Abstract
The natural history of HIV-1 infection is highly variable in different individuals, spanning from a rapidly progressive course to a long-term asymptomatic infection. A major determinant of the pace of disease progression is the in vivo level of HIV-1 replication, which is regulated by a complex network of cytokines and chemokines expressed by immune and inflammatory cells. The chemokine system is critically involved in the control of HIV-1 replication by virtue of the role played by specific chemokine receptors, most notably CCR5 and CXCR4, as cell-surface coreceptors for HIV-1 entry; hence, the chemokines that naturally bind such coreceptors act as endogenous inhibitors of HIV-1. Here, we show that the CXC chemokine CXCL4 (PF-4), the most abundant protein contained within the α-granules of platelets, is a broad-spectrum inhibitor of HIV-1 infection. Unlike other known HIV-suppressive chemokines, CXCL4 inhibits infection by the majority of primary HIV-1 isolates regardless of their coreceptor-usage phenotype or genetic subtype. Consistent with the lack of viral phenotype specificity, blockade of HIV-1 infection occurs at the level of virus attachment and entry via a unique mechanism that involves direct interaction of CXCL4 with the major viral envelope glycoprotein, gp120. The binding site for CXCL4 was mapped to a region of the gp120 outer domain proximal to the CD4-binding site. The identification of a platelet-derived chemokine as an endogenous antiviral factor may have relevance for the pathogenesis and treatment of HIV-1 infection.
Progression of HIV type 1 (HIV-1) disease results from a complex balance between viral and host factors (1). Among the latter are major histocompatibility complex alleles (2), cell-surface receptors (3), intracellular restriction elements (4), and soluble cytokines (5) and chemokines (6), all of which are subject to genotypic and/or phenotypic regulation. In particular, the chemokine system plays a critical role in the pathogenesis of HIV-1 infection due to the evolutionary choice by HIV-1 to exploit specific chemokine receptors, most notably CCR5 and CXCR4, as coreceptors for entry into susceptible cells. This choice has turned the chemokine ligands of such receptors, i.e., CCL3 (MIP-1α), CCL4 (MIP-1β), and CCL5 (RANTES) for CCR5, and CXCL12 (SDF-1) for CXCR4, into endogenous inhibitors of HIV-1 replication (6). The in vivo expression of these chemokines and their receptors is influenced by specific genetic polymorphisms (7–10) and has been implicated as a key factor affecting the outcome of HIV-1 disease. Although CCL3, CCL4, and CCL5 were initially identified as HIV-suppressive factors produced by CD8+ T cells (11), chemokines can be produced by a variety of other cells that become activated in the course of immune and inflammatory responses, including CD4+ T cells, NK cells, mononuclear phagocytic cells, endothelial cells, and platelets. The latter are specialized anucleated cells that store in their α-granules large amounts of CCL5, CXCL4 (PF-4), and the CXCL7 (NAP-2) precursor, β-thromboglobulin (12). Besides their classic role in hemostasis, platelets also participate in the inductive phase of inflammatory responses (13, 14), and it has been suggested that they play a key role in the pathogenesis of certain inflammatory disorders such as hepatitis (15) and renal glomerular disease (16).
A variety of platelet dysfunctions have been documented during the progression of HIV-1 disease (17), which in some patients lead to severe clinical manifestations such as idiopathic thrombocytopenic purpura (18) and thrombosis (19, 20). In addition, platelet alterations, including abnormal platelet activation and apoptosis, are frequently detected in HIV-1–infected individuals even with normal platelet counts (21, 22). Sustained platelet activation in these subjects may be due, at least in part, to direct binding and internalization of HIV-1 virions by platelets, mediated by the C-type lectin receptors CLEC-2 and DC-SIGN (23).
In this study, we investigated the interaction between HIV-1 and the CXC chemokine CXCL4, the most abundant protein contained within platelet α-granules, which is released at micromolar concentrations upon platelet activation (24). We found that CXCL4 is a broad-spectrum inhibitor of HIV-1, acting through an unconventional mechanism mediated by direct interaction of the chemokine with the major viral envelope glycoprotein, gp120. These findings may have implications for our understanding of HIV-1 disease pathogenesis as well as for the development of novel strategies for the therapy and prevention of HIV-1 infection.
Results
CXCL4 Inhibits Infection of CD4+ T Cells and Macrophages by Different Phenotypic Variants of HIV-1.
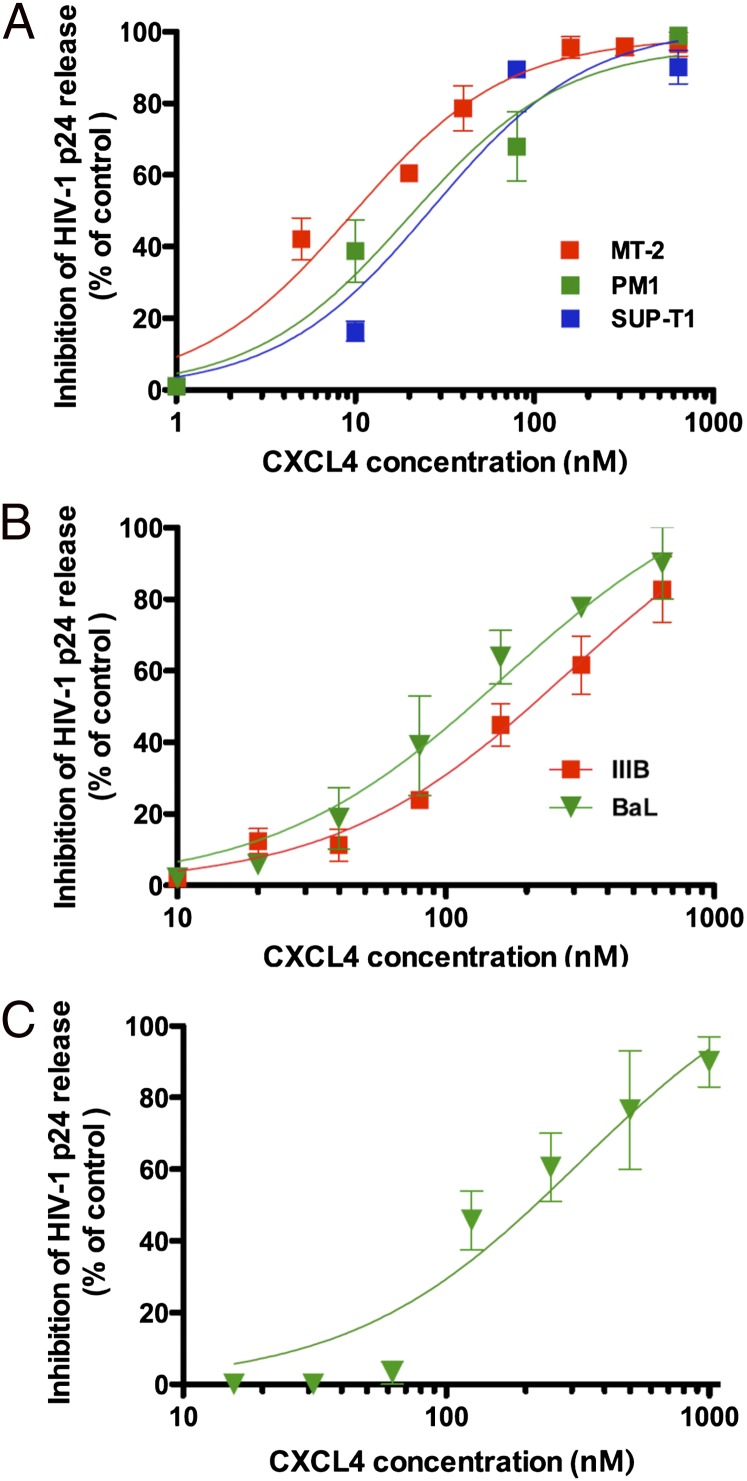
The effect of recombinant human CXCL4 on HIV-1 infection was initially evaluated on the immortalized human CD4+ T-cell lines MT-2, PM1, and Sup-T1, which express CXCR4 and are thereby susceptible to infection by CXCR4-tropic (X4) HIV-1 strains. Fig. 1A shows that CXCL4 inhibited in a dose-dependent fashion the replication of a prototypic X4 strain, HIV-1 IIIB, in all three cell lines, with half-maximal inhibitory concentrations (IC50) of 10 nM in MT-2, 20 nM in PM1, and 27 nM in Sup-T1.
Fig. 1.
Inhibition of HIV-1 replication by recombinant human CXCL4. (A) Dose-dependent inhibition of a prototypic X4 HIV-1 strain (IIIB) in the immortalized human CD4+ T-cell lines MT-2, PM1, and Sup-T1. (B) Dose-dependent inhibition of two HIV-1 variants with different coreceptor-usage phenotype [HIV-1 IIIB (X4) and HIV-1 BaL (R5)] in primary human CD4+ T cells. (C) Dose-dependent inhibition of a prototypic macrophage-tropic HIV-1 strain (BaL, R5) in primary human macrophage cultures. Although CXCL4 is predominantly a tetramer in solution, molar values were calculated on the basis of the molecular weight of the monomer (∼7.8 Kd); thus, the data represent the most conservative estimate of the antiviral potency of CXCL4. Virus replication was assessed by measuring the amount of extracellular p24 Gag protein by enzyme immunoassay. To reduce interexperimental variability, the data were normalized with respect to the level of virus replication detected in control cultures (not treated with CXCL4). All of the data represent mean values (±SD) from at least three independent experiments, each performed in duplicate.
To evaluate whether the inhibitory activity of CXCL4 was restricted to HIV-1 isolates that use CXCR4 as a coreceptor, the effect of the recombinant chemokine was evaluated on HIV-1 strains with different coreceptor specificity, one X4 (IIIB) and one CCR5-tropic (R5) (BaL), in primary human CD4+ T cells. Unlike the classic HIV-suppressive chemokines (6), CXCL4 inhibited both viral variants with similar efficiency, with IC50 values of 284 nM against IIIB, and 166 nM against BaL (Fig. 1B). Because the cells of the mononuclear phagocytic system represent another major in vivo target for HIV-1 infection, we tested the effect of CXCL4 on infection of primary human macrophages by HIV-1 BaL (R5). As shown in Fig. 1C, CXCL4 inhibited HIV-1 in a dose-dependent fashion also in these cells, with an IC50 of 134 nM.
To further validate the biological relevance of our findings, we tested the efficacy of the native CXCL4 chemokine obtained from activated human platelets, which inhibited HIV-1 with the same potency as the recombinant molecule (Fig. S1). Of note, neither the viability nor the proliferation of primary CD4+ T cells were affected by treatment with CXCL4 over a broad range of concentrations (Fig. S2), thereby excluding that inhibition of HIV-1 replication was due to nonspecific effects on cell metabolism or survival.
CXCL4 Has a Broad Spectrum of Antiviral Activity Against Primary HIV-1 Isolates.
To assess the breadth of antiviral activity of CXCL4, we tested a panel of primary HIV-1 isolates (n = 13) of different coreceptor-usage phenotype (X4, R5, and multitropic) and genetic subtype (B, C, and F) in primary human CD4+ T cells; three reference laboratory-adapted isolates, IIIB, BaL, and ADA, were studied in parallel. The majority of the primary isolates tested (10/13; 76.9%) were sensitive to CXCL4-mediated inhibition, with IC50 values ranging from <25 to 480 nM, whereas three isolates were insensitive within the CXCL4 dose range tested (IC50 > 650 nM), but there was no correlation between CXCL4 sensitivity and viral coreceptor-usage phenotype or genetic subtype (Table 1).
Table 1.
Sensitivity of primary and laboratory-adapted HIV-1 isolates to CXCL4-mediated inhibition
| Coreceptor usage |
||||||||||
| HIV-1 isolate | Genetic subtype | CXCR4 | CCR5 | CCR2b | CCR3 | CCR8 | CXCR6 | CX3CR1 | GPR15 | CXCL4 IC50, nM |
| 07USLR | B | +++ | — | — | — | — | — | — | — | >650.0 |
| 07USPC | B | +++ | ++ | — | + | ++ | + | + | — | 429.5 |
| 07USLD | B | ++ | — | — | — | — | — | — | — | 315.3 |
| 07USAR | B | ++ | +++ | — | — | — | + | — | + | 294.8 |
| 08USKD | B | — | ++ | — | — | — | — | — | — | 194.8 |
| 08USSE | B | — | ++ | + | — | — | — | — | — | 480.5 |
| 98USSG | B | +++ | +++ | + | + | +++ | ++ | +++ | — | 448.7 |
| 97IT6366 | B | — | +++ | — | — | — | — | — | — | 189.7 |
| 92HT599 | B | +++ | +++ | +++ | ++ | +++ | ++ | +++ | +++ | 26.9 |
| 92US714 | B | — | +++ | — | — | — | — | — | — | >650.0 |
| 97ZA009 | C | — | +++ | — | — | — | — | — | — | <25.0 |
| 98IN017 | C | — | ++ | + | — | — | — | — | — | >650.0 |
| 93BR019 | F | + | +++ | — | — | + | + | — | — | 384.3 |
| IIIB | B | +++ | — | — | — | + | — | — | — | 57.7 |
| BaL | B | — | +++ | — | — | — | — | — | — | 89.7 |
| ADA | B | — | ++ | — | — | — | — | — | — | 274.4 |
A panel of 13 primary HIV-1 isolates of different coreceptor-usage phenotype and genetic subtype was studied; three well-characterized laboratory-adapted HIV-1 isolates (IIIB, BaL, and ADA) were also tested in parallel as reference. Coreceptor use was tested in Ghost cells expressing human CD4 and each of the coreceptors. +++, strong replication; ++, average replication; +, weak replication; —, no replication. Half maximal inhibitory concentration (IC50) for CXCL4 in cultures of purified primary CD4+ T cells previously activated with PHA and IL-2.
To further evaluate the specificity of the anti–HIV-1 activity of CXCL4, we tested the efficacy of the recombinant chemokine against the related retroviruses HIV type 2 (HIV-2) and simian immunodeficiency virus (SIV), including three primary HIV-2 isolates and three in vivo passaged strains of SIV derived from two different primate species, as well as against an unrelated DNA virus, human herpesvirus (HHV)-6A. Table S1 shows that recombinant CXCL4 used at concentrations up to 1 μM exerted no inhibitory effects against any of these viruses. Thus, within the limitations of the small number of isolates tested, these data suggest that the antiviral activity of CXCL4 is specific for HIV-1.
CXCL4 Blocks an Early Step in the HIV-1 Infectious Cycle.
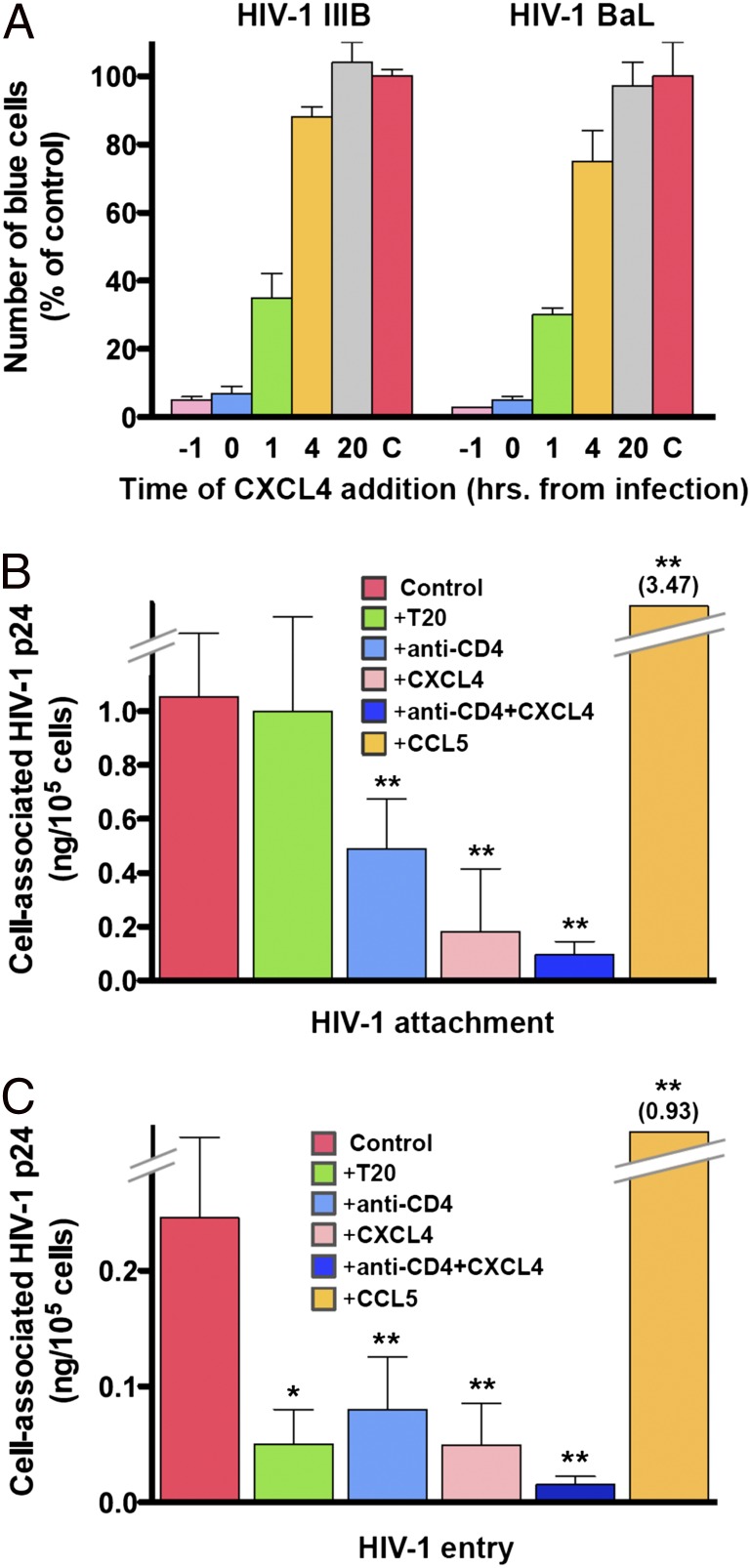
To investigate the mechanism of antiviral activity of CXCL4, initial experiments were conducted using the multinuclear activation of galactosidase indicator (MAGI) assay, which can be temporally restricted to evaluate a single HIV-1 infectious cycle. In this assay, CXCL4 potently inhibited infection by different biological variants of the virus with IC50 values of 26 nM for HIV-1 IIIB and 42 nM for HIV-1 BaL (Fig. S3). To precisely define the time window of CXCL4 sensitivity in the replication cycle of HIV-1, we performed kinetic experiments in which MAGI cells were treated with CXCL4 at different times before or after exposure to HIV-1 IIIB or HIV-1 BaL. Fig. 2A shows that CXCL4 was highly effective against both viral variants when added 1 h before or at the time of infection; however, efficacy began to wane as early as 1 h postinfection and was nearly completely lost when CXCL4 was added 4 h postinfection. These results indicated that CXCL4 blocks an early step in the viral replication cycle, whereas it appears to have no effects on later steps such as reverse transcription, gene transcription/translation, and maturation.
Fig. 2.
CXCL4 inhibits the early steps of the HIV-1 replication cycle. (A) Time dependence of CXCL4 sensitivity as evaluated using the MAGI assay. Enumeration of blue cells after 48 h denotes the completion of a single infectious cycle of HIV-1. To reduce interexperimental variability, the data were normalized with respect to the number of blue cells detected in control cultures (C) not treated with CXCL4. CXCL4 was added at different times relative to infection with HIV-1 IIIB or HIV-1 BaL, and the test was developed after 48 h of infection. Results shown are from a representative experiment performed in duplicate wells. (B) Effect of CXCL4 on HIV-1 attachment. The attachment assay was performed using activated primary human CD4+ T cells incubated with the infectious stock of a CXCL4-sensitive primary HIV-1 isolate (92HT599; R5X4). Virus attachment was measured as the total amount of cell-associated HIV-1 p24 Gag protein after subtraction of background p24 levels (measured in cells incubated with the virus at 4 °C and then treated with trypsin to remove cell surface-bound HIV-1 virions). (C) Effect of CXCL4 on HIV-1 entry. For the entry assay, at the end of the incubation period the cells were treated with trypsin to remove residual extracellular virions bound to the external surface of the cells. Virus entry was measured as the amount of trypsin-resistant, intracellular p24 protein. The recombinant chemokines and the anti-CD4 mAb (RPA-T4) were used at 5 μg/mL; peptide T20 was used at 50 μg/mL. Data represent mean values (±SD) from at least three independent experiments. *P < 0.05 and **P < 0.001 vs. untreated control by paired two-tailed Student t test.
CXCL4 Blocks HIV-1 Infection at the Level of Virus Attachment and Entry.
Having established that CXCL4 acts on the early stages of the HIV-1 infectious cycle, we tested the effects of this chemokine on viral attachment and entry using an HIV-1 entry assay based on the quantification of total vs. intracellular (trypsin resistant) HIV-1 p24 Gag protein in primary CD4+ T cells exposed to HIV-1 for 4 h at 37 °C. The experiments were performed using HIV-1 92HT599, a CXCL4-sensitive multitropic isolate, and HIV-1 07USLR, a CXCL4-resistant X4 isolate (Table 1). CXCL4 markedly reduced both attachment (Fig. 2B) and entry (Fig. 2C) of the sensitive HIV-1 isolate with a potency comparable to that of the anti-CD4 monoclonal antibody (mAb) used as positive control, whereas, as expected, the HIV-1 entry inhibitor T20 was effective only on viral entry but not on attachment; an additive inhibitory effect was observed when CXCL4 and the anti-CD4 mAb were used in combination. In contrast, as documented with other CXCR4-using HIV-1 strains (25, 26), treatment with the CC chemokine CCL5 induced an enhancement of both viral attachment and entry (Fig. 2 B and C). Consistent with the results obtained in infectivity assays, CXCL4 did not exert inhibitory effects on attachment and entry of the insensitive isolate 07USLR (Fig. S4).
CXCL4 Directly Interacts with the Major HIV-1 Envelope Glycoprotein, gp120.
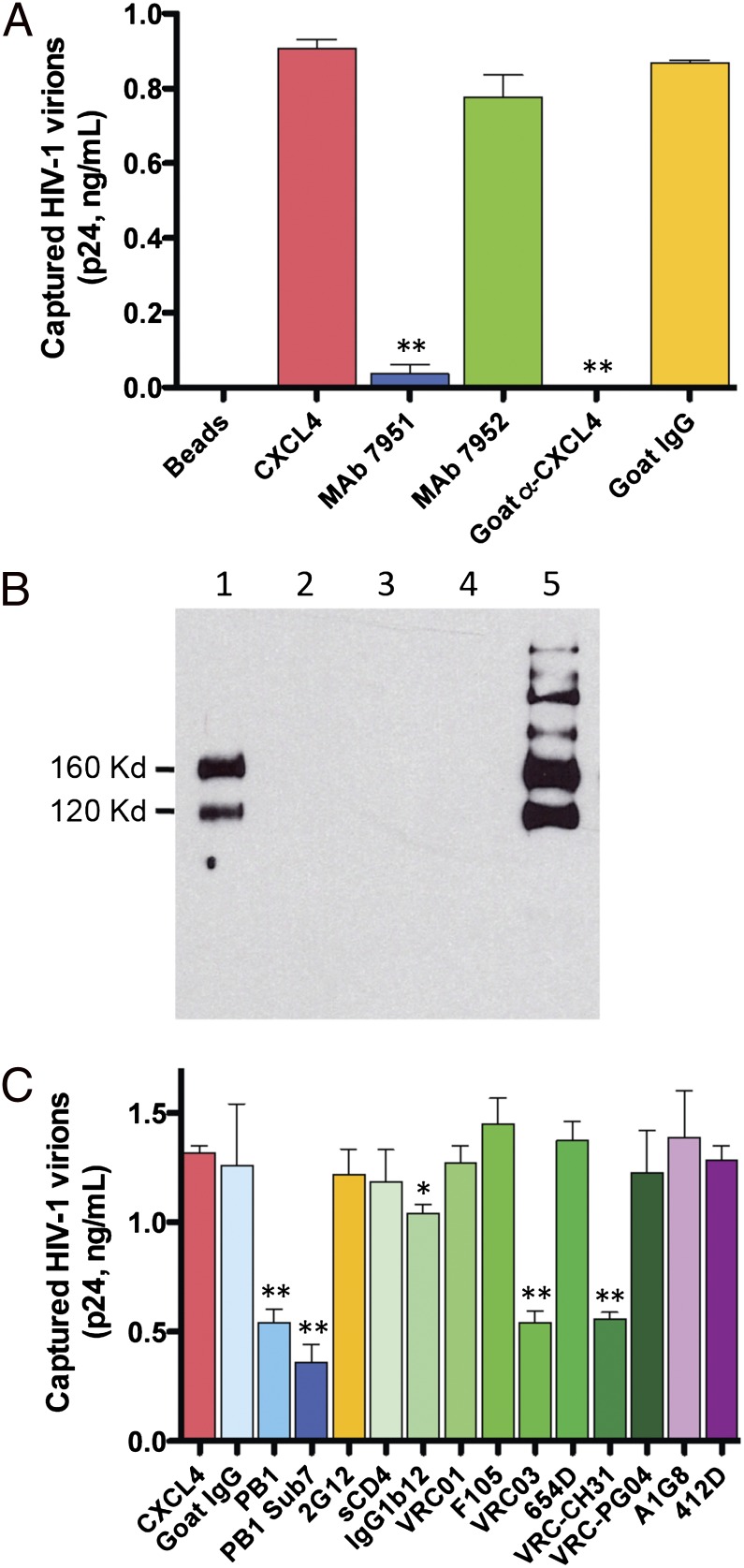
The molecular mechanism of antiviral action of CXCL4 was further investigated. The possibility that CXCL4 might directly block CD4 or the coreceptors was ruled out on the basis of the following evidence: (i) the expression of such receptors in primary CD4+ T cells was not downmodulated upon treatment with CXCL4 (Fig. S5A); (ii) a soluble form of the CD4 ectodomain (sCD4) did not bind to immobilized CXCL4 in enzyme immunoassays (Fig. S5B); (iii) CXCL4 exerted no antagonistic effects on CCR5- or CXCR4-mediated intracellular signaling (Fig. S5C); (iv) the only high-affinity receptor for CXCL4 hitherto identified, CXCR3B (27), is not prominently expressed on HIV-1 target cells and has never been shown to serve as a coreceptor for HIV-1. In light of these observations, we shifted our focus toward the viral envelope and developed an HIV-1 virion-capture assay in which recombinant CXCL4 is used as a molecular bait bound to the surface of immunomagnetic beads. Fig. 3A shows that CXCL4-armed magnetic beads efficiently captured virions of the CXCL4-sensitive isolate HIV-1 IIIB; as a proof of specificity, the uptake was abrogated by polyclonal anti-CXCL4 antibodies, as well as by one anti-CXCL4 mAb (no. 7591), but not by a second anti-CXCL4 mAb (no. 7952) directed against a distinct epitope of the chemokine. Using the same assay, the CXCL4-insensitive primary isolate HIV-1 97USLR was captured with a markedly reduced (more than eightfold) efficiency compared with the sensitive isolate HIV-1 IIIB (Fig. S6).
Fig. 3.
CXCL4 directly interacts with the major HIV-1 envelope glycoprotein, gp120. (A) Capture of HIV-1 virions (strain IIIB; X4) by CXCL4-armed immunomagnetic beads. Polyclonal goat anti-CXCL4 IgG and irrelevant goat IgG were used at 20 μg/mL; anti-CXCL4 mAbs (nos. 7951 and 7952) at 5 μg/mL. Data represent mean values (±SD) from at least three independent experiments. **P < 0.001 by paired two-tailed Student t test. (B) Coimmunoprecipitation of the HIV-1 envelope glycoproteins gp120 and gp160 by CXCL4. Assays were performed using the PM1 clone (28) persistently infected with HIV-1 IIIB (X4), which expresses on its surface both gp120 and its uncleaved precursor, gp160. Lane 1, gp120 and gp160 precipitated with CXCL4 and anti-CXCL4 rabbit antiserum; lane 2, anti-CXCL4 rabbit antiserum alone; lane 3, CCL3 with anti-CCL3 rabbit antiserum; lane 4, anti-CCL3 rabbit antiserum alone; lane 5, positive control (direct immunoprecipitation with the anti-gp120 mAb 2G12). (C) Inhibition of HIV-1 virion capture by polyclonal antisera, soluble CD4 (sCD4) and human mAbs directed against HIV-1 gp120. Polyclonal goat antisera against recombinant gp120 outer-domain fragments (PB1 and PB1 Sub 7) were used at 1:50 dilution; irrelevant goat IgG at 100 μg/mL; 4-domain sCD4 at 10 μg/mL; human anti-gp120 mAbs at 10 μg/mL. Data represent the mean values (±SD) from at least four independent experiments performed for each inhibitor. *P < 0.05 and **P < 0.01 vs. untreated control by paired two-tailed Student t test.
To verify whether HIV-1 virion capture by CXCL4 was mediated by the viral envelope glycoproteins or, conversely, by cellular proteins incorporated into HIV-1 particles, we performed coimmunoprecipitation studies using chronically infected PM1 cells, which express on their surface the HIV-1 envelope in its native trimeric conformation (28). Fig. 3B shows that CXCL4 was able to coprecipitate the external HIV-1 envelope glycoprotein, gp120, as well as its uncleaved precursor, gp160, whereas neither glycoprotein was coprecipitated by another HIV-suppressive chemokine, CCL3, which acts via blockade of the CCR5 coreceptor (6). CXCL4 was also able to coprecipitate the recombinant, monomeric form of HIV-1 gp120 (Fig. S7), thus ruling out indirect interactions mediated by other viral or cellular components expressed on the surface of infected PM1 cells.
The CXCL4-Binding Site Is Located Within the gp120 Outer Domain Proximal to the CD4-Binding Site.
To identify the CXCL4-binding site on the surface of gp120, virion-capture competition experiments were performed using a panel of anti-gp120 antibodies of defined specificity or soluble CD4 (sCD4). Fig. 3C shows that a polyclonal antiserum (PB1) raised in goats against a glycosylated recombinant protein representing the complete outer domain (amino acids 295–474) of gp120 from isolate HIV-1 IIIB significantly inhibited virion capture mediated by CXCL4; a similar antiserum (PB1 Sub7) directed against a smaller fragment of the outer domain (amino acids 350–455) also reduced virion uptake, narrowing the CXCL4-binding site to a region spanning the C3, V4, and C4 domains of gp120, inclusive of the core region of the CD4-binding domain (CD4-BD). On the basis of these results, we tested a panel of seven human mAbs directed against the CD4-BD. Two such mAbs (VRC03 and VRC-CH31) markedly reduced CXCL4-mediated capture of HIV-1 particles, whereas one (IgG1b12) showed a modest inhibitory activity and the remaining four (VRC01, VRC-PG04, F105, and 654-D) were ineffective; sCD4 itself did not interfere with CXCL4-mediated virion capture (Fig. 3C). As an additional proof of specificity, mAbs directed against a mannose-dependent epitope on the silent face of the outer domain (2G12) or against CD4-induced epitopes of gp120 (A1G8 and 412D) exerted no inhibitory effects. Altogether, these results indicated that CXCL4 interacts with a region of the gp120 outer domain located proximal to the CD4-BD, but not directly overlapping with this domain. To further confirm the lack of identity between the binding sites of CXCL4 and CD4 in gp120, we tested a mutated gp120 bearing the D368R substitution that abrogates interaction with CD4 (29) showing that it was coimmunoprecipitated by CXCL4 with similar efficiency as wild-type gp120 (Fig. S8). Of note, the two mAbs that blocked CXCL4-mediated HIV-1 virion capture (VRC03 and VRC-CH31) were previously shown to be considerably less effective than two noninhibitory mAbs (VRC01 and VRC-PG04) in competition assays with sCD4 (30), suggesting that their gp120-binding interface extends beyond the CD4-BD, to a contiguous region that presumably encompasses the CXCL4-binding site.
Discussion
Endogenous cytokines and chemokines are key factors in the in vivo control of HIV-1 replication (5, 6). In this study, we identified a unique HIV-suppressive chemokine, CXCL4, which displays an unusually broad spectrum of anti–HIV-1 activity and an unconventional mechanism of action. CXCL4 is a founding member of the CXC chemokine family (31), although it lacks the typical ELR motif and is not chemotactic for polymorphonuclear granulocytes. Similar to other known HIV-suppressive chemokines, CXCL4 interferes with the earliest events in the viral infectious cycle; unlike such chemokines, however, it has the ability to block infection by a wide variety of primary and laboratory-adapted HIV-1 isolates, regardless of their coreceptor specificity. Consistent with this observation, we found that the mechanism of action of CXCL4 is fundamentally different from that of other known HIV-suppressive chemokines because it is not mediated by interaction with a specific viral coreceptor such as CCR5 or CXCR4. Rather, we demonstrated that CXCL4 directly binds to the external viral envelope glycoprotein, gp120, hindering its initial interaction with the cellular membrane. Although CXCL4 has a remarkably high affinity for glycosaminoglycans (GAGs) expressed on the cellular membrane (31), a critical role of cell-surface polyanionic molecules in facilitating the CXCL4/gp120 interaction is unlikely because CXCL4 is able to coprecipitate gp120 even in the complete absence of GAGs. Mapping of the CXCL4-interactive region of gp120 using a panel of specific antibodies or sCD4 permitted us to narrow the CXCL4-binding site to the gp120 outer domain and, more precisely, to a region proximal to the CD4-BD. Whereas two mAbs with a nominal specificity for the CD4-BD, VRC03 and VRC-CH31, effectively competed with CXCL4 for HIV-1 virion capture, neither sCD4 nor VRC01 and VRC-PG04, two CD4-BD–specific mAbs that potently compete with sCD4 for binding to gp120 (30), showed inhibitory effects, indicating that CXCL4 does not block HIV-1 via direct steric hindrance of CD4 binding. Interestingly, VRC03 and VRC-CH31 were shown to be poor competitors with sCD4 for binding to gp120 (30), suggesting that their contact surface in gp120 overlaps with the CD4-BD, but also extends to neighboring regions. Taken together, these findings suggest that the CXCL4-binding site is located proximal to the CD4-BD within the outer domain of gp120, but it does not encompass the CD4 footprint. Further studies, including mapping of the CXCL4-binding domain of gp120, comparative sequence analysis of CXCL4-sensitive and insensitive isolates, mutagenesis studies, and structural analysis of CXCL4/gp120 molecular complexes, will help to dissect the precise molecular events that lead to HIV-1 blockade by this chemokine, paving the way toward the development of specific inhibitors mimicking the inhibitory action of CXCL4.
Although CXCL4 was first described nearly 50 y ago, many aspects of the physiology and pathology of this unique chemokine remain enigmatic. CXCL4 is primarily produced by megakaryocytes and their offspring cells, platelets, which can release it at micromolar concentrations upon activation (24). Thus, it is important to emphasize that the effective antiviral concentrations of CXCL4 against various primary HIV-1 isolates in physiologically relevant target cells such as CD4+ T cells and macrophages (∼25–500 nM) are fully compatible with the physiological levels of this chemokine. These massive amounts of CXCL4 are deemed to be necessary to trigger the initial phase of hemostasis (24); yet, CXCL4 also exerts immunomodulatory and proinflammatory effects, especially by promoting monocyte activation and differentiation (32). These properties of CXCL4 may be of particular clinical relevance when platelets are abnormally activated in the course of inflammatory processes (13–16) including HIV-1 infection where a variety of platelet anomalies have been described associated with sustained platelet activation and increased tendency toward thrombosis (17–22). Thus, CXCL4 may play a dual role in the pathogenesis of HIV-1 disease, on one side by suppressing HIV-1 replication but on the other by fostering immunologic activation, inflammation, and coagulation abnormalities. In this context, binding of HIV-1 virions to CXCL4 deposited on the surface of platelets could act as a trigger for further platelet activation and CXCL4 release, in a similar fashion to the putative effects of HIV-1 binding to CLEC-2 and DC-SIGN on the platelet membrane (23).
In vivo studies on the role of endogenous chemokines in a complex syndrome like AIDS are inherently difficult to perform and can only prove association, not causation. Nevertheless, preliminary results from our laboratory indicate that higher serum levels of CXCL4 in HIV-1–infected individuals correlate with a less advanced clinical stage, suggesting that this cytokine may play a protective role in vivo. Moreover, our identification of primary HIV-1 isolates that are insensitive to CXCL4-mediated inhibition, at least within the dose range used in this study, is compatible with the hypothesis that in vivo escape from CXCL4-mediated inhibition may occur, as previously documented for other HIV-suppressive chemokines (33). Further studies are currently underway in our laboratory to better understand the influence of endogenous CXCL4 production and/or genetic polymorphisms within the CXCL4 locus on the natural history of HIV-1 infection.
Materials and Methods
Cells and Proteins.
Details about the primary cells, immortalized cell lines, recombinant proteins, and antibodies used in this study are given in SI Materials and Methods.
Viral Isolates and Infection Assays.
The HIV-1 isolates used in this study included three laboratory strains [IIIB (X4), BaL, and ADA (R5)] and 13 primary isolates minimally passaged ex vivo exclusively in primary human T cells (97IT6366 was kindly provided by Gabriella Scarlatti (San Raffaele Scientific Institute, Milan, Italy); 92HT599, 92US714, 97ZA009, 98IN017, and 97BR019 were provided by the National Institutes of Health-AIDS Research and Reference Reagent Program (NIH-ARRRP); all of the other isolates were obtained in our laboratory from cultured patient peripheral blood mononuclear cells). The coreceptor-usage phenotype was evaluated using Ghost cells expressing CD4 and each of the indicated coreceptors (provided by the NIH-ARRRP). The three primary HIV-2 isolates used (CBL-20, CDC310342, and 7924A) were obtained from the NIH-ARRRP. The three SIV isolates were all passaged in vivo in macaques: mac251 (a gift of Ronald C. Desrosiers, New England Primate Research Center, Southborough, MA) and mac251/745 in Macaca mulatta; smE660/307 in Macaca nemestrina. HHV-6A (strain GS) is a laboratory strain extensively passaged in primary CD4+ T cells. Acute cell-free HIV-1 infection was performed by addition of the viral stocks (20–50 pg of p24 Gag antigen per well) to duplicate cultures of 105 cells in round-bottom 96-well plates (Nunc) in RPMI medium containing 10% (vol/vol) FBS; adherent macrophages and MAGI-CCR5 cells were infected in 48-well flat-bottom plates (∼5 × 104 per well). The cells were pretreated or not with CXCL4 for 30 min before exposure to the virus. The levels of HIV-1 replication were assessed by measuring the extracellular release of p24 Gag protein in cell-free culture supernatants taken between day 3 and day 7 postinfection using a commercial enzyme immunoassay (Perkin Elmers). The levels of HIV-2 or SIV replication were measured using a commercial ELISA for HIV-2 p27 Gag protein (Beckman Coulter). Infection with HHV-6A was performed in primary human CD4+ T cells activated for 72 h with anti-CD3 and anti-CD28 MAbs and exposed to the viral stock at the approximate multiplicity of infection of 0.1; viral replication was tested by real-time PCR (34) on cell-free culture supernatants harvested at day 5 postinfection. The MAGI assay was performed using MAGI-CCR5 cells, which constitutively express CXCR4 and are engineered to express human CD4 and CCR5. In this assay, single-cycle HIV-1 infection was evaluated at 48 h after exposure to the viral stocks by measuring intracellular β-galactosidase activity according to the protocol provided by the NIH-ARRRP. Half-maximal inhibitory concentrations (IC50) were computed from dose–response curves using PRISM software.
HIV-1 Attachment and Entry Assays.
The HIV-1 attachment and entry assays were performed in primary human CD4+ T cells using two primary HIV-1 isolates, one CXCL4 sensitive (92HT599, R5X4) and one CXCL4 insensitive (07USLR, X4). Primary human CD4+ T cells (106 per replicate; two replicates per treatment) purified by immunomagnetic beads and previously activated ex vivo with PHA and IL-2 for 5–7 d were preincubated for 15 min at room temperature with the inhibitors in 50 μL of serum-free PBS and then exposed to 500 μL the undiluted viral stocks (124 ng of p24/mL for 92HT599; 67 ng of p24/mL for 07USLR) for 4 h at 37 °C in the continuous presence of the inhibitors. One aliquot of untreated cells was incubated for 4 h at 4 °C and served to determine the background signal level (trypsin-insensitive despite low-temperature conditions preventing virus entry). After incubation, the cells were extensively washed with PBS to remove unbound virus and divided in two aliquots: one was treated with prewarmed bovine trypsin (Sigma) at 1.25 mg/mL for 10 min at 37 °C, followed by trypsin inactivation by addition of cold RPMI medium containing 10% (vol/vol) FBS. Both trypsin-treated and -untreated cells were then washed three times with cold PBS, and the final dry pellets were frozen at −80 °C overnight. The pellets were lysed using 100 μL of 0.5% (wt/vol) Triton X-100, and the amount of cell-associated p24 protein was quantified. The specific signal was calculated by subtracting from the p24 levels measured in each test sample the background p24 levels measured in cells incubated at 4 °C and treated with trypsin.
HIV-1 Virion-Capture Assay.
Immunomagnetic beads (4 × 104 per tube) covalently linked to a polyclonal antiserum to rabbit IgG (Invitrogen) were incubated with a polyclonal rabbit IgG antibody to human CXCL4 (Peprotech), washed with PBS containing 0.05% (wt/vol) bovine casein and then loaded with recombinant human CXCL4 (2.5 μg per tube); after removing unbound CXCL4 by repeated washings with PBS, chemokine-armed beads were incubated with 0.5 mL of the viral stocks (isolate HIV-1 IIIB, 20–30 ng of p24 Gag protein per tube; isolate 07USLR, 42 ng of p24 per tube). Inhibitors were preincubated either with the CXCL4-armed beads (anti-CXCL4 antibodies or control goat IgG) or with the viral stock (sCD4, anti-gp120 antibodies); murine anti-CXCL4 mAbs 7951 and 7952 were used at 5 μg/mL, affinity-purified goat anti-CXCL4 at 20 μg/mL, PB1 and PB1 Sub7 at 1:50 dilution, normal goat IgG at either 20 or 100 μg/mL, and sCD4 and human anti-gp120 mAbs at 10 μg/mL. After incubation for 1 h at 4 °C, the beads were washed with PBS casein to remove unbound virus particles and treated with 0.5% Triton X-100 to lyse the captured virions. The amount of captured p24 Gag protein was quantified by enzyme immunoassay.
Coimmunoprecipitation and other assays.
Details about coimmunoprecipitation and other assays are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Francesca Sironi, Monica Tolazzi, and Donald Van Ryk for technical help; John R. Mascola for recombinant HIV-1 gp120 and anti-gp120 mAbs VRC01, VRC03, VRC-CH31, and VRC-PG04; James E. Robinson for mAb 412D; George K. Lewis for mAb B13; Gabriella Scarlatti for HIV-1 isolate 97IT6366; Ronald C. Desrosiers for in vivo passaged SIV mac251; and the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), for the reagents indicated in Materials and Methods. This work was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207314109/-/DCSupplemental.
References
- 1.Fauci AS. Multifactorial nature of human immunodeficiency virus disease: Implications for therapy. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 2.Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: Surviving the fittest. Annu Rev Immunol. 2011;29:295–317. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 4.Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48–57. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 6.Lusso P. HIV and the chemokine system: 10 years later. EMBO J. 2006;25:447–456. doi: 10.1038/sj.emboj.7600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci USA. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 10.Winkler C, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, et al. Identification of RANTES, MIP-1 α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 12.Gear AR, Camerini D. Platelet chemokines and chemokine receptors: Linking hemostasis, inflammation, and host defense. Microcirculation. 2003;10:335–350. doi: 10.1038/sj.mn.7800198. [DOI] [PubMed] [Google Scholar]
- 13.May AE, Seizer P, Gawaz M. Platelets: Inflammatory firebugs of vascular walls. Arterioscler Thromb Vasc Biol. 2008;28:s5–s10. doi: 10.1161/ATVBAHA.107.158915. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava K, et al. Platelet factor 4 mediates inflammation in experimental cerebral malaria. Cell Host Microbe. 2008;4:179–187. doi: 10.1016/j.chom.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iannacone M, et al. Platelets mediate cytotoxic T lymphocyte-induced liver damage. Nat Med. 2005;11:1167–1169. doi: 10.1038/nm1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boccardo P, Remuzzi G, Galbusera M. Platelet dysfunction in renal failure. Semin Thromb Hemost. 2004;30:579–589. doi: 10.1055/s-2004-835678. [DOI] [PubMed] [Google Scholar]
- 17.Sloand E. Hematologic complications of HIV infection. AIDS Rev. 2005;7:187–196. [PubMed] [Google Scholar]
- 18.Scaradavou A. HIV-related thrombocytopenia. Blood Rev. 2002;16:73–76. doi: 10.1054/blre.2001.0188. [DOI] [PubMed] [Google Scholar]
- 19.Shen YM, Frenkel EP. Thrombosis and a hypercoagulable state in HIV-infected patients. Clin Appl Thromb Hemost. 2004;10:277–280. doi: 10.1177/107602960401000311. [DOI] [PubMed] [Google Scholar]
- 20.Lijfering WM, Sprenger HG, Georg RR, van der Meulen PA, van der Meer J. Relationship between progression to AIDS and thrombophilic abnormalities in HIV infection. Clin Chem. 2008;54:1226–1233. doi: 10.1373/clinchem.2008.103614. [DOI] [PubMed] [Google Scholar]
- 21.Holme PA, et al. Enhanced activation of platelets with abnormal release of RANTES in human immunodeficiency virus type 1 infection. FASEB J. 1998;12:79–89. doi: 10.1096/fasebj.12.1.79. [DOI] [PubMed] [Google Scholar]
- 22.Pretorius E, et al. Ultrastructural changes in platelet aggregates of HIV patients: A scanning electron microscopy study. Ultrastruct Pathol. 2008;32:75–79. doi: 10.1080/01913120802034793. [DOI] [PubMed] [Google Scholar]
- 23.Chaipan C, et al. DC-SIGN and CLEC-2 mediate human immunodeficiency virus type 1 capture by platelets. J Virol. 2006;80:8951–8960. doi: 10.1128/JVI.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slungaard A. Platelet factor 4: A chemokine enigma. Int J Biochem Cell Biol. 2005;37:1162–1167. doi: 10.1016/j.biocel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Kinter A, et al. CC-chemokines enhance the replication of T-tropic strains of HIV-1 in CD4(+) T cells: Role of signal transduction. Proc Natl Acad Sci USA. 1998;95:11880–11885. doi: 10.1073/pnas.95.20.11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trkola A, et al. The CC-chemokine RANTES increases the attachment of human immunodeficiency virus type 1 to target cells via glycosaminoglycans and also activates a signal transduction pathway that enhances viral infectivity. J Virol. 1999;73:6370–6379. doi: 10.1128/jvi.73.8.6370-6379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lasagni L, et al. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusso P, et al. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): Failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schall TJ, Bacon KB. Chemokines, leukocyte trafficking, and inflammation. Curr Opin Immunol. 1994;6:865–873. doi: 10.1016/0952-7915(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 32.Scheuerer B, et al. The CXC-chemokine platelet factor 4 promotes monocyte survival and induces monocyte differentiation into macrophages. Blood. 2000;95:1158–1166. [PubMed] [Google Scholar]
- 33.Scarlatti G, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 34.Locatelli G, et al. Real-time quantitative PCR for human herpesvirus 6 DNA. J Clin Microbiol. 2000;38:4042–4048. doi: 10.1128/jcm.38.11.4042-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.