the high-throughput nature of our study.
The search for template complexes employed TM-align (21) to perform the structural alignment of the modeled components on the 11,932 X-ray structures extracted from PDB. The structures resolution had to be < 3 Å, they had to be at least a dimeric biological unit, and the sequence identity between different structures had to be < 90% for at least one component of the dimers. Biological unit coordinates were obtained from ftp://ftpwwpdb.org/pub/pdb/data/biounit. The pairs of the structural alignments were ranked by the sum of the receptor and ligand TM-scores, and those with the TMm (the smallest of the receptor and the ligand TM-scores) > 0.4 were retained for model building, performed by applying the rigid-body transformation matrices to the component models. Models with very small interfaces (buried surface area < 50 Å2), or too many clashes (> 5% of the intermolecular atom–atom distances shorter than the sum of corresponding van der Waals radii) were rejected, in which case the next alignment on the list was used.
A publicly available Web-based resource to browse and analyze possible models of protein–protein complexes, based on the target-template structural similarity, is currently being developed in our group.
Supplementary Material
ACKNOWLEDGMENTS.
Andrej Sali, University of California, San Francisco, and Sarah Teichmann, Medical Research Council, United Kingdom for reading the manuscript and providing valuable suggestions. The study was supported by National Institutes of Health Grant R01 GM074255.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200678109/-/DCSupplemental.
References
- 1.Janin J, Bahadur RP, Chakrabarti P. Protein–protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 2.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 3.Mintseris J, et al. Integrating statistical pair potentials into protein complex prediction. Proteins. 2007;69:511–520. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 4.de Vries SJ, et al. HADDOCK versus HADDOCK: New features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 5.Wollacott AM, Zanghellini A, Murphy P, Baker D. Prediction of structures of multidomain proteins from structures of the individual domains. Protein Sci. 2007;16:165–175. doi: 10.1110/ps.062270707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha N, Kumar S, Nussinov R. Interdomain interactions in hinge-bending transitions. Structure. 2001;9:1165–1181. doi: 10.1016/s0969-2126(01)00687-6. [DOI] [PubMed] [Google Scholar]
- 7.Janin J, et al. CAPRI: A critical assessment of predicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 8.Lensink MF, Wodak SJ. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010;78:3073–3084. doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- 9.Moult J, Fidelis K, Kryshtafovych A, Rost B, Tramontano A. Critical assessment of methods of protein structure prediction—Round VIII. Proteins. 2009;77(Suppl 9):1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- 10.Levitt M. Nature of the protein universe. Proc Natl Acad Sci USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell RB, et al. A structural perspective on protein–protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH—prediction of interactions using known interfaces. Proteins. 2007;69:839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 13.Keskin O, Nussinov R, Gursoy A. PRISM: Protein–protein interaction prediction by structural matching. Meth Mol Biol. 2008;484:505–521. doi: 10.1007/978-1-59745-398-1_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 15.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein–protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korkin D, et al. Structural modeling of protein interactions by analogy: Application to PSD-95. PLoS Comput Biol. 2006;2:1365–1376. doi: 10.1371/journal.pcbi.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee S, Zhang Y. Protein–protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;13:955–966. doi: 10.1016/j.str.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrey D, et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53:430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucl Acids Res. 2005;33:2303–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, Douguet D, Tovchigrechko A, Vakser IA. DOCKGROUND system of databases for protein recognition studies: Unbound structures for docking. Proteins. 2007;69:845–851. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- 23.Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci USA. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aloy P, Russell RB. Structural systems biology: Modelling protein interactions. Nat Rev Mol Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 25.Alfarano C, et al. The biomolecular interaction network database and related tools 2005 update. Nucl Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salwinski L, et al. The database of interacting proteins: 2004 update. Nucl Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Needleman S, Wunsch CD. A general method applicable to search for similarities in amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of database programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- 31.Kundrotas PJ, Lensink MF, Alexov E. Homology-based modeling of 3D structures of protein–protein complexes using alignments of modified sequence profiles. Int J Biol Macromol. 2008;43:198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.