Abstract
The six bacteriophage T7 tail fibers, homo-trimers of gene product 17, are thought to be responsible for the first specific, albeit reversible, attachment to Escherichia coli lipopolysaccharide. The protein trimer forms kinked fibers comprised of an amino-terminal tail-attachment domain, a slender shaft, and a carboxyl-terminal domain composed of several nodules. Previously, we expressed, purified, and crystallized a carboxyl-terminal fragment comprising residues 371–553. Here, we report the structure of this protein trimer, solved using anomalous diffraction and refined at 2 Å resolution. Amino acids 371–447 form a tapered pyramid with a triangular cross-section composed of interlocked β-sheets from each of the three chains. The triangular pyramid domain has three α-helices at its narrow end, which are connected to a carboxyl-terminal three-blade β-propeller tip domain by flexible loops. The monomers of this tip domain each contain an eight-stranded β-sandwich. The exact topology of the β-sandwich fold is novel, but similar to that of knob domains of other viral fibers and the phage Sf6 needle. Several host-range change mutants have been mapped to loops located on the top of this tip domain, suggesting that this surface of the tip domain interacts with receptors on the cell surface.
Keywords: bacterial viruses, caudovirales, crystallography, infection, Podoviridae
Bacteriophages (bacterial viruses or phages) are important biological model systems and, because of the high specificity for their host bacteria, have found application in phage typing, food security, and phage therapy (1). Escherichia coli phage T7 is a member of the Podoviridae family of the Caudovirales (tailed phages) order (2). T7 is composed of an icosahedral capsid with a 20-nm short tail at one of the vertices (3, 4). The capsid is formed by the shell protein gene product (gp) 10 and encloses a DNA of 40 kb. A cylindrical structure composed of gp14, gp15, and gp16 is present inside the capsid (5), attached to the special vertex formed by the connector, a circular dodecamer of gp8 (6). Gp11 and gp12 form the tail; gp13, gp6.7, and gp7.3 have also been shown to be part of the virion and to be necessary for infection, although their location has not been established (7, 8). Although extensive electron microscopy studies have been performed on phage T7 (3–6, 9), crystallographic studies have so far been limited to its nonstructural proteins.
The main portion of the tail is composed of gp12, a large protein of which six copies are present (10); the small gp11 protein is also located in the tail (5). Attached to the tail are six fibers, each containing three copies of the gp17 protein. T7 tail fibers are elongated homo-trimers, which are responsible for initial, reversible, host cell recognition. A second, irreversible, decision-making interaction with the bacterial membrane is presumably mediated by one or more of the tail-tube proteins. DNA transfer into the host is then mediated by an extension formed by gp14–16 (7, 8, 11). Previously, we have reported the production of well-diffracting crystals of the phage-distal carboxyl-terminal domain of gp17 containing a 45-residue purification tag (12). Here we present its structure, solved using four-wavelength anomalous diffraction analysis of a mercury derivative and refined against data collected from crystals of two different forms.
Results
Structure Solution.
Crystals of gp17(371–553) belonging to two different space groups were obtained, P212121 and C2221, and a multiwavelength anomalous diffraction dataset was collected on a crystal of form P212121 derivatized with methylmercury chloride (12). The derivative was not isomorphous to the native crystal and the native dataset was not used in phase determination. Nine heavy atom sites were identified, six of which are located near the two cysteines present in each of the three chains (Cys408 and Cys499). Two sites are near Asp442 (of chains A and B) and one near His433 of chain A. After phasing and solvent flattening at 2.7 Å resolution, a readily interpretable map was obtained in which 527 residues could be automatically traced. This model was used as input in a molecular replacement with the 1.9 Å resolution dataset obtained of the P212121 crystal form. Automatic tracing at this resolution produced a model containing 528 residues, which was completed and corrected manually and to which solvent atoms were added. An intermediate protein model was used to solve the C2221 crystal form structure at 2.0 Å resolution, which was also completed. Both structures were refined to R-factors of 15% and free R-factors of 20% and have few residues in unlikely regions of the Ramachandran plot (Table S1). One amino acid, Gly522, is in a very uncommon conformation in all three chains of both structures (all have the backbone torsion angles ϕ around 165° and ψ around −90°); however, it has convincing electron density in each case and is thus likely forced into this conformation by the rest of the structure.
Overview of the Structure.
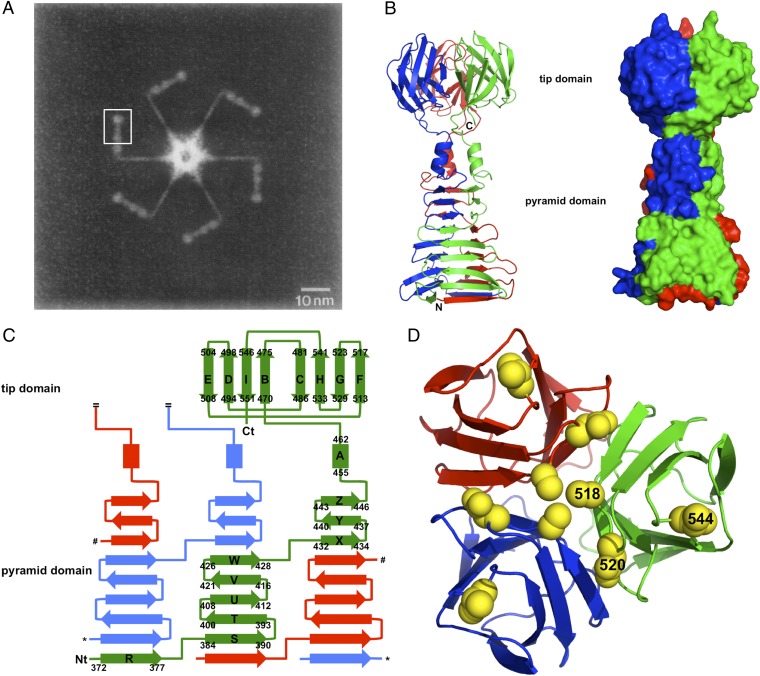
Electron micrographs revealed gp17 to be an extended protein, with a proximal rod about 16-nm long and 2 nm in diameter, a sharp kink, and a distal rod about 15-nm long and with a diameter that varies between 3 and 5 nm (Fig. 1A) (3). The distal rod can be divided into four “nodules” of unequal size, which were estimated to contain residues 268–365, 366–432, 433–456, and 466–553, respectively. The crystal structure of the gp17(371–553) fragment corresponds to the most distal three of these nodules (Fig. 1). The structure can be divided into two parts: a globular “tip” domain (residues Ala465-Glu553), corresponding to the fourth and last nodule, and a tapered interlocked mainly β-structured pyramid domain (residues Gly371 to Trp454) which, from the electron microscopy analysis, was interpreted as forming the second and third nodules. The pyramid domain contains three short α-helices (one from each monomer) at its thinner end.
Fig. 1.
Crystal structure of bacteriophage T7 gp17(371–553). (A) Composite negatively stained electron microscopy image of the tail-complex of bacteriophage T7. The region of one of the six symmetry-equivalent positions where our structure fits into this complex is boxed (reprinted from ref. 3, with permission from Elsevier, http://www.sciencedirect.com/science/journal/00222836). (B) Ribbon diagram (Left) and space-filled representation (Right) of gp17(371–553). The three monomers are colored red, green, and blue. The termini of the red monomer are indicated. (C) Topology diagram. β-Strands are shown as arrows, α-helices as rectangles. Secondary structure elements are labeled and their start and end residues are indicated for the green monomer, as are the termini. Only one of the three tip domain monomers is shown. Asterisks and hash signs indicate connections in the other monomers that were removed for clarity. (D) Top view of the tip domain with the side chains of residues Ala518, Asp520, and Val544 shown in space-filled representation (labeled in the green monomer only).
When the trimeric structures from the two crystal forms are superimposed, a rms difference of 1.2 Å between their C-α positions is observed (residues 373–553 of all chains were included in the calculation). Amino acids N-terminal to residue 373 show different conformations, probably as a result of the different crystal packing in the two structures. When individual chains are compared, another local difference becomes apparent in the region around β-turn Asp390-Arg393 in one of the chains, which forms a crystal contact in the C2221 crystal form. The tip domains alone (amino acids 465–553) superpose with an rmsd of only 0.2 Å and the pyramid domain (amino acids 373–464) superposes with an rmsd of only 0.5 Å, suggesting the short loop (Val464-Lys466) between the tip and pyramid domains is somewhat flexible. This flexibility may be of importance in the infection process. Adenovirus fiber contains a hinge region between the shaft and head domains (13); mammalian reovirus fiber (σ1 protein) has a hinge region between the last and second-to-last triple β-spiral repeats of the shaft (14). In the case of adenovirus fiber, flexibility has been shown to be important for the infection process, presumably to allow secondary receptor interaction (15). Flexibility of the T7 tail fiber may also be necessary to allow for tail conformational changes during the infection process.
Pyramid Domain.
The pyramid domain is composed of three concave nine-stranded mixed β-sheets stacked against each other, forming a tapered pyramid with a triangular cross-section. The taper is the result of the β-strands at the bottom of the pyramid being longer (up to 6 amino acids) than those near the top (down to three residues). Each β-sheet involves strands contributed by all three chains; interactions between β-strands are antiparallel between strands from the same monomer, but interaction between β-strands from neighboring monomers are parallel. Each β-sheet consists of one strand, R, from the first monomer, followed by five strands, STUVW, from the next monomer, and capped by three strands, XYZ, from the third. The loops connecting the β-strands vary from short β-turns to longer loops; all loops are well ordered in both crystal forms and interact extensively with other loops from the same or from neighboring monomers. The center of the domain contains exclusively hydrophobic and aromatic side-chains; each β-strand contributes one or two central side-chains to this core. In the loops and at the beginning and end of the β-strands, interactions are mixed and, apart from hydrophobic interactions, many polar interactions are formed. At the top of the pyramid domain three α-helices are located, one from each protein chain. Leu455 and Leu459 residues from the α-helices project into the center, forming a small hydrophobic core, capped at the top by Phe463.
Carboxyl-Terminal Globular Tip Domain.
The α-helices are connected to the tip domain by a loop (Val464-Lys466) containing both polar and apolar residues. This loop surrounds a solvent-filled central cavity between the tip and pyramid domains. The presence of this solvent-filled cavity is further indication of possible flexibility between the tip and pyramid domains.
The tip domain of each monomer (amino acids 464–553) forms a β-sandwich with the topology shown in Fig. 1C. The β-sandwich contains two sheets of four β-strands each, BIDE and CHGF. β-Strands B and C are on the outside of the trimer, E and F on the inside. A search using the program DALI (16) did not turn up β-sandwich domains with the same topology (see below). As in the β-structured pyramid domain, all loops between β-strands are well ordered in both crystal forms and interact extensively with neighboring loops. Interactions between the tip monomers are of mixed nature, involving salt-bridges, hydrogen bonds, solvent molecules, and van der Waals interactions. The end of the protein chain is locked firmly into place by two salt-bridges between Glu551 and Arg508 of a neighboring chain and between Glu553 (the very C-terminal residue) and Lys468 of the other neighboring chain. This position likely prevents attack of the C terminus by proteases and contributes to trimer stability.
Discussion
Phages belonging to the Caudovirales order attach to host bacteria with the end of their tails. Primary, generally reversible, recognition is via tail-spike proteins or tail fibers on the side of the tail; at this point the phage is not yet committed to DNA injection. Positive recognition leads to central tail proteins productively attaching to the host membrane and DNA injection. The tail-spike of the podovirus P22 has been studied extensively in terms of carbohydrate binding, hydrolysis, folding, and assembly (17, 18). Much is also known about assembly and function of the complex fibers of the myovirus T4 (19–21). Siphoviruses like T5 and λ contain less-studied side tail fibers (22, 23) which, like the T4 fibers, do not exhibit receptor-hydrolysis activity. Here we have presented unique high-resolution structural information on a podoviral tail fiber that also does not hydrolyze its receptor.
Stability and Folding of gp17(371–553).
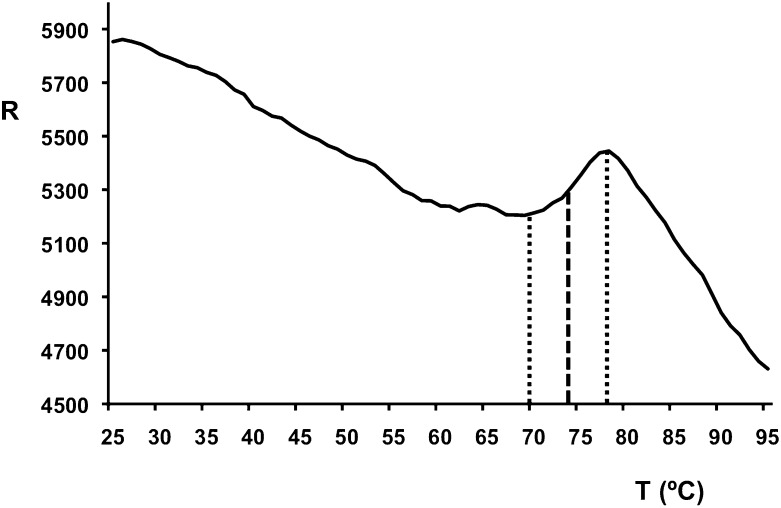
The surface area of a monomer is around 12 × 103 Å2, of which more than 40% is buried upon trimer formation (5.2 × 103 Å2). The calculated dissociation energy of the trimer is 125 kcal/mol (24) and contains more than 120 intermonomer hydrogen bonds and nearly 40 potential intermonomer salt-bridges. When the pyramid domain trimer (Gly371-Phe463) is considered, the surface area of a monomer is 7.4 × 103 Å2, of which nearly half (3.6 × 103 Å2) is buried. The calculated dissociation energy of the pyramid domain trimer is 90 kcal/mol and contains more than 90 intermonomer hydrogen bonds and 8 potential intermonomer salt-bridges. The surface area of a tip domain monomer (Val464-Glu553) is 4.7 × 103 Å2, of which 1.4 × 103 Å2 is buried (around 30%). The calculated dissociation energy of the tip domain trimer is 17 kcal/mol and contains 27 intermonomer hydrogen bonds and 30 potential intermonomer salt-bridges. The large buried surface area explains the stability of trimeric gp17(371–553), as revealed by its high melting temperature (around 74 °C) (Fig. 2) and the fact that it does not dissociate into monomers in SDS/PAGE, unless previously boiled in SDS-containing buffer (12). Our structure shows there are no disulphide bonds in the C-terminal construct, and the crystallized fragment contains the only two cysteines of the gp17 sequence.
Fig. 2.
Thermal stability assay. The relative fluorescence emission intensity (R) is plotted as a function of the temperature (x axis, °C). A melting temperature of around 74 °C was estimated (midpoint between the baseline and the point with maximum fluorescence intensity).
The C-terminal tip domain is the only domain of the current structures where the monomer has a potentially independent fold and where the three chains do not intertwine. The surface buried between monomers and calculated dissociation energy is also smaller. This finding suggests gp17 folding may begin with the spontaneous formation of a monomeric carboxyl-terminal β-barrel. Interaction of three β-barrels could then lead to trimer formation. The remaining region of the fiber would then “zip up” to form the intact trimer. This folding pathway would be the same as that proposed for adenovirus fiber (25) and consistent with the observation that amber mutants in the C-terminal domain do not incorporate truncated gp17 in the virions (10). Gp17 does not appear to require specific chaperones for folding like phage T4 fibers do (26).
Comparison with Other Proteins.
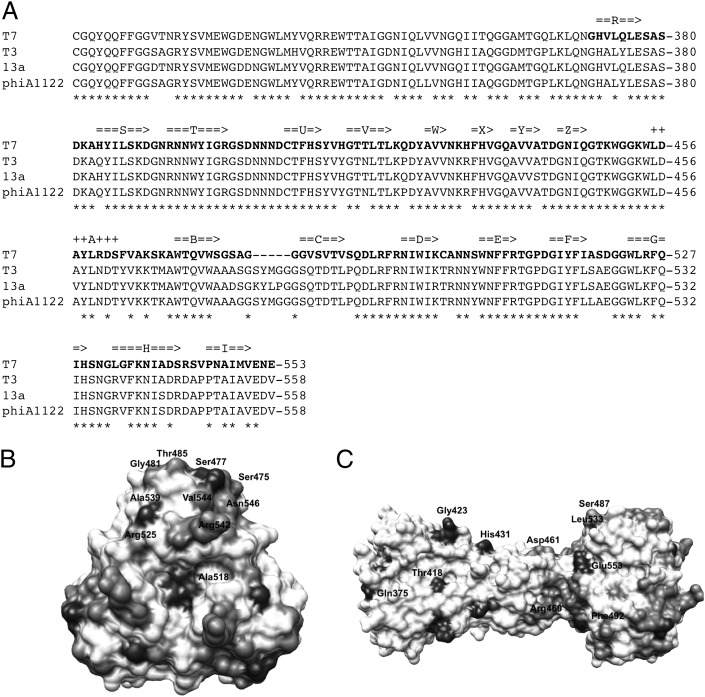
When the sequence of phage T7 gp17 is compared with protein sequences in databases, a strong similarity (> 85% sequence identity) is observed with gp17 of E. coli phage T3, Yersinia pestis phage PhiA1122, and enterobacteria phage 13a. Sequence similarity is also observed with fibers of phages from other bacterial species (Kluyvera, Salmonella, Pseudomonas, Vibrio, Klebsiella, and others). The phage attachment domain (amino acids 1–150) belongs to domain family PHA00430 and, of the gp17 domains, has the most sequence homologs in the database, consistent with the fact that many T7-like phages have the same mechanism of attaching the fiber to the phage. The tip domain has fewer sequence homologs, only T3, PhiA1122, enterobacteria phages 13a, BA14, 285P, and Yersinia phages Yep-Phi, Berlin, and Yepe, consistent with the well-known fact that many phages have evolved their C-terminal receptor attachment domain to adapt to different hosts by exchange with other phages (27). An illustrative example is E. coli phage K1F, which has a homologous phage-attachment domain but a different C-terminal domain containing endo-N-acetylneuramidase activity to digest the E. coli K1 capsular polysaccharide (28, 29). Only a few phages appear to have evolved by mutation maintaining the same structural framework. Residues 359–524 of T7 gp17 (encompassing all but the final 29 residues of our structure) have been classified in Pfam domain gp37_C, which includes a subset of the highly diverged C-terminal regions of the gp37-type tail-fiber proteins of T4-like phages, the subset found in phages AR1 and Ac3 (30). Thus, amino acid sequence comparisons suggest that the tapered triangular pyramid and part of the tip structure of T7 tail fibers may be structural building blocks used in fibers of phages of the Myoviridae as well as the Podoviridae family.
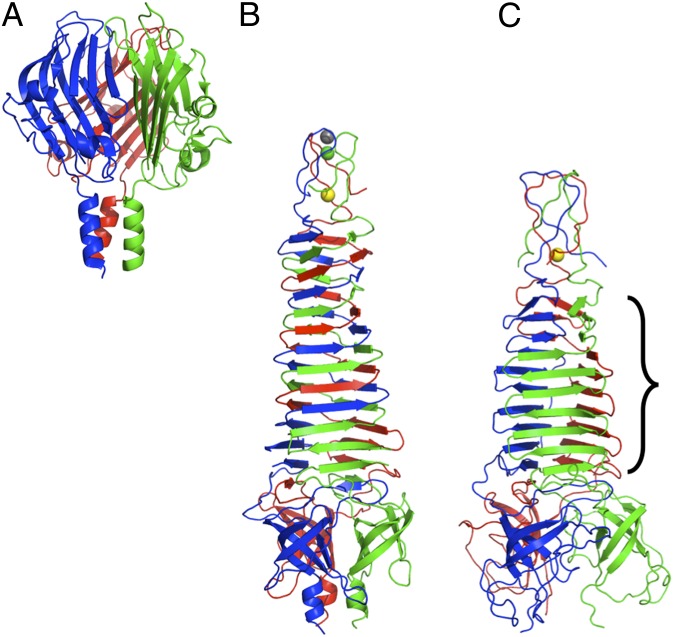
When searches are conducted either with the whole structure presented here or with separate domains, no structural homologs with the same topology are found in the PDB database. Adenovirus fiber (13), reovirus fibers (14, 31), bacteriophage PRD1 P5 (32), and phage Sf6 cell-penetrating needle (33) all have a β-structured C-terminal domain of similar size and organization as gp17 has, but with a different topology. The Sf6 cell-penetrating needle knob domain (Fig. 3A) has the most similar topology, with two small extra β-strands (C1 and C2) and a straight swap of the E and G strands compared with our structure. However, their stalk domains contain only triple β-spiral folds or a triple coiled coil (34, 35), without an intervening β-helical domain, such as the pyramid domain in the present structure. The receptor-binding proteins of lactococcus phages p2 and TP901-1 also have a similar C-terminal domain attached to a short β-helical stalk (36, 37). The bacteriophage T4 short tail-fiber (gp12) and long tail-fiber (gp37) C-terminal domains have different folds, consisting of three intertwined monomers rather than composed of individually folded monomeric domains (21, 38), and T4 fibritin (gpwac) and the phage P22 cell-penetrating needle (gp26) have a much smaller trimerisation domain (39, 40).
Fig. 3.
Structures of the cell-penetrating tail needle knob domain of phage Sf6, PDB-code 3RWN (A), gpV of phage P2, PDB entries 3QR7 and 3QR8 (B), and gp138 of phage phi92, PDB codes 3PQH and 3PQI (C). Iron, calcium, and chloride ions are shown as yellow, gray, and green balls, respectively. The region of gp138 that is topologically the same as amino acids 384–446 of phage T7 gp17 is indicated.
The pyramid domain, with its interlocked trimeric β-helix, also does not have any exact topological equivalent in the structure database. Comparable structures include the triple β-helix regions of phage T4 gp12 (41) and gp5 (42), the K1F endosialidase (43) and endo-N-acetylneuramidase (29), streptococcal (pro)phage HylP1 and Hylp2 (44, 45), the phage P22 cell penetrating needle gp26 (40), and phage Phi29 tail-spike (46). However, the most similar structures are those of phage P2 gpV (Fig. 3B) and phage Phi92 gp138 (Fig. 3C) (47, 48), which form the tip of inner tail tube and are the first proteins to pierce the membrane in these myoviruses. Both proteins contain a tapered β-helix, strongly intertwined in the case of gpV and interlocked like gp17 in the case of gp138. Eight β-strands (from sheets STUVW and XYZ) have structural equivalents in gp138. However, both gpV and gp138 contain a small apex domain with a central iron ion instead of the α-helical region, and lack a globular tip domain.
Receptor Binding.
Initial, reversible, binding of phage T7 to bacteria is mediated by the interaction of its six gp17 tail fibers with LPS (7). This interaction is presumably followed by a secondary, irreversible attachment of the tail to an unknown receptor. The relative importance of the two interactions in host-range determination is not known; gp17 may just keep the phage near the bacterial surface to make productive tail interactions with its receptor more likely (i.e., 2D diffusion vs. 3D diffusion). However, Heineman et al. (49) identified two mutants of bacteriophage T7 gp17 involved in host avoidance: a Asp520 to Glu change adapted the phage to avoid E. coli B and a Val544 to Ala change adapted the phage to avoid E. coli K12. Furthermore, Garcia et al. (50) identified a spontaneous host-range change mutant in the highly homologous phage PhiA1122 fiber in this region (Leu523 to Ser, which aligns to Ala518 of bacteriophage T7 gp17). Ala518 and Asp520 are located in the EF-loop and Val544 in the GH-loop of the gp17 tip domain; both are located on the top of the tip trimer in our structure (Fig. 1D). When the sequence of bacteriophage T7 gp17 is aligned with that of tail fibers of E. coli phage T3, Salmonella enteridis phage 13a and Y. pestis phage PhiA1122 (Fig. 4), the main differences are also observed in residues that are located in the four loops at the top of the tip domain (BC-, DE-, FG-, and HI-loops). Taken together, this information strongly suggests that the tip domain of gp17 has an important role for host-range determination, presumably by binding to a specific LPS region that differs between bacterial strains or that may be occluded in some strains and available for interactions in others.
Fig. 4.
Sequence conservation of gp17. (A) Alignment of the sequence of bacteriophage T7 gp17 with its homologs from E. coli phage T3, S. enteridis phage 13a, and Y. pestis phage PhiA1122. Amino acids present in our structures are shown in bold. Residues that are identical in all four proteins are marked with asterisks. Secondary structure elements identified in our structure are also indicated, beta-strands with arrows and α-helices with plus-signs. (B and C) Sequence conservation mapped on the structure. The color scale is from white (absolutely conserved) to black (no conservation). A top view (B) and a side view (C) are shown. Residues that are not conserved, and thus may be important for host range discrimination, are indicated.
Another possible site for receptor interaction is the concave eight-stranded β-sheet of the pyramid domain. Although there is no biochemical or mutation evidence, the concave shape and the presence of several aromatic residues is suggestive. In this region, in the P212121 crystal form, Tyr385, Tyr397, and Tyr413 interact with a trimethylamine-N-oxide molecule from the crystallization solution in each of the three monomers, but in the C2221 crystal form Tyr425 interacts with a carbonate ion in two of three monomers. Site-directed mutagenesis experiments combined with competition experiments, identification of the LPS region to which T7 binds, and cocrystallization studies with the relevant LPS fragments will be needed to identify the specific gp17-residues from the tip or pyramid domain responsible for receptor binding. If and how this initial interaction is communicated to the phage to trigger irreversible binding and DNA injection is also an interesting future subject of study.
Conclusion
We have solved the structure of the C-terminal domain of the phage T7 tail fiber, gp17(371–553). The structure contains two domains with hitherto unseen topologies, provides insight into the reason for the stability of the protein, and suggests regions that may be involved in receptor binding. The structural data may also contribute to applications of bacteriophages in, for example, bacteria typing or phage therapy, through the rational design of mutants binding different receptors or the engineering of artificial, chimeric phage fibers.
Materials and Methods
Thermal Stability Assay.
Thermal stability of the protein was measured by following the fluorescence of the dye Sypro orange in the presence of 0.01 mM gp17(371–553) as a function of temperature as described in Dupeux et al. (51). The melting temperature was estimated as the temperature corresponding to the midpoint between the baseline and the point with maximum fluorescence intensity (Fig. 2).
Crystallographic Structure Solution and Refinement.
Crystallogenesis and crystallographic data collection have been described previously (12). Datasets of the P212121 mercury derivative collected at four different wavelengths (peak, inflection point, high energy remove, low energy remote) were input into AUTOSHARP (52). Three mercury sites were identified by the SHELXC/D programs (53) and phases were refined using data between 20.0 and 2.7 Å with AUTOSHARP, which found additional sites and rejected sites, settling on a final list of nine heavy atom sites. Further solvent flattening and histogram matching was done with SOLOMON (54) and automated building proceeded using BUCCANEER (55) at 2.7 Å resolution. This model was used for molecular replacement into the higher resolution, native data using the program MOLREP (56). The model was then input into the ARP-WARP (57) auto-trace mode using the data to 1.9 Å resolution for the P212121 crystal form. Adjustment of the model, addition of extra amino acids and solvent molecules was done with COOT (58). Refinement was done using the REFMAC5 program (59). The final model contains 548 amino acids, 1 poly-ethyleneglycol fragment, 1 Tris molecule, 3 trimethylamine-N-oxide molecules, and 754 water molecules. A partially complete model refined against the high-resolution P212121 data were used to solve the C2221 structure by molecular replacement, which was extended and refined as above. This final model contains 552 amino acids (including one amino acid from the purification tag; the rest of the purification tag is apparently disordered), 2 CO3 molecules, and 910 water molecules. Data statistics were published previously (12); phasing, refinement, and model statistics can be found in Table S1. Validation was performed with MOLPROBITY (60) and protein structure figures were prepared using PYMOL (Schroedinger) and University of California at San Francisco CHIMERA (61).
Supplementary Material
Acknowledgments
We thank Ana Cuervo and José L. Carrascosa for bacteriophage T7 DNA and introducing us to T7 biology; Antonio L. Llamas-Saiz, José M. Otero, and José-Ignacio Baños-Sanz for diffraction testing of crystals; Silvia Russi and Hassan Belrhali of the European Molecular Biology Laboratory for help with data collection at BM14; Petr Leiman for careful reading of the manuscript; and Florine Dupeux, Martin Rower, Gael Seroul, Delphine Blot, and José A. Marquez for performing the thermofluor and high-throughput crystallization experiments (https://embl.fr/htxlab/). We acknowledge the use of the High-Throughput Crystallization Laboratory at the European Molecular Biology Laboratory Grenoble outstation (France), which receives funding from the European Community’s Seventh Framework Program under Contract 227764. This research was funded by research Grants BFU2008-01588 and BFU2011-24843 from the Spanish Ministry of Science and Innovation; a grant from the Bill and Melinda Gates Foundation through the Grand Challenges Exploration initiative (to M.J.v.R.); and a Formación del Profesorado Universitario predoctoral contract of the Spanish Ministry of Education (to C.G.-D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: Coordinates and structure factors for the P212121 and C2221 crystal forms have been submitted to the Protein Data Bank, www.pdb.org (PDB ID codes 4A0T and 4A0U, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119719109/-/DCSupplemental.
References
- 1.Marks T, Sharp R. Bacteriophages and biotechnology: A review. J Chem Technol Biotechnol. 2000;75:6–17. [Google Scholar]
- 2.Maniloff J, Ackermann HW. Taxonomy of bacterial viruses: Establishment of tailed virus genera and the order Caudovirales. Arch Virol. 1998;143:2051–2063. doi: 10.1007/s007050050442. [DOI] [PubMed] [Google Scholar]
- 3.Steven AC, et al. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. http://www.sciencedirect.com/science/journal/00222836. Accessed May 14, 2012. [DOI] [PubMed] [Google Scholar]
- 4.Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem. 2003;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- 5.Cerritelli ME, et al. A second symmetry mismatch at the portal vertex of bacteriophage T7: 8-Fold symmetry in the procapsid core. J Mol Biol. 2003;327:1–6. doi: 10.1016/s0022-2836(03)00117-7. [DOI] [PubMed] [Google Scholar]
- 6.Agirrezabala X, et al. Structure of the connector of bacteriophage T7 at 8A resolution: Structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347:895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Molineux IJ. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol Microbiol. 2001;40:1–8. doi: 10.1046/j.1365-2958.2001.02357.x. [DOI] [PubMed] [Google Scholar]
- 8.Serwer P, Wright ET, Hakala KW, Weintraub ST. Evidence for bacteriophage T7 tail extension during DNA injection. BMC Res Notes. 2008;1:36. doi: 10.1186/1756-0500-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ionel A, et al. Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J Biol Chem. 2011;286:234–242. doi: 10.1074/jbc.M110.187211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp P, Garcia LR, Molineux IJ. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology. 2005;340:307–317. doi: 10.1016/j.virol.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 11.Chang CY, Kemp P, Molineux IJ. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology. 2010;398:176–186. doi: 10.1016/j.virol.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Doval C, van Raaij MJ. Crystallization of C-terminal domain of the bacteriophage T7 fibre gp17. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;68:166–171. doi: 10.1107/S1744309111051049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature. 1999;401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 14.Chappell JD, Prota AE, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002;21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu E, et al. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol. 2003;77:7225–7235. doi: 10.1128/JVI.77.13.7225-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andres D, Baxa U, Hanke C, Seckler R, Barbirz S. Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection. Biochem Soc Trans. 2010;38:1386–1389. doi: 10.1042/BST0381386. [DOI] [PubMed] [Google Scholar]
- 18.Betts S, King J. There’s a right way and a wrong way: In vivo and in vitro folding, misfolding and subunit assembly of the P22 tailspike. Structure. 1999;7:R131–R139. doi: 10.1016/s0969-2126(99)80078-1. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi Y, King J. Assembly of the tail of bacteriophage T4. J Supramol Struct. 1975;3:24–38. doi: 10.1002/jss.400030104. [DOI] [PubMed] [Google Scholar]
- 20.Leiman PG, et al. Morphogenesis of the T4 tail and tail fibers. Virol J. 2010;7:355. doi: 10.1186/1743-422X-7-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartual SG, et al. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci USA. 2010;107:20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller K, Braun V. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J Virol. 1982;41:222–227. doi: 10.1128/jvi.41.1.222-227.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix RW, Duda RL. Bacteriophage lambda PaPa: Not the mother of all lambda phages. Science. 1992;258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Mitraki A, et al. Unfolding studies of human adenovirus type 2 fibre trimers. Evidence for a stable domain. Eur J Biochem. 1999;264:599–606. doi: 10.1046/j.1432-1327.1999.00683.x. [DOI] [PubMed] [Google Scholar]
- 26.Bartual SG, Garcia-Doval C, Alonso J, Schoehn G, van Raaij MJ. Two-chaperone assisted soluble expression and purification of the bacteriophage T4 long tail fibre protein gp37. Protein Expr Purif. 2010;70:116–121. doi: 10.1016/j.pep.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Veesler D, Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev. 2011;75:423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petter JG, Vimr ER. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J Bacteriol. 1993;175:4354–4363. doi: 10.1128/jb.175.14.4354-4363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulz EC, et al. Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF. J Mol Biol. 2010;397:341–351. doi: 10.1016/j.jmb.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Yu SL, Ko KL, Chen CS, Chang YC, Syu WJ. Characterization of the distal tail fiber locus and determination of the receptor for phage AR1, which specifically infects Escherichia coli O157:H7. J Bacteriol. 2000;182:5962–5968. doi: 10.1128/jb.182.21.5962-5968.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guardado Calvo P, et al. Structure of the carboxy-terminal receptor-binding domain of avian reovirus fibre sigmaC. J Mol Biol. 2005;354:137–149. doi: 10.1016/j.jmb.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 32.Merckel MC, Huiskonen JT, Bamford DH, Goldman A, Tuma R. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol Cell. 2005;18:161–170. doi: 10.1016/j.molcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Bhardwaj A, Molineux IJ, Casjens SR, Cingolani G. Atomic structure of bacteriophage Sf6 tail needle knob. J Biol Chem. 2011;286:30867–30877. doi: 10.1074/jbc.M111.260877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guardado-Calvo P, Fox GC, Llamas-Saiz AL, van Raaij MJ. Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre. J Gen Virol. 2009;90:672–677. doi: 10.1099/vir.0.008276-0. [DOI] [PubMed] [Google Scholar]
- 35.Reiter DM, et al. Crystal structure of reovirus attachment protein σ1 in complex with sialylated oligosaccharides. PLoS Pathog. 2011;7:e1002166. doi: 10.1371/journal.ppat.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spinelli S, et al. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol. 2006;13:85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- 37.Spinelli S, et al. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J Biol Chem. 2006;281:14256–14262. doi: 10.1074/jbc.M600666200. [DOI] [PubMed] [Google Scholar]
- 38.Thomassen E, et al. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J Mol Biol. 2003;331:361–373. doi: 10.1016/s0022-2836(03)00755-1. [DOI] [PubMed] [Google Scholar]
- 39.Tao Y, Strelkov SV, Mesyanzhinov VV, Rossmann MG. Structure of bacteriophage T4 fibritin: A segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 40.Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope–penetrating needle. Nat Struct Mol Biol. 2007;14:1221–1226. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- 41.van Raaij MJ, Schoehn G, Burda MR, Miller S. Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J Mol Biol. 2001;314:1137–1146. doi: 10.1006/jmbi.2000.5204. [DOI] [PubMed] [Google Scholar]
- 42.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 43.Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol. 2005;12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- 44.Smith NL, et al. Structure of a group A streptococcal phage-encoded virulence factor reveals a catalytically active triple-stranded beta-helix. Proc Natl Acad Sci USA. 2005;102:17652–17657. doi: 10.1073/pnas.0504782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra P, et al. Polysaccharide binding sites in hyaluronate lyase—Crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose. FEBS J. 2009;276:3392–3402. doi: 10.1111/j.1742-4658.2009.07065.x. [DOI] [PubMed] [Google Scholar]
- 46.Xiang Y, et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol Cell. 2009;34:375–386. doi: 10.1016/j.molcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamashita E, et al. The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:837–841. doi: 10.1107/S1744309111005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG. Phage pierces the host cell membrane with the iron-loaded spike. Structure. 2012;20:326–339. doi: 10.1016/j.str.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Heineman RH, Springman R, Bull JJ. Optimal foraging by bacteriophages through host avoidance. Am Nat. 2008;171:E149–E157. doi: 10.1086/528962. [DOI] [PubMed] [Google Scholar]
- 50.Garcia E, et al. The genome sequence of Yersinia pestis bacteriophage phiA1122 reveals an intimate history with the coliphage T3 and T7 genomes. J Bacteriol. 2003;185:5248–5262. doi: 10.1128/JB.185.17.5248-5262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupeux F, Röwer M, Seroul G, Blot D, Márquez JA. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr D Biol Crystallogr. 2011;67:915–919. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- 52.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 53.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 54.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 55.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 56.Vagin AA, Isupov MN. Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr D Biol Crystallogr. 2001;57:1451–1456. doi: 10.1107/s0907444901012409. [DOI] [PubMed] [Google Scholar]
- 57.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.