Abstract
Choice of host plants by phytophagous insects is essential for their survival and reproduction. This choice involves complex behavioral responses to a variety of physical and chemical characteristics of potential plants for feeding. For insects of the order Hemiptera, these behavioral responses involve a series of steps including labial dabbing and probing using their piercing mouthparts. These initial probing and feeding attempts also elicit a rapid accumulation of phytohormones, such as jasmonic acid (JA), and the induced defense metabolites they mediate. When Nicotiana attenuata plants are rendered JA deficient by silencing the initial committed step of the JA biosynthesis pathway, they are severely attacked in nature by hemipteran leafhoppers of the genus Empoasca. By producing N. attenuata plants silenced in multiple steps of JA biosynthesis and perception and in the biosynthesis of the plant’s three major classes of JA-inducible insecticidal defenses, we demonstrate that the choice of plants for feeding by Empoasca leafhoppers in both nature and the glasshouse is independent of the accumulation of major insecticidal molecules. Moreover, this choice is independent of the presence of Candidatus Phytoplasma spp. and is not associated with detectable changes in plant volatiles but instead depends on the plant´s capacity to mediate JA signaling. We exploited this trait and used Empoasca leafhoppers to reveal genetic variation in JA accumulation and signaling hidden in N. attenuata natural populations.
Plants provide a variety of resources, such as food, mating and oviposition sites, and shelter for a majority of phytophagous insect species. Host-plant selection by insects involves complex behavioral responses to a variety of physical and chemical characteristics of the host plant that operate at different spatial scales and include long-range olfactory (e.g., plant-derived volatiles perceived by odor receptors) and visual (e.g., plant shape, size, and color) cues and short-range chemotactic and gustatory (e.g., surface metabolites perceived by chemoreceptors) cues (1–3). The physical and chemical characteristics of plants that insects use for host selection depend on the feeding guild and the dietary behavior (e.g., polyphagy or oligophagy) of the insect species (4). For example, Drosophila melanogaster flies (order Diptera) use a wide range of olfactory cues such as methyl-, ethyl-, and propyl esters of short-chain fatty acids generated by microorganisms growing on decaying fruit (5), whereas Drosophila sechellia flies use a specific molecule (methyl hexanoate) emitted by its exclusive food plant, Morinda citrifolia (6). Insects with mouthparts capable of piercing plant tissues and sucking out liquids (e.g., order Hemiptera) use labial dabbing and probing to perceive chemical cues (e.g., waxes, terpenoids, acyl sugars, and alkaloids) on tissue surfaces or internal cellular layers (1–3, 7). Interestingly, it has been shown that insects also can perceive phytohormones. Helicoverpa zea (order Lepidoptera) larvae can perceive jasmonic acid (JA) (8), which accumulates in the food plants during attack and induces de novo synthesis of plant defense metabolites (9, 10). Thus, one possible scenario is that insects can select plants for feeding based on the plant’s capacity to produce JA (or to signal JA-mediated responses) by eavesdropping on the defensive capacity of a potential host plant.
In some cases insects can suppress the accumulation of plant defense metabolites as a mechanism of food plant selection (11, 12). These suppression mechanisms often are associated with the alteration of phytohormone biosynthesis or signaling pathways and may involve specific enzymes (e.g., glucose oxidase) produced by the insect (13) or vectored microorganisms (14, 15). Leafhoppers of the genus Empoasca are hemipterans (family Cicadellidae) that feed on phloem and cell contents of a broad range of host plants (16, 17). During feeding, the leafhoppers may induce “hopper burn” in the plant tissue, damage that is characterized by the yellowing of the tissue around the feeding site (18). Empoasca leafhoppers can also vector viruses, bacteria, and fungi and transmit them efficiently to plants as a consequence of their ingestion–egestion feeding behavior (18). For example, cell wall-lacking bacteria of the species Candidatus Phytoplasma (hereafter, Ca. Phytoplasma) can be transmitted by Empoasca leafhoppers (19, 20). Interestingly, the transmission of Ca. Phytoplasma spp. into the plant can affect the interaction of the plant with the transmitting insect via the modification of direct or indirect plant defenses. It has been shown that Malus domestica trees infected by Ca. Phytoplasma mali emit larger amounts of β-caryophyllene, a volatile that lures insect vectors to infected plants, than do noninfected trees, (14). A recent laboratory study performed with Arabidopsis thaliana and Macrosteles quadrilineatus leafhoppers showed that effector proteins produced by Ca. Phytoplasma asteris interfere with the activation of JA biosynthesis in the plant and thereby reduce the induction of JA-mediated defense responses (15).
Nicotiana attenuata (Solanaceae), an annual tobacco plant native to the southwestern United States, germinates after fires from long-lived seed banks to form monocultures and must cope with an unpredictable insect community (21). Populations of N. attenuata plants are known to harbor significant genetic diversity among individuals, and the genetic diversity frequently is larger within populations than among populations (22), probably because of the plant’s fire-dependent germination and long-lived seed banks (21). In their natural environment, as well as in the glasshouse, N. attenuata plants respond strongly and specifically to attacks by insects of different feeding guilds (23, 24). A large number of these responses are governed by a strong burst of JA, which is amplified by elicitors in the insect’s oral secretions (OS) (10, 25–27). The initial steps of JA biosynthesis involve the release of trienoic fatty acids [e.g., α-linolenic acid (18:3)] from membrane lipids in the chloroplast by the action of glycerolipases [GLA1 in N. attenuata (28, 29)]. The released trienoic fatty acids are oxidized by 13-lipoxygenases (LOX3 in N. attenuata) to form (13S)-hydroperoxy-18:3 (30). This molecule is the substrate for allene oxide synthase (AOS) that forms (9Z, 13S, 15Z)-12, 13-epoxy-18:3, which subsequently is cyclized to an isomeric mixture of 12-oxo-phytodienoic acid (OPDA) by allene oxide cyclase (AOC) (31). OPDA is transported into the peroxisome where the C10–C11 double bond in (9S, 13S)-OPDA is reduced by an OPDA reductase (OPR) (32). The reduced OPDA (OPC-8:0) undergoes three cycles of β-oxidation involving acyl-CoA transferase (ACX) (33) and finally forming (3R, 7S)-JA. JA can be modified, e.g., by jasmonyl-O-methyl transferase (JMT) to form methyl-jasmonic acid (MeJA) (34) or by JASMONATE RESISTANT (JAR) that conjugates isoleucine to form JA-Ile (35, 36). JA-Ile activates the SCFCOI1–JAZ complex (37), which transcriptionally activates genes involved in the biosynthesis of defense molecules, among other responses (38, 39).
Previously, we have reported that N. attenuata plants rendered deficient in JA biosynthesis by silencing NaLOX3 (as-lox3) are heavily damaged by Empoasca leafhoppers in nature (40). Here, we used a set of 11 N. attenuata transgenic lines deficient in specific steps of JA biosynthesis and perception and deficient in the accumulation of major insecticidal molecules to disentangle the mechanisms underlying the selection of plants for feeding by Empoasca leafhoppers. In addition, we used these insects to discover genetic variations in JA accumulation and signaling in natural N. attenuata populations.
Results
Ca. Phytoplasma Species Are Not Found in Empoasca Leafhoppers or the Plants They Attack in the Field.
A previous study showed that Ca. Phytoplasma asteris transmitted by Macrosteles quadrilineatus leafhoppers can affect JA biosynthesis in infected A. thaliana plants in the laboratory (15). Thus, we first examined whether the interaction between Empoasca leafhoppers and N. attenuata plants involved Ca. Phytoplasma species. For this purpose we carried out two different sets of experiments. In the 2009 field season, we observed that N. attenuata plants silenced in the expression of COI1 (ir-coi1) were severely attacked by adult Empoasca leafhoppers. These adult leafhoppers originated from Cucurbita foetidissima plants growing adjacent to our field plot and are referred to hereafter as “Empoasca spp.” Empoasca spp. adults and leaves from Empoasca spp.–damaged and undamaged ir-coi1 plants were collected for analysis (first set). In parallel, adult Empoasca leafhoppers were collected and used to establish a colony in our glasshouse (see Materials and Methods for details). This colony was composed of a single Empoasca species and is referred to hereafter as “Empoasca sp.” Adult leafhoppers from this in-house colony were used to challenge inverted repeat (ir)-coi1 plants for 7 d under glasshouse conditions. Adult leafhoppers and attacked and unattacked (control) leaves were collected and used for analysis (set 2). The presence of Ca. Phytoplasma spp. was analyzed in adult Empoasca leafhoppers and leaf samples from both sets. A PCR approach using universal primers for the amplification of 16S rRNA (Table S1) (14, 41, 42) was used for the detection of Ca. Phytoplasma spp. (Fig. S1). Ca. Phytoplasma spp. were detected in the positive control samples (isolated genomic DNA from Ca. Phytoplasma asteris and Callistephus chinensis leaves infected with Ca. Phytoplasma asteris) but not in the negative control samples or in samples collected from field and glasshouse experiments (Fig. S1; see SI Materials and Methods for details). These results demonstrate that the interaction between Empoasca leafhoppers and N. attenuata plants was independent of the presence of Ca. Phytoplasma spp.
Generation of a Toolbox of Transformed Plants to Examine Empoasca Leafhopper Plant-Choice Mechanisms.
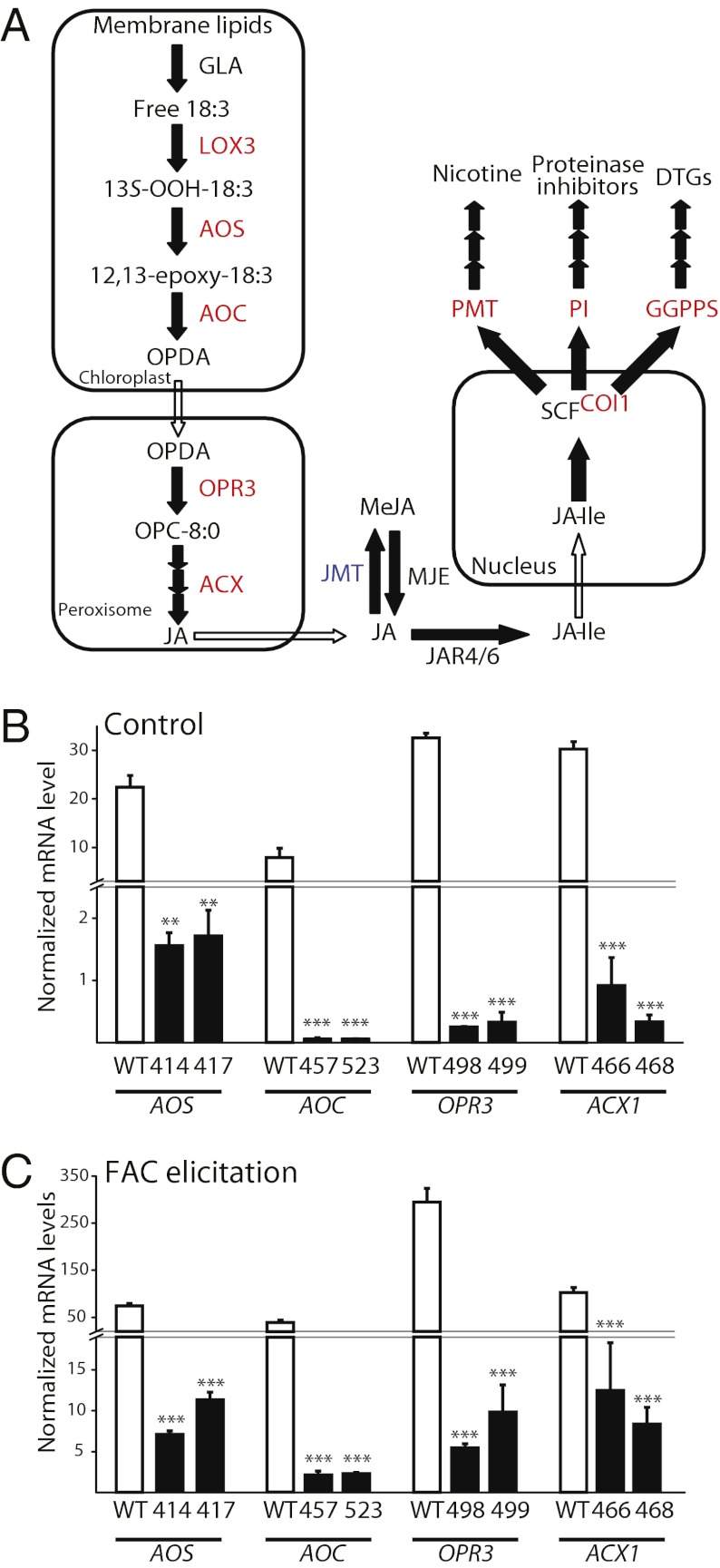
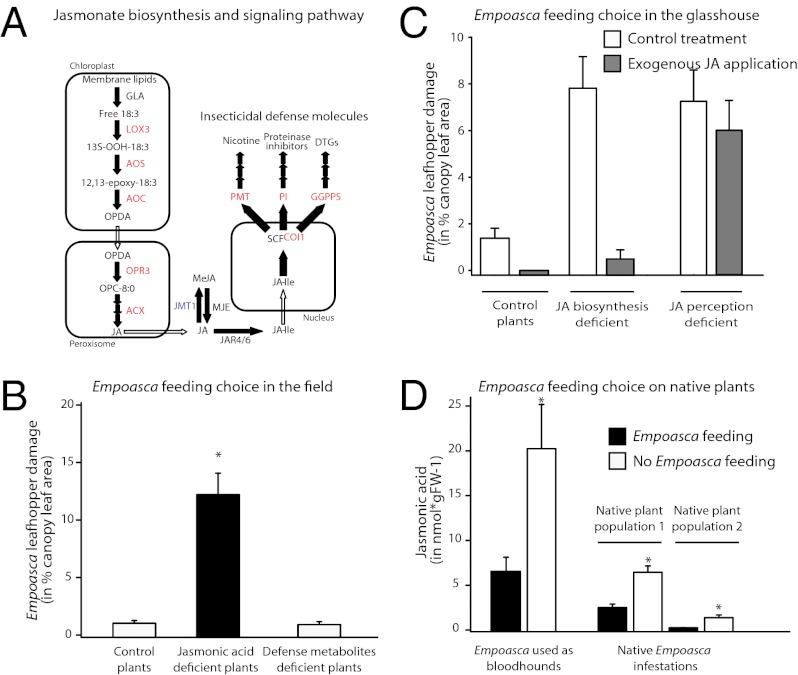
To examine the mechanisms underlying plant choice by Empoasca leafhoppers, we transformed N. attenuata plants with RNAi ir constructs to silence (i) specific steps of JA biosynthesis [ir-lox3 (43), ir-aos, ir-aoc, ir-opr3, and ir-acx1; see SI Materials and Methods for details]; (ii) JA perception (ir-coi1) (44); and (iii) accumulation of JA-dependent defense molecules [i.e., nicotine (ir-pmt); trypsin proteinase inhibitor (PI) (ir-pi); nicotine and PIs (ir-pmt/pi) (45); and diterpene glycosides (ir-ggpps) (46)] (Fig. 1A). Additionally, we ectopically expressed JA methyl transferase 1 (JMT1; 35S-jmt1) to deplete jasmonate accumulation metabolically by redirecting the flux of JA into methyl-JA (MeJA) (47). Compared with control plants, 35S-jmt1 plants have reduced levels of JA-Ile after simulated herbivory, and therefore their COI1-mediated JA-signaling capacity is reduced (47).
Fig. 1.
Lines of transformed plants generated to dissect the mechanisms of Empoasca spp. feeding choice. (A) Enzymes of the jasmonate biosynthetic and signaling pathway and of jasmonate-regulated direct defenses in N. attenuata. All enzymes in red font were silenced by RNAi. JMT (blue font) was expressed ectopically to generate a toolbox of transformed plants to examine the feeding choice of Empoasca spp. in nature. RNAi lines were generated by transforming N. attenuata plants with constructs harboring an ir fragment of each gene (Table S2). (B and C) Silencing efficiency of two independently transformed homozygous ir lines with a single transfer-DNA insertion was determined by qPCR analysis in control leaves (B) and leaves harvested 60 min after FAC elicitation (C). Transcript levels were quantified by comparing the levels of corresponding genes with the eukaryotic elongation factor 1A-α (NaEF1A-α) (average ± SE, n = 3). Asterisks indicate statistically significant differences in WT plants versus the ir line. *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t test with Welch correction.
Plants silenced in the expression of the AOS (ir-aos), AOC (ir-aoc), OPR3 (ir-opr3), and ACX1 (ir-acx1) genes were newly generated for this study, and two homozygous, independently transformed lines harboring a single transfer DNA insertion (Table S2 and Fig. S2) were selected for each genotype. In unelicited leaves, the transcript levels of NaAOS, NaAOC, NaOPR3, and NaACX1 were silenced by 10- to 100-fold in the respective lines compared with WT (Fig. 1B). In leaves treated with synthetic fatty acid–amino acid conjugate (FAC) to amplify the induction of the JA biosynthesis pathway (28), the target transcripts were reduced by five- to 100-fold in the respective lines compared with WT at 60 min after treatment (Fig. 1C). Consistent with the reduced expression of the JA biosynthesis genes, all lines showed significantly reduced levels of JA and JA-Ile after FAC treatment (Fig. S3). ir-acx1 plants lost their capacity to suppress JA accumulation in the third generation, and therefore one line (line A466) was used as a control in addition to the plants transformed with the empty vector (EV) construct.
This set of transgenic lines, deficient in JA accumulation, JA perception, and JA-dependent defense molecules, allowed us to study in detail the steps of the JA biosynthesis and signaling pathways that are responsible for feeding-choice decisions of Empoasca leafhoppers.
Defense Molecules or Volatiles Do Not Direct Initial Empoasca Leafhopper Feeding in Nature.
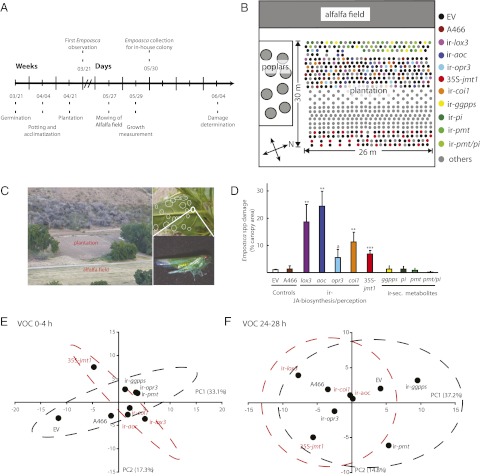
To examine Empoasca leafhopper plant choice in nature, all the transgenic N. attenuata lines mentioned above were grown in a fully randomized design in a field plot in the Great Basin Desert during the 2009 field season (Fig. 2 A and B). ir-aos plants did not survive transplantation to the field and were not included in the analysis. An adjacent alfalfa (Medicago sativa) field served as a source of Empoasca spp. adults that were encouraged to move into the N. attenuata plantation by mowing the alfalfa field (Fig. 2C). Eight days after mowing, we quantified Empoasca spp. attack as the percentage of total canopy area damaged (Fig. 2D). Importantly, although the degree of damage was expressed per total canopy area for normalization, we observed that 80–90% of the damage inflicted by Empoasca leafhoppers occurred on stem leaves.
Fig. 2.
Empoasca spp. select plants deficient or modified in JA accumulation or perception for feeding in the field, independently of released HIPVs. (A) Timeline of experimentation during the 2009 Utah field season. (B) Design of the field plot and the location of the different N. attenuata genotypes within the field plot. (C) (Left) In the field, N. attenuata plants were grown adjacent to an alfalfa (M. sativa) field, a source of Empoasca spp. (Right) Empoasca spp. damage on N. attenuata was determined as the percentage of canopy area exhibiting the characteristic damage (white circles) resulting from Empoasca spp. attack. (D) Eight days after the mowing of the alfalfa field, damage was quantified on EV and A466 controls, lines silenced in JA biosynthesis (ir-lox3, ir-aoc, ir-opr3) and perception (ir-coi1), lines silenced in JA-dependent defense molecules [i.e., diterpene glycosides (ir-ggpps), trypsin PIs (ir-pi), nicotine (ir-pmt), and both nicotine and PIs,(ir-pmt/pi)], and lines ectopically expressing a jasmonic acid methyl transferase (35S-jmt1). Asterisks indicate significant differences compared with control plants (n = 7–10). *P < 0.05; **P < 0.01; ***P < 0.001; a, P = 0.07; Student’s t test. (E and F) After Empoasca leafhoppers were caged on single leaves, HIPVs were collected for 4 h during two different periods: immediately after the leafhoppers were caged (0–4 h) (E) and 24 h after the leafhoppers were caged (24–28 h) (F). Of 197 detected volatiles released with Empoasca spp. feeding, 83 HIPVs had a fold change (FC) of 1.5 ≤FC ≤0.66 (P value <0.05) compared with volatiles released from unattacked plants of the same genotype. These 83 HIPVs were used for PCA analysis in which the two volatile-trapping periods were analyzed individually. N. attenuata plants were grouped in two classes based on Empoasca spp. damage (black indicates no significant differences in Empoasca spp. damage compared with controls; red indicates significant differences), and PCs 1 and 2 of the transgenic lines were plotted against each other.
The ir-lox3, ir-aoc, 35S-jmt1, and ir-coi plants were heavily damaged by Empoasca spp. (Fig. 2D). Interestingly, opr3 plants had WT levels of Empoasca spp. damage (Fig. 2D) even though the levels of JA and JA-Ile in these plants were strongly reduced (Fig. S3). The reduced levels of Empoasca spp. damage on ir-opr3 plants can be explained by their reduced number of stem leaves resulting from decelerated growth in the field (Fig. S4). Thus, when the experiment was conducted, the canopy of ir-opr3 plants was dominated by rosette leaves. An alternative explanation for these observations is that OPDA may play a role in the mechanisms underlying the interaction between Empoasca leafhoppers and N. attenuata plants. However, this possibility was ruled out by additional experiments (see below). The damage to plants with reduced levels of JA-dependent defense molecules (ir-pmt, ir-pi, ir-pmt/pi, and ir-ggpps) was not distinguishable from that of control plants (Fig. 2D). These results demonstrated that Empoasca leafhoppers feed preferentially on plants with reduced jasmonate accumulation and reduced ability to perceive feeding rather than on plants with reduced accumulation of nicotine, PIs, and diterpene glycosides (DTGs) (ir-pmt, ir-pi, ir-pmt/pi, and ir-ggpps).
To test whether Empoasca leafhopper feeding choice in nature was driven by constitutively emitted volatiles from N. attenuata plants, we first trapped leaf volatiles from unattacked ir-lox3, ir-aoc, ir-opr3, 35S-jmt1, ir-coi1, ir-pmt, ir-ggpps, EV, and A466 plants grown in the field. Ultra-high-resolution GC analysis detected 197 volatiles constitutively released from these plants. These 197 volatiles were subjected to principal component analysis (PCA) (48) for which the genotypes were grouped in two classes based on their significant differences in Empoasca spp. damage (Fig. 2D). Principal components (PCs) 1 and 2 explained 44% of the variation but did not separate the two plant classes (Fig. S4C). Second, we tested whether herbivory-induced plant volatiles (HIPVs), which consist of green leaf volatiles released immediately upon insect attack and terpenoids released during the following photoperiod (48), affected Empoasca spp. feeding choices. For this purpose, we caged three adult Empoasca leafhoppers on leaves of the genotypes mentioned above and trapped HIPVs for 4 h during two different periods, immediately after the leafhoppers were caged (0–4 h) and 24 h after the leafhoppers were caged (24–28 h). HIPVs differentially emitted by Empoasca spp. feeding from the different transgenic lines were defined as compounds with a fold change (FC) of 1.5 ≤FC ≤0.66 and a P value <0.05 compared with unattacked plants of the same genotype. Of the 197 volatiles detected, 83 HIPVs were identified as accumulating differentially among all the genotypes (Table S3). These 83 HIPVs were used for PCA analysis (48) in which the two volatile-trapping periods were analyzed individually. As mentioned above, the genotypes were grouped in two classes based on their significant differences in Empoasca spp. damage (Fig. 2D). PCs 1 and 2 explained 50% of the variation within the first trapping period and 52% of the variation within the second trapping period, but in neither trapping period did PC1 and PC2 separate the two plant classes (Fig. 2 E and F). In summary, these results demonstrate that with an ultra-high-resolution analysis of plant volatiles neither the detectable constitutively released volatiles nor HIPVs were associated with Empoasca spp. feeding preferences in the field.
Empoasca Leafhopper Damage Correlates with Reduced Levels of OPDA and JA Accumulation.
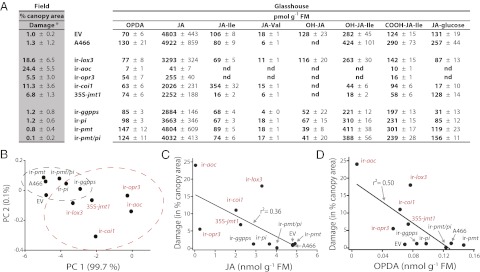
The field observations showed clearly that Empoasca spp. preferentially attacked plants with reduced jasmonate accumulation and signaling capacity. To evaluate more directly the association between the Empoasca leafhopper feeding preferences observed in the field (Fig. 2D) and the jasmonate (OPDA, JA, and JA derivatives)-producing capacities of the transgenic N. attenuata lines used, we first assessed the capacity of EV, A466, ir-lox3, ir-aoc, ir-opr3, ir-coi1, 35S-jmt1, ir-ggpps, ir-pi, ir-pmt, and ir-pmt/pi plants to accumulate jasmonates after standardized mechanical wounding under glasshouse conditions (Fig. 3A). PCA separated the transgenic N. attenuata lines deficient in JA accumulation and perception from controls and from lines deficient in JA-dependent defense molecules (Fig. 3B). PC 1 explained almost all (99.7%) variance present in the data and was influenced positively by jasmonate levels (loading factors, 0.2–0.4) but negatively by Empoasca spp. damage (loading factor, −0.34) (Fig. 3B). A correlation analysis between the amount of Empoasca spp. damage quantified in the field and the capacity of the plants to accumulate jasmonates after standardized mechanical wounding revealed that damage correlated negatively with the capacity of the plants to accumulate OPDA and JA (JA vs. damage: Pearson’s R2 = 0.36, P = 0.03; OPDA vs. damage: Pearson’s R2 = 0.50, P = 0.01) (Fig. 3 C and D). We therefore hypothesized that the extent of initial Empoasca leafhopper feeding on N. attenuata plants depended either on OPDA or JA accumulation or on their respective signaling capacities.
Fig. 3.
Wound-elicited JA and OPDA levels correlate with Empoasca spp. damage. (A) All lines analyzed in the field were grown in the glasshouse, and jasmonate accumulations in treated leaves were analyzed 1 h after wounding. Numbers represent average ± SE, n = 5. (*Damage data are the same as shown in Fig. 2D and have been included here for comparison and to assist with the understanding of the results in this figure.) (B) PCA separated the transgenic N. attenuata lines deficient in JA accumulation and perception from controls and from lines deficient in JA-dependent defense molecules. (C and D) Empoasca spp. damage observed in the field correlated negatively with the wound-induced JA and OPDA levels observed in the glasshouse (Pearson correlation; JA vs. damage: Pearson’s R2 = 0.36, P = 0.03; OPDA vs. damage: R2 = 0.50, P = 0.01).
Empoasca Leafhopper Feeding Preferences Depend on the Plants’ Capacity to Mediate JA Signaling.
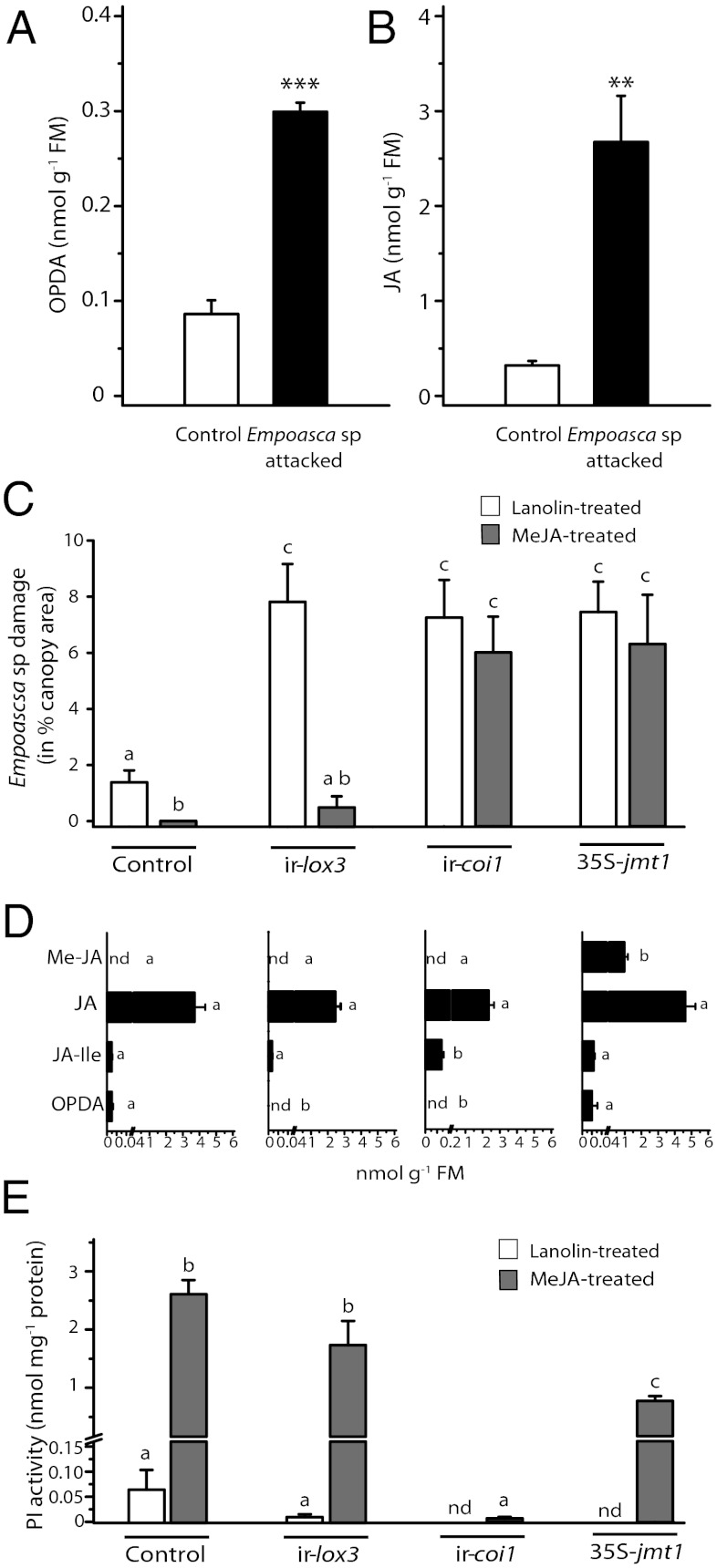
To examine which jasmonate triggers initial feeding of Empoasca leafhoppers on N. attenuata plants, we first analyzed the induction of jasmonate biosynthesis in WT leaves after Empoasca sp. feeding. For this experiment, 25 adult Empoasca sp. from the glasshouse colony were caged on single leaves of N. attenuata WT plants. After 24 h, the levels of OPDA, JA, JA derivatives, and MeJA were quantified. The levels of OPDA, JA, JA-Ile, 11/12-hydroxy-JA-Ile, and 11/12-carboxy-JA-Ile were increased significantly (three- to eightfold) in leaves attacked by Empoasca sp. compared with control leaves (Fig. 4 A and B and Table S4A). The 11/12-hydroxy-JA, MeJA, and JA-amino acid conjugates other than JA-Ile were not detected in leaves. Thus, the induction of OPDA and JA accumulation by Empoasca sp. feeding is consistent with a potential role of these two jasmonates in the plant-selection process.
Fig. 4.
Initial Empoasca sp. feeding induces jasmonate accumulation and is triggered by JA signaling capacities. (A and B) Twenty-five Empoasca sp. were caged on N. attenuata WT leaves, and jasmonate accumulation was measured. OPDA and JA levels were measured in control and attacked leaves 24 h after Empoasca sp. feeding. Bars represent average ± SE; n = 4. Asterisks represent significant differences compared with control plants. **P < 0.01; ***P < 0.001; Student’s t test. (C and D) In glasshouse bioassays, MeJA treatment of leaves enhances the resistance to Empoasca sp. attack of WT (control) and ir-lox3 plants but not of ir-coi1 and 35S-jmt1 plants. Leaves of WT, ir-lox3, ir-coi1, and 35S-jmt1 plants were treated with lanolin or with lanolin containing MeJA. (C) Two days after the treatment, plants were challenged with 150 Empoasca sp. adults, and damage was recorded after 7 d. Damage is shown as the percentage of canopy area; data shown are average ± SE; n = 15–18. Different letters indicate statistically significant differences. P < 0.05; Student’s t test. (D) Levels of MeJA, JA, JA-Ile, and OPDA were quantified in MeJA-treated leaves of WT, ir-lox3, ir-coi1, and 35S-jmt1 plants 3 d after the treatment (Table S4). Data shown are average ± SE; n = 4–9. Different letters indicate statistical significance among lines and within analytes. P < 0.05; Student’s t test. nd, not detected. (E) To analyze JA signaling capacities, we determined the levels of trypsin PI activity in lanolin- and MeJA-treated leaves 3 d after the treatment (average ± SE; n = 6). Different letters indicate statistically significant differences. P < 0.05; Student’s t test. nd, not detected.
To determine the predominant factor (i.e., OPDA or JA accumulation or mediated signaling) influencing plant selection by Empoasca sp., we performed feeding-choice experiments using N. attenuata WT (control), ir-lox3, ir-coi1, and 35S-jmt1 plants in the glasshouse. Leaves were treated either with lanolin (control treatment) or with lanolin containing MeJA. Two days after the treatment, the plants were challenged with 150 adult Empoasca sp., and the percentage of leaf damage was determined after 7 d (Fig. S5A). Lanolin-treated ir-lox3, ir-coi1, and 35S-jmt1 plants were damaged significantly more than lanolin-treated WT plants by Empoasca sp. (Fig. 4C). Treatment with MeJA decreased Empoasca sp. damage ir-lox3 plants significantly, by eightfold compared with lanolin treatment, but did not decrease damage in ir-coi1 and 35S-jmt1 plants (Fig. 4C).
To evaluate the accumulation of jasmonates after external MeJA application, the levels of OPDA, JA, JA derivatives, and MeJA were quantified in unchallenged WT, ir-lox3, ir-coi1, and 35S-jmt1 plants treated with lanolin or MeJA (Table S4B). None of the jasmonates analyzed were detected in lanolin-treated leaves. JA levels accumulated to 2.5–4.6 nmol/g fresh mass (FM) in MeJA-treated leaves of WT, ir-lox3, ir-coi1, and 35S-jmt1 plants (Fig. 4D and Table S4B), indicating that the JA levels did not affect feeding damage by Empoasca sp.. JA-Ile was detected in low amounts (0.01–0.02 nmol/g FM) in MeJA-treated leaves of WT, ir-lox3, and 35S-jmt1 plants but was detected in significantly higher amounts in MeJA-treated ir-coi1 leaves (0.14 nmol/g FM), as is consistent with the lower metabolism of JA-Ile in ir-coi1 plants (44, 49, 50). Thus, because Empoasca sp. feeding damage was similar on lanolin- and MeJA-treated ir-coi1 plants, JA-Ile levels did not affect Empoasca sp. feeding choice directly. In MeJA-treated leaves, MeJA was detected only in the leaves of 35S-jmt1 plants (1 nmol/g FM) (Fig. 4D and Table S4B) but did not directly affect Empoasca sp. feeding. OPDA was detected in similar amounts in MeJA-treated leaves of control and 35S-jmt1 plants (0.01–0.02 nmol/g FM) (Fig. 4D) but was not detected in ir-lox3 and ir-coi1 plants, as is consistent with the activation of the JA biosynthetic pathway by exogenous MeJA (23). Although OPDA levels in MeJA-treated 35S-jmt1 plants were similar to those of MeJA-treated WT leaves, Empoasca sp. feeding on 35S-jmt1 was not affected by MeJA treatment. This result demonstrated that OPDA or its associated signaling cascade was not involved in the plant-selection process by Empoasca sp. The results revealed that Empoasca sp. feeding on N. attenuata can be reduced by external MeJA application to lines in which JA biosynthesis is silenced but not in lines in which JA perception is silenced (ir-coi1) or in plants in which the flux of JA is redirected to an inactive jasmonate (35S-jmt1).
To provide independent evidence of the deficiency in JA-mediated defense signaling in ir-coi1 and 35S-jmt1 plants, the induction of trypsin PI activity [as an indicator of defenses induced by JA-mediated COI1 signaling (51)], was quantified in WT, ir-lox3, ir-coi1, and 35S-jmt1 plants treated with MeJA. Compared with lanolin-treated leaves, PI activity was induced to an activity of 2–2.5 nmol/mg protein in MeJA-treated leaves of control and ir-lox3 plants but was not induced in leaves of ir-coi1 plants and was reduced by 60% (0.9 nmol/mg protein) in 35S-jmt1 plants (Fig. 4E). Although MeJA treatment of 35S-jmt1 plants partially induced JA-mediated defenses (i.e., PI activity), damage by Empoasca sp. on MeJA-treated 35S-jmt1 plants was similar to that on lanolin-treated 35S-jmt1 plants. These experiments demonstrated that MeJA treatment induced defense responses (i.e., PI activity) in N. attenuata and were consistent with the results obtained from the field with N. attenuata ir-pi lines (Fig. 2D) (i.e., the choice of plants for feeding by Empoasca leafhoppers was independent of PI levels).
Finally, to evaluate if the Empoasca sp. in-house colony used for these experiments was free of Ca. Phytoplasma spp, we analyzed the presence of Ca. Phytoplasma spp. in severely Empoasca sp-damaged leaves from WT, ir-lox3, ir-coi1, and 35S-jmt1 plants and in 10 adult Empoasca leafhoppers from the colony. Phytoplasma were not detected in either leaf or leafhopper samples (Fig. S5B; see SI Materials and Methods for details).
Empoasca Leafhopper Attack Identified Variations in JA Accumulation in Nature.
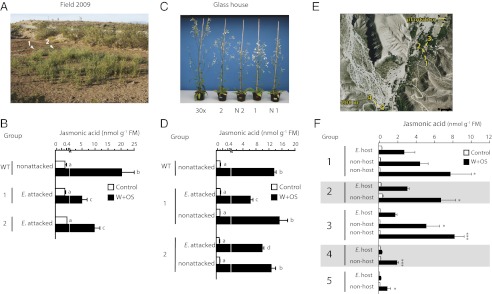
The work presented above demonstrated that Empoasca spp. damage could be used to identify genetically modified N. attenuata plants deficient in JA accumulation and signaling in the field and glasshouse. We next asked if Empoasca spp. attack also could identify natural variation in JA accumulation and signaling. During the 2009 field season, a natural population of 100 N. attenuata plants was screened using Empoasca spp. (collected from C. foetidissima plants) to discover genetic variation in JA accumulation hidden in natural populations. Two of the plants in the population showed Empoasca spp. damage; when JA was elicited by treating leaves with Manduca sexta OS in the field, these plants accumulated less JA than neighboring unattacked plants of the same population (Fig. 5 A and B). Elicitation by OS provides a standard method to assess the maximal capacity of the plants to produce JA after an insect-associated response. Moreover, because the OS was collected from M. sexta larvae, the analysis allowed us to exclude the possibility that factors (other than phytoplasma) introduced by Empoasca spp. attack were responsible for the suppression of the JA burst. Self-pollinated seeds from the two plants showing Empoasca spp. damage and their closest undamaged neighbors were collected and grown in the glasshouse. Consistent with the field results, elicitation of JA through treatment of the leaves with M. sexta OS elicited smaller JA bursts in the progeny of the two plants previously showing Empoasca spp. damage than in the progeny of previously unattacked plants (Fig. 5 C and D).
Fig. 5.
Empoasca spp. identified natural variation in JA accumulation in genetically diverse native N. attenuata populations. (A) During the 2009 field season, ∼50 Empoasca spp. were released into a native N. attenuata population (ca. 100 plants), and Empoasca spp. damage on all plants was determined after 2 d. Two plants (arrowheads marked I and 2) with detectable Empoasca spp. damage were found. (B) Leaves from both plants and from a nonattacked control plant were treated with M. sexta OS to elicit JA and were harvested for jasmonate analysis after 60 min. Both plants showing Empoasca spp. damage had lower OS-elicited amounts of JA than did control plants (average ± SE, n = 3–4). Bars sharing same the letters are not significantly different. P < 0.05; Student’s t test. (C) Self-pollinated seeds from these plants (1 and 2) and from neighboring undamaged plants (N1, N2) were grown in the glasshouse, treated with OS to elicit JA, and harvested for jasmonate analysis after 60 min. (D) JA accumulations in plants from two accessions showing Empoasca spp. damage in the field were significantly lower than in undamaged neighbors (average ± SE, n = 4). Bars sharing same the letters are not significantly different. P < 0.05; Student’s t test. (E) During the 2011 field season, two areas with ∼400 and 200 native N. attenuata plants, respectively, were surveyed to identify Empoasca spp.-damaged plants. We found five plants with increased Empoasca spp. damage compared with neighboring plants in the same developmental stage. (F) Leaves of these five plants and undamaged neighbor plants were treated with OS to elicit JA and harvested for JA analysis after 60 min. All plants with Empoasca spp. damage had lower elicited JA values than did their undamaged neighbors (average ± SE, n = 3). Asterisks indicate statistical significance of the plants attacked by Empoasca spp. compared with their respective neighbors. *P < 0.05; ***P < 0.001; Student’s t test.
A second screening of native N. attenuata populations was carried out during the 2011 field season. In this case, and in contrast to 2009, we used natural infestations of Empoasca spp. originating from C. foetidissima plants growing within natural N. attenuata populations. We screened two populations consisting of ∼400 and 200 N. attenuata plants, respectively, (Fig. 5E) and found three plants in one population and two plants in the other that were highly damaged by Empoasca spp. compared with neighboring plants at similar growth stages (Fig. S5C). Undamaged leaves from the five plants showing Empoasca spp. damage and from undamaged neighbor plants were treated with M. sexta OS for JA elicitation and were harvested 60 min after the treatment. All five plants showing Empoasca spp. damage accumulated significantly lower JA levels than their undamaged neighbors within the same population (Fig. 5F). Again, the analysis of phytoplasma in leaves and Empoasca spp. collected in both plant populations was negative (Fig. S5D; see SI Materials and Methods for details). These results demonstrated that the initial Empoasca leafhopper feeding choice can be used to identify variations in JA accumulation or signaling in N. attenuata populations. In addition, these experiments allowed us to exclude any role of Empoasca spp. feeding in the suppression of the JA signaling in these plants.
Discussion
Volatile Release Is Not Associated with Empoasca Leafhopper Damage.
Plant selection by Empoasca leafhoppers can be guided by perceiving nonvolatile molecules during the first feeding but also by perceiving specific volatile cues released from plants appropriate for feeding. Several studies have highlighted the fundamental role of plant volatiles in plant selection by herbivore insects (52, 53). For example, Manduca moths can distinguish N. attenuata plants already infested by M. sexta larvae and preferably lay eggs on uninfested plants (54). Furthermore, Empoasca fabae leafhoppers are more arrested by volatiles emitted from an alfalfa genotype susceptible to E. fabae than by volatiles from a resistant alfalfa genotype (55). Additionally, phytopathogen infections can cause dramatic changes in volatile emissions, and these emissions can lure insect vectors to infected plants (14). Our studies focused on the initial selection of plants for feeding by Empoasca leafhoppers. We therefore analyzed N. attenuata volatile emissions before and during the first stages of leafhopper attack rather than tracking long-term changes. Changes in plant volatiles released from both unattacked and Empoasca spp-attacked plants and analyzed by ultra-high-resolution GCxGC-ToF mass spectrometry were not associated with Empoasca spp. feeding preferences. Although we cannot exclude the possibility that either unmeasured volatiles or nonlinear behavioral responses to emitted volatiles might play a role, our results are consistent with the conclusion that the choice of plants for feeding by Empoasca leafhoppers is based on plant characteristics perceived during the initial probing process (see below).
Empoasca Leafhopper Plant Choice Is Not Associated with the Presence of Ca. Phytoplasma Species in Nature or in the Glasshouse.
Recent laboratory studies have shown that Ca. Phytoplasma asteris infection reduces JA biosynthesis in A. thaliana plants (15) and that Ca. Phytoplasma mali infections induce Malus domestica to emit higher amounts of β-caryophyllene to lure insect vectors (14). In nature, the proportion of leafhoppers infected by Ca. Phytoplasma spp. is low [∼2% (56)], and in our study the choice of N. attenuata plants by Empoasca leafhoppers was not associated with the presence of Ca. Phytoplasma spp. in either natural and laboratory environments.
Major Defense Molecules of N. attenuata Do Not Affect Initial Empoasca Leafhopper Feeding.
Since Fraenkel (57) first argued that plant secondary metabolites have a defensive function, many laboratory studies have described defensive metabolites induced by herbivore attack (7). We demonstrate that Empoasca leafhopper feeding on N. attenuata plants is independent of the accumulation of the three major classes of insecticidal compounds in N. attenuata (i.e., nicotine, trypsin PIs, and DTGs). The biosynthesis of these molecules is regulated by JA signaling, and they are known to have a strong influence on the herbivore community on N. attenuata in field experiments (45, 46, 58, 59). Although the total herbivore damage on ir-pmt, ir-pi, ir-pmt/pi, and ir-ggpps plants was significantly higher than on control plants (Fig. S4D), damage by Empoasca spp. was similar to that on control plants (Fig. 2D) in the field. Furthermore, in the glasshouse, MeJA treatment of 35S-jmt1 plants significantly induced the activity of PIs, but Empoasca leafhoppers attacked these plants and lanolin-treated 35S-jmt1 plants similarly. Empoasca leafhopper performance may be affected by nicotine, PIs, or DTGs, but the initial feeding choice of Empoasca leafhoppers (as reflected by the initial feeding damage) clearly is not affected by these molecules.
JA Signaling Mediates Plant Choice by Empoasca Leafhoppers.
In nature, Empoasca leafhoppers preferentially select N. attenuata plants for feeding when the plants are rendered deficient in JA accumulation or JA perception (Fig. 2D). Eleven ir lines deficient in JA accumulation, JA perception, and JA-regulated defense molecules were analyzed in the field to disentangle the mechanisms responsible for the feeding preferences of the Empoasca leafhopper. We also reassessed as-aos plants, which previously were shown to be attacked similarly to WT plants by Empoasca spp. (Fig. S6A) (40). We found that the wound-induced accumulation of JA was similar to that in EV control plants (Fig. S6B), providing a likely explanation for why Empoasca spp. did not attack as-aos plants in the 2003 field season (40). Our results demonstrate that initial Empoasca spp. feeding on N. attenuata plants in the field correlates negatively with the accumulation of jasmonates (Fig. 3). To examine these correlations, wound-induced jasmonate accumulation rather than Empoasca spp.-induced JA accumulation was used because (i) Empoasca spp. feed differentially on the different genotypes analyzed, and (ii) Empoasca spp. feed at irregular times. Therefore, we used a single wounding elicitation to provide rigorous quantitative measures for comparisons of JA bursts among genotypes.
The results demonstrated that in the glasshouse and in the field the plant’s capacity to induce defense responses mediated by JA signaling dictates the initial feeding choice of Empoasca spp. In ir-coi1 plants, treatment with MeJA elicits the accumulation of JA but not of JA-mediated defenses and therefore did not decrease Empoasca spp. feeding damage compared with lanolin-treated plants (Fig. 4). In 35S-jmt1 plants, MeJA treatment induces JA-mediated defenses, but attack from Empoasca spp. was not decreased (Fig. 4). Thus, initial Empoasca spp. feeding can be mediated either directly by the plant’s JA signaling capacity or, most likely, by JA-dependent responses via COI1. The identification of single molecules or a combination of molecules (including signaling complexes) that Empoasca spp. perceives in the plant will require experiments in which the response of Empoasca spp. to the exposure of these molecules can be determined. These responses can range from behavioral responses (e.g., attraction, repulsion) to the recording of electrical signals from the precibarial sensilla (60) that likely are used by Cicadellidae leafhoppers in assessing the JA bursts elicited by their ingestion–egestion mode of feeding. Imaging techniques, e.g., those commonly used to study calcium signaling in Drosophila melanogaster neurons upon excitation with specific molecules, can be developed for Empoasca spp. (5). These experiments must be coupled with the isolation of molecules from the plant and, in later steps, with the generation of transgenic plants deficient in the accumulation of specific molecules.
JA is a chemical and functional analog of eicosanoids in animals, and during the divergence of plants and animals the function of these oxygenated derivatives of fatty acids in mediating defense responses against sucking insects has been conserved. Hematophagous insects induce the accumulation of eicosanoids in the bite zone that elicits inflammation and defensive behavioral responses in the host (61). Thus, as for blood-feeding dipterans, the responses induced by phytophagous hemipterans are associated with the production of oxygenated forms of fatty acids. During the course of evolution, Empoasca spp. have acquired the capacity to select appropriate hosts with diminished defensive response by indirectly perceiving JA signaling (e.g., the SCFCOI1-JA-Ile complex) after feeding. As an evolutionary counterresponse, animal and plant hosts may have amplified their attack-elicited signaling to function as an aposematic signal, warning these eavesdropping potential grazers of impending defense responses.
In summary, we demonstrate that Empoasca spp. leafhoppers select plants for feeding in nature by eavesdropping on JA-mediated signaling. Given their high mobility, Empoasca spp. leafhoppers may probe plants at random in the field and settle on those with lower levels of JA-mediated signaling for sustained feeding. This behavior can be used to identify natural variation in JA accumulation in native populations. This native tobacco uses fire to synchronize its germination from long-lived seed banks to grow in dense populations characterized by intense intraspecific competition and variable herbivore pressures (21). Hence, we hypothesize that growth–defense tradeoffs for this plant likely are severe, and these tradeoffs likely provide the selective pressure to maintain these JA-signaling mutants in native populations, despite the clear disadvantages of being defense impaired. Once we have completed the sequencing of the N. attenuata genome, we will characterize in greater detail these JA-signaling mutants that Empoasca leafhoppers have identified for us from natural populations.
Materials and Methods
Plant Material and Growth.
Seeds of the 30th and 31st generations of an inbred line of N. attenuata were used as the WT genotype in all experiments. The inbred line originated from seeds collected in 1988 from a natural population at the DI Ranch in southwestern Utah. Seeds were germinated and plants were grown as described (62).
We used plants harboring an EV construct [line A-03–9-1 (63)] as controls in all field experiments. We used the stably transformed lines [ir-lox3 line A-03–562-2 (43), ir-aos, ir-aoc, ir-opr3, ir-acx1, ir-coi1 line A-04–249-A-1 (44), and 35S-jmt1 line A-07–291-2 (47)] as plants that are silenced to various degrees in their JA production and JA signaling and perception capabilities and lines silenced in the expression of JA-mediated direct defenses, namely, DTGs [ir-ggpps line A-07–231-3 (46)], nicotine [r-pmt line A-03–108-3 (45)], PIs [ir-pi line A-04–186-1 (45)], and nicotine and PIs together [ir-pmt/pi line A-04–103-3 (45)].
Plant Treatments.
In the glasshouse, tissue was collected from the first fully elongated leaf at nodes +1 (64) of rosette-stage (∼30-d-old) N. attenuata plants. Wounding was performed by rolling a fabric pattern wheel three times on each side of the midvein. To analyze differences in jasmonate accumulation between plants, leaves were treated with water, synthetic FAC, or Manduca sexta OS. For FAC elicitation, the wounds were supplied immediately with 0.6 pmol of synthetic N-linolenoyl-glutamic acid [18:3-Glu; 20 µL of a 0.03 nmol/mL solution in 0.02% (vol/vol) Tween 20/water]. For OS elicitation we used M. sexta OS, stored under argon at −20 °C immediately after collection until use. The wounds were supplied with 20 µL of 1:5 (vol/vol) diluted OS. Leaf tissue was collected at different times and was frozen immediately in liquid nitrogen for subsequent analysis.
For the screening of native N. attenuata plants in the field, the least damaged nonsenescing leaves available on each plant were used. The leaves were chosen randomly as either control or elicited leaves. Wounding and OS elicitation were performed as described above for the glasshouse treatments. Tissue was collected 60 min after elicitation and was frozen immediately between dry ice blocks.
For MeJA treatment, 34.6 µL of pure MeJA (Sigma-Aldrich) was dissolved in 5 mL of pure lanolin (Roth) to attain a final concentration of 7.5 µg/µL. Twenty microliters of pure lanolin or MeJA-containing lanolin were applied to the abaxial side of the bases of the first three fully elongated leaves (positions +1, +2, +3) (64) of rosette-stage plants. Choice experiments were performed 2 d after treatments. For analysis of jasmonate and PI activity, tissue was collected from the untreated leaf portion 3 d after the treatment and was frozen immediately in liquid nitrogen.
To analyze the changes in jasmonate levels after Empoasca sp. attack, 25 adult leafhoppers were caged on WT leaves in two 50-mL plastic containers (Huhtamaki); two empty 50-mL plastic containers were placed on WT leaves as controls. After 24 h, the plastic containers together with leaves and insects were removed from the plant and flushed with CO2 for 15 s to anesthetize the leafhoppers. All Empoasca sp. were removed, and the leaves were frozen immediately in liquid nitrogen. Control leaves were treated similarly in the absence of Empoasca sp.
Insect Collection and Treatment.
For all experiments in the field, Empoasca leafhoppers were collected from infested C. foetidissima plants growing adjacent to the field plot at the Lytle Ranch Preserve in southwest Utah (N 37.146301, W 114.019795) during the 2009 and 2011 field seasons. For field experiments, we collected Empoasca spp. adults on the day of the experiments, between 4:00 AM and 6:00 AM, when the leafhoppers are relatively immobile and are easier to collect. Leafhopper adults were kept in plastic containers until the start of the experiment.
To establish a glasshouse colony of Empoasca sp, ∼400 adults were collected from C. foetidissima growing at the Lytle Ranch Preserve on May 30, 2009 and on June 16, 2011, were placed in an adapted Plexiglas container (1 × 2 × 1.5 m) in the glasshouse, and were reared on Cucurbita moschata, Cucurbita maxima, and Cucurbita pepo plants, which were replaced weekly. (So far, we have not been able to identify the Empoasca species used in this study because the genus is poorly understood (18). We assume that the species is Empoasca fabae, but further corroboration is required to ascertain the actual species. We would greatly appreciate expert knowledge regarding the identity of the leafhopper species. We can provide males as voucher specimens, stored in 80% (vol/vol) ethanol/water, as well as living individuals as long as our in-house colony remains viable) For the glasshouse experiments, Empoasca sp. adults were collected from the colony immediately before use, anesthetized with CO2 for 15 s, and placed in 50-mL plastic containers in different numbers, according to the experiment.
Field Experiments.
Seeds of the N. attenuata genotypes EV, ir-lox3, ir-aos, ir-aoc, ir-opr3, A466 (ir-acx1), ir-coi1, 35S-jmt1, ir-ggpps, ir-pi, ir-pmt, and ir-pmt/pi, were imported under US Department of Agriculture Animal and Plant Health Inspection Service (APHIS) notification number 07–341-101n, and the field experiments were conducted under notification number 06–242-3r-a2. All transformed N. attenuata genotypes mentioned above were used for experiments in the experimental field plot at the Lytle Ranch Preserve near Santa Clara, Utah in 2009.
For germination, seeds were treated with 1 mM gibberellic acid (GA3) in 1:50 (vol/vol) diluted liquid smoke (House of Herbs), germinated on agar plates containing Gamborg’s B5 medium (Duchefa), and grown in a shade house. After 2 wk, young seedlings were transplanted into Jiffy 703 pots (Jiffy Products), fertilized with ∼300 µg Borax (7.5 mg/L Na2[B4O5(OH)4]*8 H2O in water), and grown outdoors for 2 wk. At 28–30 d after germination, plants were planted in the field plot, and a plastic label carrying the APHIS identification number of each genotype was buried under the roots of each plant to ensure unambiguous genotype identifications. A 10-cm bamboo stick with the APHIS identification code was placed in the soil beside each plant. The field plot consisted of 26 rows separated by open irrigation troughs which allowed plants to be watered every second day until they were established rosette-stage plants.
Size-matched EV, ir-pi, ir-pmt, ir-pmt/pi, and ir-ggpps plants (10–15 plants per genotype) were planted in a fully randomized design. A466, ir-lox3, ir-aos, ir-aoc, ir-opr3, and ir-coi1 plants were planted in a randomized design, each paired with a size-matched EV plant (15 pairs per genotype). 35S-jmt1 plants were planted pairwise with a size-matched EV plant (26 pairs). All plants were monitored daily during reproductive growth, and all flowers of each genotype were removed before their corollas opened and could release pollen. Rosette diameter and stem length were determined continuously between May 15–29, 2009 (24–38 d after plantation).
An alfalfa field adjacent to the field plot provided a source of Empoasca spp. leafhoppers. We mowed this field on May 27, 2009 to encourage movement of leafhoppers to the N. attenuata plantation. We quantified the damage caused by Empoasca spp. feeding as the percentage of the leaf area damaged normalized to the total plant area 8 d after the mowing of the alfalfa field (44 d after transplantation).
Jasmonate Analysis.
The analysis of OPDA, JA, JA-Ile, MeJA, JA-Val, 11/12-hydroxy-JA, 11/12-hydroxy-JA-Ile, and 11/12-carboxy-JA-Ile was performed as previously described (29, 47). The PCA (Fig. 3B) was performed using the Metaboanalyst software (65, 66). The grouping of the transgenic N. attenuata lines, necessary for PCA analysis, was done by separating lines deficient in JA accumulation and perception from controls and from lines deficient in JA-dependent defense molecules. For this analysis levels of jasmonates and Empoasca spp. damage were normalized using autoscaling.
Analysis of PI Activity.
Leaf tissue from 40-d-old ir-lox3, ir-coi1, 35S-jmt1, and WT plants was harvested 2 d after treatment with either lanolin or MeJA containing lanolin. The analysis of PI activity was performed as previously described (67).
Empoasca Species Choice Experiments.
Six leaves from three plants each of WT, ir-lox3, ir-coi1, and 35S-jmt1 were treated with pure lanolin or with MeJA containing lanolin (7.5 µg/µL) as described above. Two days after the treatment, the plants were placed in a fully randomized design within a containment cage and were challenged with 150 Empoasca sp. adults. After 7 d, the percentage of canopy leaf area damaged by Empoasca sp. of treated leaves (Fig. S5A) was determined.
Identification of Natural Variation in Capacity for JA Accumulation in Native Populations of N. attenuata.
Native N. attenuata plants growing in the Great Basin Desert, southwestern Utah, were selected for analysis (22). In 2009 we selected one population consisting of ∼100 plants (Fig. 5A, N 37.08844, W 113.93228), and in 2011 we selected two populations consisting of ∼200 and 400 plants, respectively (Fig. 5E and Fig. S5C).
In 2009, all plants in the population were inspected carefully; plants showed no evidence of Empoasca spp. feeding damage or the presence of Empoasca spp. adults or nymphs on the plants. Fifty Empoasca spp. adults were collected from infested C. foetidissima plants growing adjacent to our field plot and were released into the native population. Two days after this release, the plants were rescreened for Empoasca spp.-damaged leaves, and two plants that had been attacked were found. Leaves from these two plants and from an unattacked control plant were treated with OS to elicit JA as described, harvested 60 min after elicitation, and frozen immediately on dry ice for subsequent jasmonate analysis. Flowers from these plants were bagged to exclude flower visitors, and seed capsules were collected from the self-pollinated flowers of both Empoasca spp.-attacked plants and from their closest undamaged neighbors. Seeds from each plant and from 30× inbred WT plants were grown in the glasshouse. Leaves were left untreated (control) or were treated with OS to elicit JA as described above and were harvested after 60 min for jasmonate analysis.
In 2011, we screened all 600 plants of the two populations and found five plants with visible Empoasca spp. damage. Leaves from these plants and from their nearest undamaged neighbors either were left untreated or were treated with OS as described, harvested 60 min after elicitation, and frozen immediately on dry ice for subsequent jasmonate analysis.
Supplementary Material
Acknowledgments
We thank the Brigham Young University for use of their field station; Danny Kessler, Celia Diezel, and Heriberto Madrigal for help during the field seasons; Drs. Georg Jander, David Heckel, and Saskia Hogenhout for helpful comments on earlier versions of this manuscript; and Dr. Saskia Hogenhout for providing Ca. Phytoplasma asteris control samples.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 9240 (volume 109, number 24).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200363109/-/DCSupplemental.
References
- 1.Thorsteinson AJ. Host selection in phytophagous insects. Annu Rev Entomol. 1960;5:193–218. [Google Scholar]
- 2.Bernays EA, Chapman RF. Host-Plant Selection by Phytophagous Insects. New York: Chapman and Hall; 1994. p 312. [Google Scholar]
- 3.Powell G, Tosh CR, Hardie J. Host plant selection by aphids: Behavioral, evolutionary, and applied perspectives. Annu Rev Entomol. 2006;51:309–330. doi: 10.1146/annurev.ento.51.110104.151107. [DOI] [PubMed] [Google Scholar]
- 4.Prokopy RJ, Owens ED. Visual generalist with visual specialist phytophagous insects – host selection behavior and application to management. Entomol Exp Appl. 1978;24(3):609–620. [Google Scholar]
- 5.Hansson BS, Knaden M, Sachse S, Stensmyr MC, Wicher D. Towards plant-odor-related olfactory neuroethology in Drosophila. Chemoecology. 2010;20:51–61. doi: 10.1007/s00049-009-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekker T, Ibba I, Siju KP, Stensmyr MC, Hansson BS. Olfactory shifts parallel superspecialism for toxic fruit in Drosophila melanogaster sibling, D. sechellia. Curr Biol. 2006;16:101–109. doi: 10.1016/j.cub.2005.11.075. [DOI] [PubMed] [Google Scholar]
- 7.Dicke M. Chemical ecology of host-plant selection by herbivorous arthropods: A multitrophic perspective. Biochem Syst Ecol. 2000;28:601–617. doi: 10.1016/s0305-1978(99)00106-4. [DOI] [PubMed] [Google Scholar]
- 8.Li XC, Schuler MA, Berenbaum MR. Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature. 2002;419:712–715. doi: 10.1038/nature01003. [DOI] [PubMed] [Google Scholar]
- 9.Dicke M, Baldwin IT. The evolutionary context for herbivore-induced plant volatiles: Beyond the ‘cry for help’. Trends Plant Sci. 2010;15:167–175. doi: 10.1016/j.tplants.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Howe GA, Jander G. Plant immunity to insect herbivores. Annu Rev Plant Biol. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- 11.Will T, Tjallingii WF, Thönnessen A, van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutti NS, et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc Natl Acad Sci USA. 2008;105:9965–9969. doi: 10.1073/pnas.0708958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musser RO, et al. Herbivory: Caterpillar saliva beats plant defences. Nature. 2002;416:599–600. doi: 10.1038/416599a. [DOI] [PubMed] [Google Scholar]
- 14.Mayer CJ, Vilcinskas A, Gross J. Phytopathogen lures its insect vector by altering host plant odor. J Chem Ecol. 2008;34:1045–1049. doi: 10.1007/s10886-008-9516-1. [DOI] [PubMed] [Google Scholar]
- 15.Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc Natl Acad Sci USA. 2011;108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gyrisco GG. Forage insects and their control. Annu Rev Entomol. 1958;3:421–448. [Google Scholar]
- 17.Carter W. Injuries to plants caused by insect toxins 2. Bot Rev. 1952;18(10):680–721. [Google Scholar]
- 18.Backus EA, Serrano MS, Ranger CM. Mechanisms of hopperburn: An overview of insect taxonomy, behavior, and physiology. Annu Rev Entomol. 2005;50:125–151. doi: 10.1146/annurev.ento.49.061802.123310. [DOI] [PubMed] [Google Scholar]
- 19.Pérez KA, et al. Transmission of the phytoplasma associated with bunchy top symptom of papaya by Empoasca papayae Oman. J Phytopathol. 2010;158(3):194–196. [Google Scholar]
- 20.Galetto L, Marzachì C, Demichelis S, Bosco D. Host plant determines the phytoplasma transmission competence of Empoasca decipiens (Hemiptera: Cicadellidae) J Econ Entomol. 2011;104:360–366. doi: 10.1603/ec10174. [DOI] [PubMed] [Google Scholar]
- 21.Preston CA, Baldwin IT. Positive and negative signals regulate germination in the post-fire annual, Nicotiana attenuata. Ecology. 1999;80(2):481–494. [Google Scholar]
- 22.Bahulikar RA, Stanculescu D, Preston CA, Baldwin IT. ISSR and AFLP analysis of the temporal and spatial population structure of the post-fire annual, Nicotiana attenuata, in SW Utah. BMC Ecol. 2004;4:12. doi: 10.1186/1472-6785-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voelckel C, Baldwin IT. Herbivore-induced plant vaccination. Part II. Array-studies reveal the transience of herbivore-specific transcriptional imprints and a distinct imprint from stress combinations. Plant J. 2004;38:650–663. doi: 10.1111/j.1365-313X.2004.02077.x. [DOI] [PubMed] [Google Scholar]
- 24.Diezel C, von Dahl CC, Gaquerel E, Baldwin IT. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol. 2009;150:1576–1586. doi: 10.1104/pp.109.139550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stork W, Diezel C, Halitschke R, Gális I, Baldwin IT. An ecological analysis of the herbivory-elicited JA burst and its metabolism: Plant memory processes and predictions of the moving target model. PLoS ONE. 2009;4:e4697. doi: 10.1371/journal.pone.0004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, 3rd, Teal PE. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA. 2009;106:653–657. doi: 10.1073/pnas.0811861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol. 2010;152:96–106. doi: 10.1104/pp.109.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonaventure G, Schuck S, Baldwin IT. Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: A specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant Cell Environ. 2011;34:1507–1520. doi: 10.1111/j.1365-3040.2011.02348.x. [DOI] [PubMed] [Google Scholar]
- 30.Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003;36:794–807. doi: 10.1046/j.1365-313x.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- 31.Schaller F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot. 2001;52:11–23. [PubMed] [Google Scholar]
- 32.Breithaupt C, et al. Crystal structure of 12-oxophytodienoate reductase 3 from tomato: Self-inhibition by dimerization. Proc Natl Acad Sci USA. 2006;103:14337–14342. doi: 10.1073/pnas.0606603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen L, Henriksen A. Acyl-CoA oxidase 1 from Arabidopsis thaliana. Structure of a key enzyme in plant lipid metabolism. J Mol Biol. 2005;345:487–500. doi: 10.1016/j.jmb.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 34.Seo HS, et al. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA. 2001;98:4788–4793. doi: 10.1073/pnas.081557298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suza WP, Staswick PE. The role of JAR1 in Jasmonoyl-L: -isoleucine production during Arabidopsis wound response. Planta. 2008;227:1221–1232. doi: 10.1007/s00425-008-0694-4. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, et al. Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta. 2007;226:159–167. doi: 10.1007/s00425-007-0477-3. [DOI] [PubMed] [Google Scholar]
- 37.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 38.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 39.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 40.Kessler A, Halitschke R, Baldwin IT. Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science. 2004;305:665–668. doi: 10.1126/science.1096931. [DOI] [PubMed] [Google Scholar]
- 41.Lorenz KH, Schneider B, Ahrens U, Seemuller E. Detection of the apple proliferation and pear decline phytoplasma by PCR amplification of ribosomal and nonribosomal DNA. Phytopathology. 1995;85:771–776. [Google Scholar]
- 42.Lee IM, Bartoszyk IM, Gundersen DE, Mogen B, Davis RE. Nested-PCR assays for ultrasensitive detection of potato ring rot bacterium, Clavibacter michiganensis subsp. sepedonicus. Appl Environ Microbiol. 1997;63:2625–2630. doi: 10.1128/aem.63.7.2625-2630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allmann S, Halitschke R, Schuurink RC, Baldwin IT. Oxylipin channelling in Nicotiana attenuata: Lipoxygenase 2 supplies substrates for green leaf volatile production. Plant Cell Environ. 2010;33:2028–2040. doi: 10.1111/j.1365-3040.2010.02203.x. [DOI] [PubMed] [Google Scholar]
- 44.Paschold A, Halitschke R, Baldwin IT. Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the Role of herbivore movement in avoiding defenses. Plant J. 2007;51:79–91. doi: 10.1111/j.1365-313X.2007.03119.x. [DOI] [PubMed] [Google Scholar]
- 45.Steppuhn A, Baldwin IT. Resistance management in a native plant: Nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol Lett. 2007;10:499–511. doi: 10.1111/j.1461-0248.2007.01045.x. [DOI] [PubMed] [Google Scholar]
- 46.Heiling S, et al. Jasmonate and ppHsystemin regulate key Malonylation steps in the biosynthesis of 17-Hydroxygeranyllinalool Diterpene Glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell. 2010;22:273–292. doi: 10.1105/tpc.109.071449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stitz M, Gase K, Baldwin IT, Gaquerel E. Ectopic expression of AtJMT in Nicotiana attenuata: Creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. Plant Physiol. 2011;157:341–354. doi: 10.1104/pp.111.178582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaquerel E, Weinhold A, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphigidae) and its natural host Nicotiana attenuata. VIII. An unbiased GCxGC-ToFMS analysis of the plant’s elicited volatile emissions. Plant Physiol. 2009;149:1408–1423. doi: 10.1104/pp.108.130799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VanDoorn A, Bonaventure G, Schmidt DD, Baldwin IT. Regulation of jasmonate metabolism and activation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 2011;190:640–652. doi: 10.1111/j.1469-8137.2010.03622.x. [DOI] [PubMed] [Google Scholar]
- 50.Koo AJK, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA. 2011;108:9298–9303. doi: 10.1073/pnas.1103542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Dam NM, Baldwin IT. Heritability of a quantitative and qualitative protease inhibitor polymorphism in Nicotiana attenuata. Plant Biol. 2003;5:179–185. [Google Scholar]
- 52.Baldwin IT. Plant volatiles. Curr Biol. 2010;20:R392–R397. doi: 10.1016/j.cub.2010.02.052. [DOI] [PubMed] [Google Scholar]
- 53.Bruce TJA, Wadhams LJ, Woodcock CM. Insect host location: A volatile situation. Trends Plant Sci. 2005;10:269–274. doi: 10.1016/j.tplants.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Allmann S, Baldwin IT. Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles. Science. 2010;329:1075–1078. doi: 10.1126/science.1191634. [DOI] [PubMed] [Google Scholar]
- 55.Ranger CM, et al. Discrimination by the potato leafhopper (Hemiptera: Cicadellidae) of host volatiles from resistant and susceptible alfalfa, Medicago sativa L. Environ Entomol. 2005;34:271–280. [Google Scholar]
- 56.Hill GT, Sinclair WA. Taxa of leafhoppers carrying phytoplasmas at sites of ash yellows occurrence in New York state. Plant Dis. 2000;84:134–138. doi: 10.1094/PDIS.2000.84.2.134. [DOI] [PubMed] [Google Scholar]
- 57.Fraenkel GS. Rasion detre of secondary plant substances. Science. 1959;129:1466–1470. doi: 10.1126/science.129.3361.1466. [DOI] [PubMed] [Google Scholar]
- 58.Ussuf KK, Laxmi NH, Mitra R. Proteinase inhibitors: Plant-derived genes of insecticidal protein for developing insect-resistant transgenic plants. Current Sci India. 2001;80:847–853. [Google Scholar]
- 59.Zavala JA, Patankar AG, Gase K, Baldwin IT. Constitutive and inducible trypsin proteinase inhibitor production incurs large fitness costs in Nicotiana attenuata. Proc Natl Acad Sci USA. 2004;101:1607–1612. doi: 10.1073/pnas.0305096101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Backus EA, McLean DL. The sensory systems and feeding behavior of leafhoppers. II. A comparison of the sensillar morphologies of several species (Homoptera: Cicadellidae) J Morphol. 1983;176:3–14. doi: 10.1002/jmor.1051760102. [DOI] [PubMed] [Google Scholar]
- 61.Alvarenga PH, et al. The function and three-dimensional structure of a thromboxane A2/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite. PLoS Biol. 2010;8:e1000547. doi: 10.1371/journal.pbio.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology. 2002;12:177–183. [Google Scholar]
- 63.Bubner B, Gase K, Berger B, Link D, Baldwin IT. Occurrence of tetraploidy in Nicotiana attenuata plants after Agrobacterium-mediated transformation is genotype specific but independent of polysomaty of explant tissue. Plant Cell Rep. 2006;25:668–675. doi: 10.1007/s00299-005-0111-4. [DOI] [PubMed] [Google Scholar]
- 64.Pluskota WE, Qu N, Maitrejean M, Boland W, Baldwin IT. Jasmonates and its mimics differentially elicit systemic defence responses in Nicotiana attenuata. J Exp Bot. 2007;58:4071–4082. doi: 10.1093/jxb/erm263. [DOI] [PubMed] [Google Scholar]
- 65.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652-60. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 67.Gilardoni PA, Hettenhausen C, Baldwin IT, Bonaventure G. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell. 2011;23:3512–3532. doi: 10.1105/tpc.111.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]