Blood consists of eight principal cell types of diverse function. They have in common a limited life span and an inability to replicate; they therefore must be replaced continuously throughout life. A small pool of pluripotent hemopoietic stem cells that reside in the bone marrow gives rise to all blood cell types through a process of simultaneous lineage commitment, cell proliferation, and differentiation (Fig. 1). In the last two decades, numerous cytokines have been identified as essential extracellular factors in this process (1–4). Some, like stem cell factor, interleukin 3, or granulocyte–macrophage colony-stimulating factor (GM-CSF), contribute to the progeny of many blood cell types. Others, like Epo, G-CSF, or thrombopoietin, exert their principal action on progenitors of a single lineage. Mice in which the genes for lineage- specific cytokines or their receptors are disrupted show a severe deficiency in the blood cells that constitute the progeny of their physiological targets (5, 6). For example, disruption of the EpoR gene results in embryonic lethality caused by the absence of red blood cells; similarly, disruption of the interleukin 7 receptor leads to severe lymphocyte deficiency. Therefore, lineage-restricted cytokine receptors each play an essential and specific role in differentiation. What is it that makes their signaling essential? The report in this issue of Proceedings by Goldsmith et al. (7) builds on a series of reports in the last year (8–11) that is fundamentally altering our view on the role of cytokine receptors in hematopoietic differentiation.
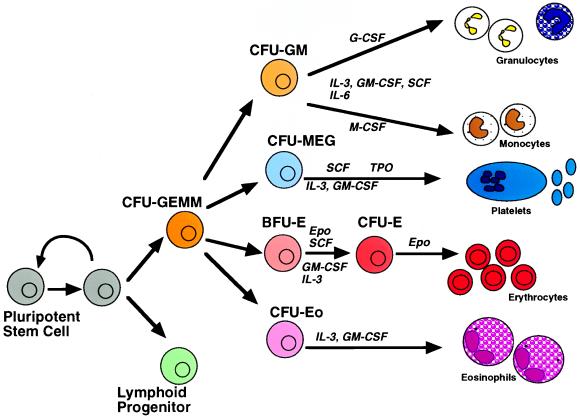
Figure 1.
Structure of the hematopoietic compartment. Bone marrow pluripotent stem cells may either self-renew or give rise to eight different hematopoietic lineages through a gradual process of commitment and differentiation. Some of the cytokines involoved in supporting this process are illustrated. M-CSF, Macrophage colony stimulating factor; SCF, stem cell factor; Epo, erythropoietin; Tpo, thrombopoietin; CFU-GEMM, CFU granulocyte-erythroid-monocyte-megakaryocyte; CFU-GM, CFU granulocyte-monocyte; CFU-me, CFU megakaryocyte; CFU-E, CFU erythroid; CFU-Eo, CFU eosinophil; BFU-E, burst-forming unit erythroid.
Two diametrically opposed hypotheses have been proposed to explain the requirement for cytokine receptor signaling. The stochastic hypothesis suggests that commitment of a progenitor to a particular lineage is a stochastic event, subsequent to which cell differentiation proceeds along a pre-determined program; growth factors are merely required to ensure the survival and proliferation of committed progenitors (12–16). In contrast, the inductive, or instructive, hypothesis, attributes to growth factors a direct role in cell differentiation, predicting that cell fate will be determined by the type of growth factor acting on the cell (17–19). Hybrid hypotheses also have been proposed (20).
The stochastic hypothesis was first proposed by Till, McCulloch, and colleagues in the early 1960s (12). They injected irradiated mice with hematopoietic tissue containing multipotent progenitor cells known as CFU-S (or colony-forming unit-spleen). These seeded the spleen, and over the next 2 weeks each gave rise to a colony containing differentiated blood cells as well as a small number of undifferentiated CFU-S cells that arose as a result of CFU-S self-renewal. If rigid control mechanisms were governing the process of CFU-S cell differentiation and self-renewal, then CFU-S cells of similar genotype in a similar environment should give rise to colonies with similar cell content. However, the number of CFU-S cells per colony was found to be highly variable, with most colonies containing very few, and a few containing many. The frequency of CFU-Ss was best fitted by the γ distribution, which describes a process in which either “birth” (self-renewal) or “death” (differentiation) occur as probabilistic events (ref. 12; Fig. 2). On the strength of their data, Till and McCulloch suggested that the process of commitment for differentiation is stochastic, not closely regulated for individual cells, but occurring with a given probability for the population as a whole (12, 14). A similar analysis suggested that lineage choices subsequent to the commitment of stem cells to differentiation also occurred stochastically (13). The stochastic model was also most consistent with the finding that “twin” colonies, arising after two daughter cells from a single progenitor were separated after cell division and re-plated under identical growth conditions, showed highly variable frequencies of blood cells from different lineages (15).
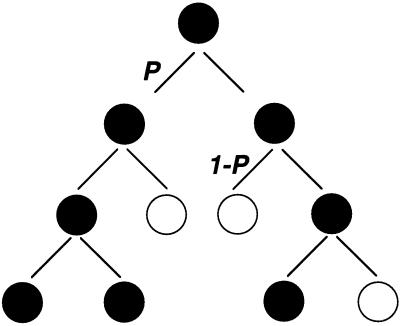
Figure 2.
The stochastic model for cell fate determination. A possible sequence of events or “family tree” in which a single progenitor gives rise to a hematopoietic colony containing a mixture of differentiated and undifferentiated cells. “Birth” (self-renewal, black circles) or “death” (differentiation, white circles) occur as probabilistic events (11). The probability P or “1-P” for each outcome is the same at each generation.
The stochastic model was conceived before identification of most of the currently known cytokines. The purification and cloning of cytokines and their receptors made it possible to test their effects on differentiation directly. Experiments using primary cell cultures give a largely uniform answer: addition of cytokines to the growth media (18), or ectopic expression of their receptors, does not alter the differentiation potential of stem cells. Thus, recombinant expression of either a constitutively active form of EpoR or of the receptor for macrophage growth factor CSF-1 in multi-potent progenitor cells did not bias their differentiation in favor of the erythroid or the macrophage lineages, respectively; instead, there was an increase in the proliferative potential of progenitors (21). Similarly, transgenic expression in mice of a truncated (hypersensitive) EpoR at the ubiquitously expressed HPRT locus has no effect on baseline hematopoietic parameters; when supplemented with exogenous Epo, the mice exhibit a marked increase in multipotential progenitors but no erythrocytosis (22).
In contrast, expression of lineage-restricted cytokines or their receptors in established hematopoietic cell lines sometimes results in apparent lineage-specific gene induction. Thus, ectopic expression of GM-CSF in FDCP-1 cells results in their differentiation into granulocytes and macrophages (20); and, expression of the EpoR in pre-B Ba/F3 cells leads to induction of the erythroid protein glycophorin (23) and to β-globin gene transcription (24, 25). For several cytokine receptors, specific domains are essential for generating a differentiation signal but dispensable for mitogenesis (26–29). Thus, a C-terminal region of the G-CSF receptor cytoplasmic domain is essential for induction of the neutrophil protein myeloperoxidase but not for transducing a mitogenic signal (26), and the absence of this C-terminal region is thought to account for some cases of severe congenital neutropenia (Kostmann syndrome) (30–32).
However, the bulk of the data demonstrating inductive effects by cytokine receptors uses cell line models, which often have an uncertain lineage status and poor differentiation responses; for this reason, they should be interpreted with caution. Important to note, intense study of cytokine receptor signaling has not identified candidate signal transduction proteins that could serve as differentiation-specific signals. For example, signaling molecules recruited and activated by the EpoR include JAK2, STAT5, Grb2, SHC, ras, raf, and mitogen-activated protein kinase, the phosphatases SHP1 and SHP2, protein kinase C, phosphatidylinositol-3 (PI-3) kinase, and phospholipase C-γ (PLC-γ). None of these is unique to the EpoR, and most are activated by a large number of other cytokine receptors as well as other types of growth factor receptors (5, 6).
Recently, two different approaches were taken to test directly whether lineage-restricted cytokine receptors exert essential inductive influences in differentiation. In the first, the native cytokine receptor was replaced with an heterologous receptor. Primary erythroid progenitors were infected with retroviral constructs encoding the prolactin receptor, a nonhematopoietic receptor, and were cultured in vitro in the presence of prolactin and in the absence of Epo. Surprisingly, the prolactin receptor efficiently supported their differentiation into red blood cells (8). The prolactin receptor was similarly able to rescue erythroid progenitors from Epo receptor knockout mice (M.S. and H.F.L., unpublished work). Therefore, there is no requirement for an essential, EpoR-unique signal in erythropoiesis, rejecting an instructive role for the EpoR. The paper in this issue of Proceedings by Goldsmith et al. (7) shows that the cytoplasmic domain of three other receptors in the same subfamily—the growth-hormone receptor, G-CSF receptor, and c-mpl (the receptor for thrombopoietin)—similarly replace the requirement for EpoR in erythroid differentiation in vitro.
Two principal questions arise. First, the cytokine receptors able to replace successfully the EpoR belong to the homodimeric subfamily of cytokine receptors of which EpoR is a member. Which other receptors can replace the EpoR? Apparently not all, because the tyrosine kinase receptor for CSF-1 is not able to efficiently support erythropoiesis in vitro (33).
Second, it is important to define the ability of these heterologous cytokine receptors to replace the EpoR in a living animal. The in vivo assay in the report by Goldsmith et al. (7) provides a partial answer by showing that expression of constitutively activated chimeric cytokine receptors results in extensive erythrocytosis. However, these studies were performed in a wild-type mouse expressing the wild-type EpoR as well as Epo. Erythrocytosis may reflect expansion of a progenitor pool by expression of constitutively activated receptors (22, 34, 35) with subsequent terminal differentiation supported by the wild-type EpoR. A rigorous test for the ability of any heterologous receptor to replace the EpoR in vivo will require its expression in EpoR−/− progenitors.
The ability to substitute for the native cytokine receptor is not unique to the erythroid lineage. Recently, a “knockin” mouse was generated in which the cytoplasmic domain of c-mpl was replaced with that of the G-CSF receptor. Mice homozygous for the resulting chimeric receptor had near-normal platelet counts, indicating that signals emanating from the G-CSF receptor are functionally equivalent to those of c-mpl in supporting growth and terminal differentiation of megakaryocytes (36).
The second approach makes use of transgenic expression of the anti-apoptotic protein bcl-2 on a background of mice mutant for specific cytokine receptors. Mice mutant for the interleukin 7 receptor show severe lymphocyte deficiency; transgenic expression of bcl-2 in T cells of these mice rescued T cell lymphocyte development and reversed the lymphopenic phenotype (9, 10). Similarly, the low macrophage and osteoclast cell numbers found in the op/op (M-CSF deficient) mouse can be restored by transgenic expression of bcl-2 in macrophage progenitors (11). The essential function of these cytokine receptors is therefore to ensure survival of progenitors while lineage commitment and maturation events occur by other means. Of interest, transgenic expression of bcl-2 in erythroid progenitors did not result in Epo-independence (37) which may indicate that the signals generated by EpoR, although not unique, are not exclusively concerned with progenitor cell survival. Alternatively, additional anti-apoptotic pathways may be operating in erythroid cells.
Of interest, this lack of specificity for differentiation appears to have been exploited by leukemogenic oncoproteins. Epo-independent erythroid colony formation is a diagnostic feature of myeloproliferative disorders, and we have demonstrated recently that the BCR/ABL oncoprotein of chronic myeloid leukemia efficiently supports the differentiation of EpoR−/− erythroid progenitors into red blood cells (Ghaffari, S., Gerlach, M., H.F.L., and G.Q.D., unpublished work).
Taken together, these recent studies suggest that the specificity of lineage-restricted cytokines is a result of the unique expression pattern of each receptor by progenitors of a given lineage. Cytokine receptor expression is the result, rather than the cause, of lineage commitment; it allows specific cytokines to selectively rescue and amplify progenitors of a particular lineage according to physiological need. The unique outcome of cytokine receptor signaling is a result of the unique cellular environment in committed progenitors. Differentiation of committed progenitors apparently proceeds along a predetermined program, supported by cytokine receptor-activated “generic” signals common to many cytokine receptors.
The absence of instructive signaling by lineage-restricted cytokine receptors raises the question of how lineage commitment is determined. We cannot exclude a role for as yet unknown extracellular inductive factors in this process. Alternatively, cell fate determination may occur stochastically. The biochemical events that might result in a stochastic lineage choice are completely unknown; similarly, it is not clear whether the probability of a stochastic event may be altered by external factors.
Programs of lineage-specific gene expression are thought to be induced by master regulator transcription factors. SCL is a master regulator of hemopoiesis, without which no blood cells are formed (38–40). GATA-1 may play a similar role in the erythroid lineage because in its absence no red cells are formed (41). Although it is not known how these transcription factors are regulated, it now seems unlikely that cytokine receptor signaling plays an essential role in this process.
Last, if different cytokine receptors activate largely similar signaling molecules, why are their cytoplasmic domains so divergent? The principal function of cytokine receptors appears to be the regulation of cell numbers of a particular lineage; their cytoplasmic domains therefore may be uniquely adapted to provide the required “gain” in response to a proliferative stimulus. Indeed, negative signals that presumably feed back and dampen receptor signaling are known to emanate from cytokine receptors (42–46), which also is demonstrated by several families with hereditary erythrocytosis who carry a truncated EpoR (47, 48). Cytokine receptors may therefore each generate a uniquely different quantitative, rather than qualitative, response, in line with the physiological processes they each regulate.
Acknowledgments
We thank Drs. Stefan Constantinescu and Saghi Ghaffari for discussion and comments on the manuscript. Supported by Grant HL 32262 from The National Institutes of Health and by a grant from Amgen Corporation to H.F.L., National Cancer Institute Grant CA76418–01, and the Burroughs-Wellcome Fund to G.Q.D. and in part by a Howard Hughes postdoctoral fellowship for physicians to M.S.
ABBREVIATIONS
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- CFU-S
colony-forming unit-spleen
Footnotes
The companion to this commentary is published on pages 7006–7011.
References
- 1.Metcalf D. Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 2.Metcalf D. Trends Biochem Sci. 1992;17:286–289. doi: 10.1016/0968-0004(92)90436-d. [DOI] [PubMed] [Google Scholar]
- 3.Clark S C, Kamen R. Science. 1987;236:1229–1237. doi: 10.1126/science.3296190. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa M. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 5.Watowich S S, Wu H, Socolovksy M, Klingmuller U, Constaninescu S N, Lodish H F. Annu Rev Cell Dev Biol. 1996;12:91–128. doi: 10.1146/annurev.cellbio.12.1.91. [DOI] [PubMed] [Google Scholar]
- 6.Socolovsky, M., Constantinescu, S. N., Bergelson, S., Sirotkin, A. & Lodish, H. F. (1998) Adv. Protein Chem., in press. [DOI] [PubMed]
- 7.Goldsmith M A, Mikami A, Yun Y, Liu K D, Thomas L, Pharr P, Longmore G D. Proc Natl Acad Sci USA. 1998;95:7006–7011. doi: 10.1073/pnas.95.12.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Socolovsky M, Dusanter-Fourt I, Lodish H F. J Biol Chem. 1997;272:14009–14013. doi: 10.1074/jbc.272.22.14009. [DOI] [PubMed] [Google Scholar]
- 9.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman I L. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 10.Maraskovsky E, O’Reilly L A, Teepe M, Corcoran L M, Peschon J J, Strasser A. Cell. 1997;89:1011–1019. doi: 10.1016/s0092-8674(00)80289-5. [DOI] [PubMed] [Google Scholar]
- 11.Lagasse E, Weissman I L. Cell. 1997;89:1021–1031. doi: 10.1016/s0092-8674(00)80290-1. [DOI] [PubMed] [Google Scholar]
- 12.Till J E, McCulloch E A, Siminovitch L. Proc Natl Acad Sci USA. 1964;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korn A P, Henkelman R M, Ottensmeyer F P, Till J E. Exp Hemat. 1973;1:362–375. [PubMed] [Google Scholar]
- 14.Nakahata T, Gross A J, Ogawa M. J Cell Physiol. 1982;113:455–458. doi: 10.1002/jcp.1041130314. [DOI] [PubMed] [Google Scholar]
- 15.Suda T, Suda J, Ogawa M. Proc Natl Acad Sci USA. 1984;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairbairn L J, Cowling G J, Reipert B M, Dexter T M. Cell. 1993;74:823–832. doi: 10.1016/0092-8674(93)90462-y. [DOI] [PubMed] [Google Scholar]
- 17.Borzillo G V, Ashmun R A, Sherr C J. Mol Cell Biol. 1990;10:2703–2714. doi: 10.1128/mcb.10.6.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metcalf D. Proc Natl Acad Sci USA. 1991;88:11310–11314. doi: 10.1073/pnas.88.24.11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukunaga R, Ishizaka-Ikeda E, Nagata S. Cell. 1993;74:1079–1087. doi: 10.1016/0092-8674(93)90729-a. [DOI] [PubMed] [Google Scholar]
- 20.Just U, Stocking C, Spooncer E, Dexter T M, Ostertag W. Cell. 1991;64:1163–1173. doi: 10.1016/0092-8674(91)90271-y. [DOI] [PubMed] [Google Scholar]
- 21.Pharr P N, Ogawa M, Hofbauer A, Longmore G D. Proc Natl Acad Sci USA. 1994;91:7482–7486. doi: 10.1073/pnas.91.16.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirby S L, Cook D N, Walton W, Smithies O. Proc Natl Acad Sci USA. 1996;93:9402–9407. doi: 10.1073/pnas.93.18.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jubinsky P T, Nathan D G, Wilson D J, Sieff C A. Blood. 1993;81:587–591. [PubMed] [Google Scholar]
- 24.Liboi E, Carrol M, D’Anderea A D, Mathey-Prevot B. Proc Natl Acad Sci USA. 1993;90:11351–11355. doi: 10.1073/pnas.90.23.11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiba T, Nagata Y, Machide M, Kishi A, Amanuma H, Sugiyama M, Todokoro D. Nature (London) 1993;362:646–648. doi: 10.1038/362646a0. [DOI] [PubMed] [Google Scholar]
- 26.Fukunaga R, Ishizaka-Ikeda E, Nagata S. Cell. 1993;74:1079–1087. doi: 10.1016/0092-8674(93)90729-a. [DOI] [PubMed] [Google Scholar]
- 27.Yoshikawa A, Murakami H, Nagata S. EMBO J. 1995;14:5288–5296. doi: 10.1002/j.1460-2075.1995.tb00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson S E, Starr R, Novak U, Hilton D J, Layton J E. J Biol Chem. 1996;271:26947–26953. doi: 10.1074/jbc.271.43.26947. [DOI] [PubMed] [Google Scholar]
- 29.Porteu F, Rouyez M-C, Cocault L, Benit L, Charon M, Picard F, Gisselbrecht S, Souyri M, Dusanter-Fourt I. Mol Cell Biol. 1996;16:2473–2482. doi: 10.1128/mcb.16.5.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong F, Hoefsloot L H, Schelen A M, Broeders L C A M, Meijer Y, Veerman A J P, Touw I P, Lowenberg B. Proc Natl Acad Sci USA. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong F, Hoefsloot L H, Schelen A M, Broeders C A, Meijer Y, Veerman A J, Touw I P, Lowenberg B. Proc Natl Acad Sci USA. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong F, Brynes R K, Tidow N, Welte K, Lowenberg B, Touw I P. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 33.McArthur G A, Rohrschneider L R, Johnson G R. Blood. 1994;83:972–981. [PubMed] [Google Scholar]
- 34.Longmore G D, Lodish H F. Cell. 1991;67:1089–1102. doi: 10.1016/0092-8674(91)90286-8. [DOI] [PubMed] [Google Scholar]
- 35.Pharr P N, Hankins D, Hofbauer A, Lodish H F, Longmore G D. Proc Natl Acad Sci USA. 1993;90:938–942. doi: 10.1073/pnas.90.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoffel R, Ledermann B, de Sauvage F J, Skoda R C. Blood. 1997;90:535. (abstr.). [Google Scholar]
- 37.Lacronique V, Varlet P, Mayeux P, Porteu A, Gisselbrecht S, Kahn A, Lacombe C. Blood. 1990;90:3050–3056. [PubMed] [Google Scholar]
- 38.Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey R P, Metcalf D, Begley C G. Proc Natl Acad Sci USA. 1995;92:7075–7079. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shivdasani R A, Mayer E L, Orkin S H. Nature (London) 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 40.Mead P E, Kelley C M, Hahn P S, Piedad O, Zon L I. Blood. 1997;90:2563. (abstr.). [Google Scholar]
- 41.Shivdasani R A, Orkin S H. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 42.Klingmuller U, Lorenz U, Cantley L C, Neel B G, Lodish H F. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 43.Yoshimura A, Ohkubo T, Kiguchi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starr R, Willson T A, Viney E M, Murray L J L, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, et al. Nature (London) 1997;387:917–922. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 45.Endo T, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. Nature (London) 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 46.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Nature (London) 1997;387:924–2929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 47.de la Chapelle A, Sistonen P, Lehvaslaiho H, Ikkala E, Juvonen E. Lancet. 1993;341:82–84. doi: 10.1016/0140-6736(93)92558-b. [DOI] [PubMed] [Google Scholar]
- 48.de la Chapelle A, Traskelin A-L, Juvonen E. Proc Natl Acad Sci USA. 1993;90:4495–4499. doi: 10.1073/pnas.90.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]