ABSTRACT
FAM110C belongs to a family of proteins that regulates cell proliferation. In the present study, the spatiotemporal expression pattern of FAM110C and its potential role were examined during the periovulatory period. Immature female rats were injected with equine chorionic gonadotropin (eCG) followed by human chorionic gonadotropin (hCG) and ovaries or granulosa cells were collected at various times after hCG administration (n = 3/time point). Expression levels of Fam110c mRNA and protein were highly induced both in intact ovaries and granulosa cells at 8 to 12 h after hCG treatment. In situ hybridization analysis demonstrated Fam110c mRNA expression was induced in theca and granulosa cells at 4 h after hCG, primarily localized to granulosa cells at 8 h and 12 h, and decreased at 24 h after hCG. There was negligible Fam110c mRNA detected in newly forming corpora lutea. In rat granulosa cell cultures, hCG induced expression of Fam110c mRNA was inhibited by RU486, whereas NS398 and AG1478 had no effect, suggesting that Fam110c expression is regulated in part by the progesterone receptor pathway. Promoter activity analysis revealed that an Sp1 site was important for the induction of Fam110c expression by hCG. Overexpression of FAM110C promoted granulosa cells to arrest at the G1 phase of the cell cycle but did not change progesterone levels. In summary, hCG induces Fam110c mRNA expression in granulosa cells by activation of an Sp1-binding site and the actions of progesterone. Our findings suggest that FAM110C may control granulosa cell differentiation into luteal cells by arresting cell cycle progression.
Keywords: differentiation, granulosa cell, ovary, ovulation, progesterone, SP1
Human chorionic gonadotropin induces Fam110c mRNA expression in granulosa cells, which promotes their arrest at the G1 phase of the cell cycle; this suggests that FAM110C may control granulosa cell differentiation into luteal cells.
INTRODUCTION
Cellular proliferation is regulated by controlling the structural machinery of DNA replication as well as by altering the delicate balance between stimulatory and inhibitory factors that control the transition through the checkpoints of the cell cycle. Factors that control the transition through the checkpoints of the cell cycle include cyclins and cyclin-dependent kinases (cdk). Cyclins and cdks are essential for early transition through the G1 phase of the cell cycle [1]. Inhibitors of cdks (or CKIs) can arrest the cell cycle in the G1 phase by inhibiting the activity of cyclin-cdk complexes. In the ovary, granulosa cell proliferation, cell cycle arrest, and terminal differentiation are critical for normal follicular growth, ovulation, and luteinization [2]. In the preovulatory follicle, the midcycle luteinizing hormone (LH) surge results in an arrest of the cell cycle in granulosa cells which in turn cease dividing and begin to differentiate into luteal cells [3, 4]. The mechanisms controlling granulosa cell proliferation and differentiation remain to be fully elucidated; however, information from studies in mice lacking cell cycle regulators has begun to provide insight into this complex process. Granulosa cells from mice lacking cyclin D2 are unable to proliferate in response to follicle-stimulating hormone (FSH), and ovulation fails to occur [5]. Likewise, deletion of p27, a protein that under normal cell cycle conditions prevents premature DNA replication, results in mice that are sterile due to the failure of luteinization in response to LH [6, 7]. Previously, we reported that the B cell translocation genes (BTGs) regulate the exit of granulosa cells from the cell cycle and promote their survival, which would direct their differentiation into luteal cells [8]. The present study was undertaken to further investigate cell cycle regulators in the ovary, specifically a newly identified centrosome/spindle pole-associated protein, family with sequence similarity 110C (FAM110C).
The centrosome, along with the spindles and associated microtubules, form the structural components required for mitosis and cytokinesis. The centrosome serves to establish the polarity and orientation of microtubules during interphase at the microtubule organization center and directs spindle assembly during mitosis [9, 10]. Centrosomes also provide a structural scaffold for the coordination of cell cycle progression [9, 10]. In addition, the centrosome is the site of enzymes involved in control of cell cycle progression phases G1 to S and DNA replication (reviewed in ref. 10). These observations have led to the proposal that the centrosome acts as a catalytic core and/or a functional lattice for the integration of different pathways that control the cell division cycle [10, 11]. The novel centrosome/spindle pole-associated protein (CSPP) was described by Patzke and coworkers [9] while screening genes involved in the progression from low-grade to high-grade malignancies. This CSPP is associated with the centrosome/microtubule cytoskeleton throughout the cell cycle and controlled progression from G1 to S phase and microtubule organization/spindle formation during mitosis [9, 11]. In a search for CSPP-associated proteins, FAM110 was identified as a family consisting of three genes, Fam110a, Fam110b, and Fam110c [12]. Cellular localization analysis showed that FAM110 proteins localized to centrosomes and accumulated at the microtubule organization center during cell cycle progression. Functionally, ectopic expression of the FAM110C protein impaired cell proliferation [12].
Currently, there are little to no data about the expression of the FAM110C protein in the ovary and its potential role during the periovulatory period. We hypothesized that the LH surge would induce expression of FAM110C and that this induction would facilitate luteinization of granulosa cells through its action as a cell cycle regulator. Therefore, we examined the expression pattern, regulation, and potential function of FAM110C during the granulosa-luteal cell transition period in the rat ovary.
MATERIALS AND METHODS
Materials and Reagents
All chemicals and reagents were purchased from Sigma Chemical Co. (St. Louis, MO), except where otherwise noted. Molecular biological enzymes, molecular size markers, oligonucleotide primers, pCRII-TOPO vector, culture medium, and TRIzol were from Invitrogen Life Technologies, Inc. (Carlsbad, CA).
Tissue Collection
Immature female Sprague Dawley rats (15 days old) were obtained from Harlan, Inc. (Indianapolis, IN). Animals were kept in environmentally controlled conditions under the supervision of a veterinarian. All animal procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee. Between 0900 and 1000 h on Days 22–23, rats were injected s.c. with 10 IU of pregnant mare's serum equine chorionic gonadotropin (eCG) to induce follicular development. Forty-eight hours later, animals were injected with human chorionic gonadotropin (hCG; 5 IU s.c.). Ovaries were collected at 0 h (i.e., time of hCG administration) and at 4, 8, 12, or 24 h after hCG injection (n = 3–4 animals/time point). Ovaries were removed, cleaned, weighed, and stored at −70°C for later isolation of total RNA or protein or placed in Tissue-Tek OCT compound (VWR Scientific, Atlanta, GA), snap frozen, and stored at −70°C until sectioned and processed for in situ hybridization analyses. Granulosa cells isolated from ovaries at different time points were snap frozen for later isolation of total RNA or protein.
In Situ Hybridization of Fam110c
In situ hybridization was performed using plasmids containing rat Fam110c cDNA as previously described. Oligonucleotide primers corresponding to rat Fam110c cDNA (forward, 5′-AGCCTCTATGTTACCACCTCC-3′; reverse, 5′-AGCTCCTCTTCTTGCAACC-3′) were designed using PRIMER3 software [13]. Antisense and sense cRNA probes were synthesized from the corresponding linearized plasmid and labeled with α-35S-uridine 5′-triphosphate, using a Maxiscript in vitro RNA transcription kit from Ambion (Austin, TX). After cRNA synthesis, probes were purified using G-50 Sephadex Quick Spin columns (Roche Molecular Biochemicals, Indianapolis, IN). Ovaries were sectioned at 10 μm and mounted on Probe-On Plus slides (Fisher Scientific, Pittsburgh, PA). Each cRNA probe was allowed to hybridize overnight in hybridization buffer containing 1 × 106 cpm of probe per slide at 55°C. Approximately 18–20 h later, slides were washed extensively to remove nonspecifically bound cRNA, and then treated with RNase A (0.025 mg/ml in Tris-EDTA buffer). Slides were again washed extensively, dehydrated in ethanol, and air dried. Sections were processed for autoradiography using Kodak NTB2 emulsion (Eastman Kodak, Rochester, NY) and stored at 4°C. For visualization of the in situ reaction product, slides were developed in Kodak D19 (1:1 dilution) and stained with Gill 2 hematoxylin solution (Fisher Scientific). A sense cRNA probe, used as a control for nonspecific binding, was included for each time point. One ovary each from a minimum of three animals per time point was used for in situ hybridization.
Rat Granulosa Cell Culture
To isolate granulosa cells, we collected ovaries from rats 48 h after eCG administration as described previously [14]. Briefly, granulosa cells were isolated by follicle puncture, filtered, pelleted by centrifugation at 200 × g for 5 min, and resuspended in Opti-MEM containing 0.05 mg/ml of gentamicin and 1× solution of insulin-transferrin-selenium (ITS). Granulosa cells were cultured in the absence or presence of various reagents, as discussed in detail below, for different time points at 37°C in a humidified atmosphere of 5% CO2. When reagents were dissolved in dimethyl sulfoxide (DMSO), the same concentration of DMSO was added to medium for the control cells (0.1% final concentration). Forskolin (FSK) and phorbol 12 myristate 13-acetate (PMA) were added to mimic hCG action. Pelleted cells were prepared for isolation of total RNA and protein.
Real-Time PCR Quantification of mRNA
Total RNA was isolated from ovaries or granulosa cells, using TRIzol reagent according to the manufacturer's protocol. Real-time PCR was used to measure expression of Fam110c mRNA in vitro and in vivo, as well as genes associated with steroidogenesis, Hsd3b (3 beta-hydroxysteroid dehydrogenase) and Cyp11a1 (P450-mediated side chain cleavage enzyme). Oligonucleotide primers corresponding to cDNA for rat Fam110c (forward, 5′-TGAGTCTGACACCTTCTTCC-3′; reverse, 5′-GAAGTAGCCATGCTCACG-3′), rat Hsd3b (forward, 5′-TGGTGGCACATTGCATACTT-3′; reverse, 5′-TAGCTTTGGTGAGGGGTGTC-3′), rat Cyp11a1 (forward, 5′-GCTGGAAGGTGTAGCTCAGG-3′; reverse, 5′-CACTGGTGTGGAACATCTGG-3′), and rat Rpl32 (forward, 5′-GAAGCCCAAGATCGTCAAAA-3′; reverse, 5′-AGGATCTGGCCCTTGAATCT-3′) were designed using PRIMER3 software. The specificity for each primer set was confirmed both by electrophoresis of the PCR products on a 2.0 % agarose gel and by analyzing the melting (dissociation) curve in the MxPro real-time PCR analysis program (Stratagene, La Jolla, CA) after each real-time PCR reaction. PCR products were sequenced before use. Real-time PCR reaction mixtures contained 10% of the RT reaction product, 0.4 μM forward and reverse primers, 0.3 μl of ROX reference dye (1:10 dilution; provided with SYBR GreenER quantitative PCR SuperMix Universal kit, Invitrogen), and SYBR Green SuperMix. Thermal cycling steps included 2 min at 50°C to permit optimal AmpErase uracil-N-glycosylase activity, 10 min at 95°C for initial denaturation, and then 30 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C for 40 cycles, followed by 1 min at 95°C, 30 sec at 58°C, and then 30 sec at 95°C for ramp dissociation. The relative amount of each gene transcript was calculated using the 2−ΔΔCt method and normalized to the endogenous Rpl32 reference gene.
Western Blot Analysis
Whole ovaries from different time points were homogenized in a radioimmunoprecipitation assay (RIPA) buffer-protease inhibitor cocktail (Santa Cruz Biotechnology, Santa Cruz, CA). Granulosa cells were resuspended in RIPA buffer-protease inhibitor cocktail for 30 min on ice. Tissue and cell lysates were then centrifuged at 12 000 × g for 10 min. Supernatants were stored at −70°C until use. Twenty micrograms of protein, measured by the Lowry method [15], was separated by 15% SDS-PAGE and transferred to a nitrocellulose membrane (Whatman, Sanford, ME). Western blotting was performed by blocking nonspecific binding with 5% dry milk in Tris-buffered saline buffer containing 0.1% Tween-20 for 1 h. Blots were incubated with the primary antibody for FAM110C (1:200 dilution; Santa Cruz Biotechnology) or β-actin (1:1000 dilution; Cell Signaling Technology, Danvers, MA) overnight at 4°C on a rocking platform. Blots were washed three times with PBS plus 0.1% Tween and incubated with respective secondary antibodies linked to horseradish peroxidase for 1 h. After blots were washed, they were analyzed using an enhanced chemiluminescence detection system (GE Healthcare, Piscataway, NJ) and exposed to x-ray film.
Immunoassay of Progesterone
Concentrations of progesterone in conditioned culture medium following granulosa cell culture were assayed using an Immulite kit on an Immulite 1000 machine (Siemens Healthcare Diagnostics, Los Angeles, CA). Assay sensitivity was 0.2 ng/ml, and the intra-assay and interassay coefficients of variation were 6.3% and 9.1%, respectively.
Generation of Luciferase Reporter Vector and Granulosa Cell Transfection
Genomic DNA was isolated from rat tail samples, using an Easy-DNA kit (Invitrogen). Fragments of the Fam110c gene, a 900-bp (−874/+26), a 542-bp (−516/+26), a 164-bp (−138/+26), and a 75-bp (−49/+26) fragment, were amplified using the primers attached with restriction enzyme sites (KpnI and HindIII). Fragments for the Fam110c gene promoter were cloned into the pGL3 basic vector (Promega, Madison, WI) as described previously [16].
A QuickChange II site-directed mutagenesis kit (Stratagene) was used to generate site-directed point mutation of the Fam110c promoter. The sequence of the oligonucleotide primer used to generate the Fam110c promoter containing the Sp1 binding site mutation (shown in lowercase) was 5′-GGCGTGTTTTCCCCGaaCCTTGCGGACTTCGG-3′. Granulosa cells were isolated from immature rats (48 h after eCG administration) as described above. Granulosa cells were transfected with respective firefly luciferase reporter plasmids (pGL3-Fam110c promoter constructs) and Renilla luciferase vector (pRL-TK vector) using Lipofectamine 2000 reagent (Invitrogen). The next day, granulosa cells were treated with FSK (10 μM), PMA (20 nM), or FSK plus PMA for 6 h. The cells were harvested to measure firefly and Renilla luciferase activities by using a dual-luciferase reporter assay system (Promega), and each reaction was monitored for 10 sec by the luminescence system in the Tecan Infinite 200 microplate reader (Tecan US, Durham, NC). Firefly luciferase activities were normalized by Renilla luciferase activities, and each experiment was performed in triplicate at least three times
Generation of Recombinant Adenovirus Vector
To explore the function of FAM110C protein, adenovirus vectors were constructed to overexpress FAM110C. The Fam110c cDNA was amplified by PCR using total RNA isolated from rat ovary. Specific primers for rat Fam110c (forward, 5′-TTGGTACCACCATGCGCGCCCTGCCCACC-3′; reverse, 5′-TTGCGGCCGCTCAGGCAGGTTCCTGCAACTTC-3′) contained the KpnI and KpnI sites. The PCR product was subcloned into pShuttle-CMV vector (Stratagene). AdEasy XL adenovirus vector system was used to generate recombinant Ad-Fam110c vectors. The resultant plasmids carrying the Fam110c gene under the control of a cytomegalovirus promoter were linearized using Pme1 and then cotransformed into electro-competent BJ5183 bacteria with pAdEasy-1, which contains the viral backbone. The recombinant plasmid was selected on kanamycin lysogeny broth plates. Recombinant adenovectors were linearized using PacI and transfected to Ad293 cells, where viral particles were further amplified. Adenovectors were purified, and then the titer was determined by using the AdEasy viral titer kit (Stratagene).
Flow Cytometric Analysis of Granulosa Cells
To determine the impact of FAM110C overexpression on cell cycle kinetics, rat granulosa cells were collected at 48 h after eCG priming as detailed above. Granulosa cells were cultured on 6-well plates in Opti-MEM supplemented with 0.05 mg/ml gentamicin and 1× ITS solution for 4 h before addition of the adenovirus vector containing Fam110c (Ad-Fam110c) or a control green fluorescent protein (GFP)-labeled adenovirus (Ad-GFP) vector. Granulosa cells were exposed to Ad-Fam110c or Ad-GFP at a multiplicity of infection of 50 plaque-forming units (pfu)/cell for 2 h. We routinely observed an infection efficiency of Ad-GFP in granulosa cells of approximately 70%. Medium was replaced with fresh Opti-MEM. After 2 days of culture, granulosa cells were suspended using 3% trypsin and then stained for DNA content [17]. Briefly, ribonuclease A (final concentration of 0.1 mg/ml) was added to 1 × 106 cells and incubated at 37°C for 30 min. Afterward, granulosa cells were resuspended in 50 μg/ml propidium iodide and incubated for 1 h in the dark at 4°C. The cell cycle distribution along with the percentage of cells with degraded DNA were determined at an excitation wavelength of 488 nm, using a FACS Calibur flow cytometer (Becton Dickinson) at the core flow cytometry laboratory at the University of Kentucky. Cell cycle histograms were obtained from three determinations, each with a total of 100 000 cells/treatment.
Statistical Analyses
All data are presented as means ± SEM. Data were tested for homogeneity of variance, and if data were not normally distributed, the data were log transformed. One-way ANOVA was used to test differences in Fam110c mRNA expression across time of culture or among treatments in vitro. If ANOVA revealed significant effects of time of tissue collection, time of culture, or treatment, the means were compared using the Duncan test, with a P value of <0.05 considered significant. A Student t-test was used to test for differences in FAM110C protein expression levels, as appropriate, with a P value of <0.05 considered significant.
RESULTS
hCG Induced Fam110c Expression in Periovulatory Rat Ovaries and Granulosa Cells
To determine whether hCG induced ovarian Fam110c mRNA and protein expression, expression patterns from intact ovaries or granulosa cells collected at different times after hCG administration were analyzed. In the intact ovary, the levels of mRNA for Fam110c reached the highest expression at 8 h after hCG and remained elevated at 12 h before declining to control values at 24 h (Fig. 1A). In the granulosa cell compartment, the level of Fam110c mRNA expression was similar to its expression level in the whole ovary. Fam110c mRNA was highly induced at 8 h and remained elevated at 12 h before declining at 24 h after hCG treatment in granulosa cells collected in vivo (Fig. 1B). Western blot results showed that FAM110C expression increased at 8 h and remained high until 24 h after hCG treatment in whole ovaries (Fig. 1C) and granulosa cells (Fig. 1D).
FIG. 1. .
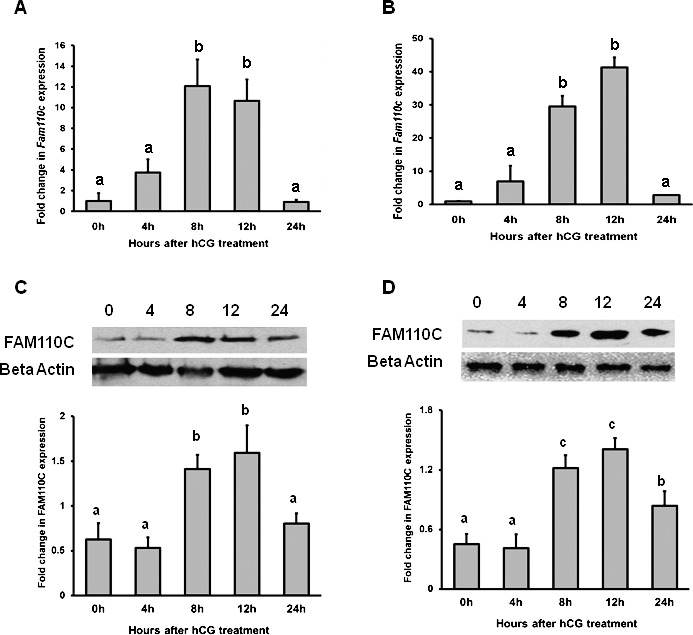
Stimulation of Fam110c expression by hCG in whole ovaries and granulosa cells in vivo. Real-time PCR analysis shows expression of Fam110c mRNA in periovulatory ovaries (A) and granulosa cells in vivo (B) after hCG administration. eCG-primed rats were injected with hCG, and ovaries or granulosa cells were collected at 0, 4, 8, 12, or 24 h after treatment. Relative levels of Fam110c mRNA were normalized to those of Rpl32 in each sample (mean ± SEM; n = 3 independent culture experiments). Western blot analysis shows FAM110C protein levels at different time points after hCG treatment in intact rat ovaries (C) and granulosa cells in vivo (D). Upper panel shows a representative Western blot; lower panel shows relative quantitative levels of FAM110C protein normalized to that of β-actin in each sample (mean ± SEM; n = 3 independent culture experiments). Bars with no common letters are significantly different (P < 0.05).
Localization of Fam110c mRNA in the Ovary
In situ hybridization of Fam110c mRNA revealed that the expression of this gene was low in the ovary at 0 h (Fig. 2, A and B). Fam110c mRNA was localized to theca-interstitial cells and granulosa cells of certain follicles at 4 h after hCG administration (Fig. 2, C, D, K, and L). At 8 and 12 h after hCG administration, there was a marked expression of Fam110c mRNA in the granulosa cell compartment, while the signal in theca-interstitial cells decreased to almost undetectable levels (Fig. 2, E–H). After ovulation, Fam110c mRNA expression was similar to that of the 0 h ovary, with little detectable signal in the forming corpus luteum (Fig. 2, I and J). There was no signal detected on the sections that were hybridized to the sense probe of Fam110c (data not shown).
FIG. 2. .
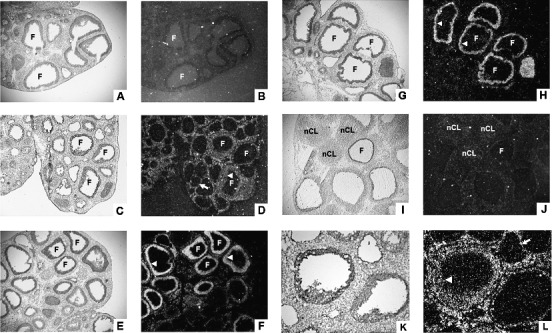
Cellular localization of Fam110c mRNA in periovulatory rat ovaries. Sections of rat ovaries obtained at 0 h (48 h post-eCG) (A, B), 4 h (C, D, K, L), 8 h (E, F), 12 h (G, H), or 24 h (I, J) after hCG injection were hybridized with antisense probes for Fam110c mRNA. Representative bright-field (A, C, E, G, I, K) and corresponding dark-field (B, D, F, H, J, L) photomicrographic views are shown. Higher magnification of the area in C is shown in K and L. Arrows indicate the theca layer. Arrowheads indicate the granulosa cell layer. F, follicle; nCL, newly formed corpus luteum. Original magnification ×40 (A–J) and ×100 (K and L).
hCG Induces Expression of Fam110c mRNA in Granulosa Cells In Vitro
The hCG-induced increase in Fam110c mRNA expression was observed in granulosa cells collected in vivo; therefore, studies were conducted to determine whether the increase in Fam110c mRNA in vivo could be mimicked in vitro by treatment with hCG. Real-time PCR data showed that hCG induced an expression pattern of Fam110c mRNA in cultured granulosa cells (Fig. 3A) similar to that in cells collected in vivo (Fig. 1B), except that the induction was advanced in vitro. In the cultured cells, Fam110c mRNA expression was increased at 4 h and declined at 12 and 24 h after hCG induction (Fig. 3A).
FIG. 3. .
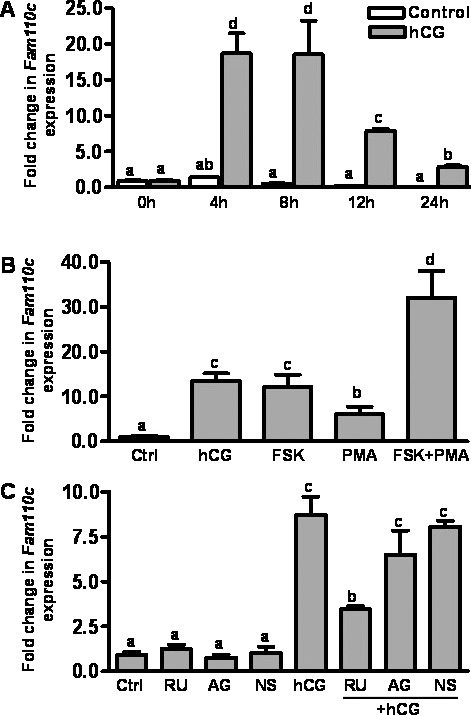
Expression and regulation of Fam110c mRNA expression by hCG in rat granulosa cells in vitro. Real-time PCR analysis demonstrates expression of Fam110c mRNA in granulosa cells. A) Fam110c mRNA expression in granulosa cells obtained from rat preovulatory ovaries (48 h post-eCG) and cultured in medium alone (Control) or with hCG (1 IU/ml) for 0, 4, 8, 12, or 24 h. B) Expression of Fam110c mRNA in granulosa cells from rat preovulatory ovaries (48 h post-eCG) cultured in medium alone (Ctrl) or with hCG (1 IU/ml), FSK (10 μM), or PMA (20 nM) for 8 h. C) Regulation of Fam110c mRNA expression in granulosa cells in vitro. Expression of Fam110c mRNA in granulosa cells from rat preovulatory ovaries (48 h post-eCG) cultured in medium alone (Ctrl) or with hCG (1 IU/ml) in the absence or presence of the progesterone receptor antagonist RU486 (1 μM), the prostaglandin-endoperoxide synthase 2 inhibitor NS398 (1 μM), or the EGF receptor tyrosine kinase selective inhibitor AG1478 (1 μM) for 8 h. In all experiments, the relative level of mRNA for Fam110c was normalized to that of Rpl32 in each sample (mean ± SEM; n = 3 independent culture experiments). Bars with no common letters are significantly different (P < 0.05). Note, data for the 8-h control (A) appear as a black bar due to the low values for this time point.
Regulation of Fam110c mRNA Expression
Both the PKA and the PKC signaling pathways are known to be activated by hCG in preovulatory granulosa cells [18, 19]. To determine which signaling pathway(s) is involved in the up-regulation of Fam110c mRNA expression in response to hCG stimulation, we cultured granulosa cells from rat preovulatory ovaries (48 h post-eCG) with hCG, FSK (which is an activator of adenylate cyclase or an activator of protein kinase C), and PMA for 8 h. Both FSK and PMA stimulated Fam110c mRNA expression (Fig. 3B), suggesting that both signaling pathways are involved in the induction of Fam110c by hCG.
hCG activates epidermal growth factor (EGF) receptor signaling and induces expression of prostaglandin synthase 2 (PTGS2) and progesterone receptor (PGR) [20]. We tested whether the up-regulation of Fam110c mRNA was mediated by hCG-induced activation of these signaling pathways by using RU486 to block the actions of progesterone receptors, NS398 to block PTGS2 synthesis, and AG1478, to prevent EGF receptor signaling. RU486 treatment reduced the hCG-induced expression of Fam110c mRNA in vitro (Fig. 3C), whereas NS398 and AG1478 had no effect (Fig. 3C). These findings suggest that activation of the progesterone receptor mediates hCG induction of Fam110c mRNA expression.
Mutation of the Sp1 Binding Site Reduces Fam110c Promoter Reporter Activity in Preovulatory Granulosa Cells
To investigate the transcriptional regulation of Fam110C, four rat Fam110c promoter reporter constructs (bp −874/+26; bp −516/+26; bp −138/+26; and bp −49/+26) were transfected into preovulatory granulosa cells. After 24 h of equilibration, granulosa cells were stimulated with or without FSK plus PMA for 6 h. Treatment with FSK plus PMA increased the luciferase activities of bp −874/+26, bp −516/+26, and bp −138/+26 reporter constructs compared with that of cells cultured without FSK plus PMA (Fig. 4). In contrast, FSK plus PMA had no effect on the activity of the reporter construct bp fragment −49/+26. Nuclear transcription factor SP1 is induced by the LH surge and is known to regulate gene expression in periovulatory granulosa cells [21, 22]. As there is a consensus Sp1 binding site at bp −72 of the Fam110c promoter, we determined whether the SP1 transcription factor was important for the induction of the Fam110c promoter activity by mutating the Sp1 binding site. Luciferase assays demonstrated that mutation of the Sp1 binding site decreased induction of the Fam110c promoter by FSK plus PMA, which suggests that SP1 is involved in Fam110c transcriptional regulation.
FIG. 4. .
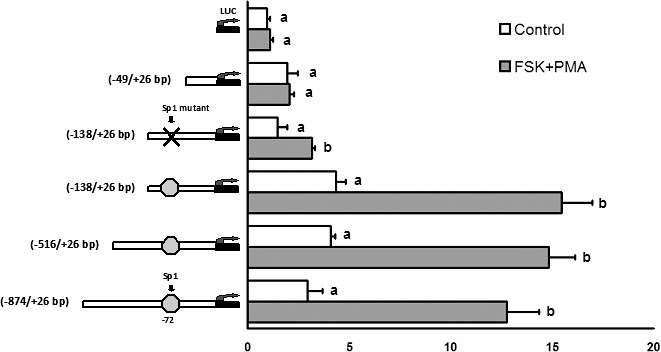
Activity of Fam110c promoter reporter constructs in cultured granulosa cells. Granulosa cells isolated from preovulatory ovaries (48 h after eCG) were transiently transfected with empty luciferase reporter vector (LUC), bp fragments −874/+26, −516/+26bp, −138/+26, and −138/+26, Sp1 mutant, or bp fragment −49/+26 Fam110c promoter luciferase reporter constructs and treated with FSK (10 μM) plus PMA (20 nM) for 6 h. Firefly luciferase activity was normalized to Renilla luciferase activity. Each experiment was performed in triplicate, and the experiment was repeated at least three times. Bars with no common letters within a reporter construct are significantly different (P < 0.05).
Cell Cycle Analysis of Granulosa Cells After Overexpression of Rat Fam110c
To investigate the potential function of FAM110C, a Fam110c overexpression adenovirus vector was constructed. Granulosa cells from preovulatory ovaries were infected with either Ad-Fam110c or Ad-GFP control adenovirus. Western blot analysis results confirmed that FAM110C was highly expressed in the Ad-Fam110c vector-infected granulosa cells (Fig. 5A). Overexpression of FAM110C inhibited granulosa cells entering the cell cycle (Fig. 5B). The percentage of cells in the G1 phase increased markedly in granulosa cells expressing FAM110C, whereas the percentage of cells in the S phase decreased (Fig. 5B). These data for the cell cycle suggest that FAM110C inhibits the transition of granulosa cells from the G1 to the S phase of the cell cycle.
FIG. 5. .
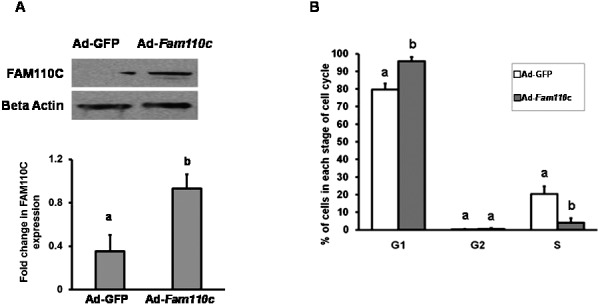
Analysis of granulosa cell cycle kinetics after overexpression of rat FAM110C. Granulosa cells from preovulatory ovaries were infected with either Ad-Fam110c or an Ad-GFP control adenovirus. A) Western blot analysis demonstrates expression of FAM110C and the endogenous control β-actin. Upper panel shows a representative Western blot; lower panel shows relative quantitative levels of FAM110C protein normalized to β-actin in each sample. B) Cell cycle kinetics using flow cytometry. Data demonstrate percentages of cells in different stages of cell cycles, G1 (G0–G1), G2, and S phases, in cells overexpressing FAM110C. Results represent means ± SEM for at least 3 individual experiments. Bars that do not share a letter or number designation are significantly different (P < 0.05).
Analysis of Granulosa Cell Steroidogenesis After Overexpression of Rat Fam110c
To explore the potential role of FAM110C in steroidogenesis, granulosa cells from preovulatory ovaries were infected with either Ad-Fam110c or Ad-GFP control and then treated with FSK plus PMA for 6 h. As expected, FSK plus PMA treatment stimulated progesterone production, increased expression of Cyp11a1 and decreased expression of Hsd3b (Fig. 6, A–C). Interestingly, overexpression of FAM110C did not affect progesterone levels or expression of Cyp11a1 or Hsd3b mRNA (Fig. 6, A–C). Fam110C mRNA was highly expressed in the Ad-Fam110c vector-infected granulosa cells and was induced by FSK plus PMA treatment (Fig. 6D).
FIG. 6. .
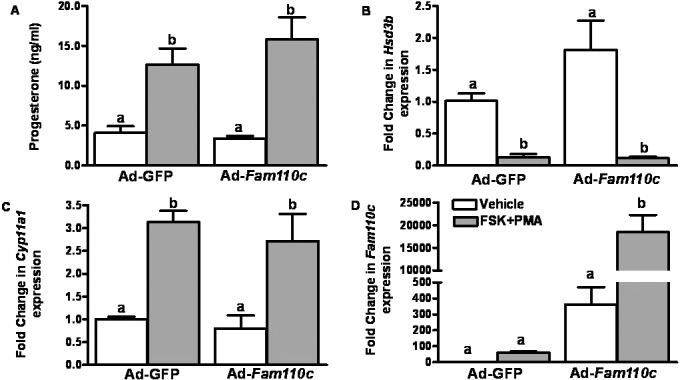
Analysis of granulosa cell steroidogenesis after overexpression of rat FAM110C. Granulosa cells from preovulatory ovaries were infected with either Ad-Fam110c or an Ad-GFP control adenovirus and then treated with vehicle (open bars) or with FSK plus PMA (closed bars) for 6 h. A) Progesterone levels were induced by FSK plus PMA in the conditioned medium. B) Hsd3b mRNA decreased after FSK plus PMA treatment but was unchanged in granulosa cells infected with the Ad-Fam110C vector. C) Cyp11a1 mRNA was induced by FSK plus PMA but was unaffected by overexpression of FAM110C. D) FAM110C is highly expressed in granulosa cells infected with the Ad-Fam110c vector, and this expression was stimulated by FSK plus PMA. Values represent means ± SEM; n = 3 independent culture experiments. Bars with no common letters are significantly different (P < 0.05).
DISCUSSION
The ovulatory gonadotropin surge initiates the cascade of events associated with follicle rupture and the formation of the corpus luteum. During this process, granulosa cells of the preovulatory follicle irreversibly cease dividing as they transition to luteal cells. This loss of granulosa cell proliferation during the process of luteinization is the result of a change in the checkpoints of the cell cycle, which control cellular proliferation and differentiation [23]. It has been shown that granulosa cells exit from the cell cycle by down-regulation of cyclin D2, down-regulation of cyclin E, and up-regulation of the CKIs p21 and p27 [2, 23, 24]. In this study, we demonstrate for the first time that FAM110C was induced after an LH/hCG stimulus and that overexpression of FAM110C impacted granulosa cell cycle kinetics by inhibiting transition from the G1 to the S phase of the cell cycle.
Members of the FAM110 family were originally identified by a yeast two-hybrid screen for CSPP-associated proteins [12]. Northern blot analysis was used to screen tissue-specific expression, and all members of the FAM110 family were detected in the thyroid, stomach, prostate, and testis. In the ovary, the overall expression of the FAM110 members was low, with levels of Fam110c mRNA that were higher than the expression levels of Fam110a or Fam110b mRNA, although the stage of the ovarian cycle was not delineated [12]. We confirmed that ovarian expression of Fam110c was elevated compared to the other Fam110 members, using our rat ovarian gene expression database [16] prior to initiating the present study.
Administration of hCG in the current study stimulated the expression of Fam110c mRNA in both the granulosa and theca cell compartments. Initially, Fam110c mRNA was induced in the theca cells and the granulosa cells of certain follicles at 4 h, as seen by in situ hybridization analysis. Fam110c mRNA expression then switched to the granulosa cell compartment of antral follicles by 8 and 12 hours after hCG but was lost in the newly forming corpora luteum. The physiologic significance of this pattern of expression where Fam110c mRNA switches from the theca to the granulosa cells is unclear. However, this expression pattern was also found for the B cell translocation gene 1 product [8], a protein involved in inhibiting cellular proliferation and regulating differentiation [25], specifically arresting granulosa cells at the G0–G1 phase of the cell cycle [8]. We speculate that this switch from theca to granulosa cell compartment reflects the rate at which theca and granulosa cells respond to hCG induction of gene expression.
FAM110C impairs progression through the G1 phase of the cell cycle, resulting in cell cycle arrest in human embryonic kidney cells [12]. In the ovary, granulosa cells quickly exit the cell cycle after the LH surge [23]. Thus, we postulated that the marked induction of Fam110c mRNA in granulosa cells might facilitate the process of luteinization, specifically the transition from a proliferative granulosa cell to a differentiated luteal cell. Support for such a postulate is based upon the findings of Hauge and colleagues [12], who explored the function of FAM110C. The investigators noted that at low levels of FAM110C expression in HEK293T cells, the protein was localized at the centrosome/microtubule organization center during interphase. At mitosis, FAM110C was associated with the microtubules of the spindle apparatus [12]. Of note for the present study was the previous observation that overexpression of FAM110C was able to impair cell cycle progression from the G1 to the S phase in HEK293T cells [12]. Although the precise mechanism by which FAM110C impairs cell cycle progression is unknown, it was proposed that FAM110C could impact microtubule assembly necessary for cell entry into mitosis [12]. Irrespective of the exact mechanism, these previous findings of FAM110C localization and function led to our hypothesis that FAM110C might regulate the switch from proliferation to differentiation during the luteinization process. To determine whether FAM110C impacted cell cycle kinetics, we overexpressed FAM110C in granulosa cells and observed an increase in the number of granulosa cells in the G1 stage of the cell cycle. These findings support the hypothesis for a potential role of FAM110C in the control of granulosa cell cycle. Of interest, however, was the finding that overexpression of FAM110C did not influence progesterone levels in the conditioned medium nor expression of two of the steroidogenic enzymes involved in granulosa cell steroidogenesis. We interpret these findings to indicate that FAM110C impacts cell cycle kinetics but that this is insufficient to cause luteal cell differentiation, at least as assessed by its failure to affect mRNA levels for 3BHSD and P450scc.
Experiments were performed to delineate the signaling pathways controlling the hormonal induction of Fam110c. Fam110c mRNA was induced by hCG treatment of cultured granulosa cells, which could be mimicked by FSK, highlighting the importance of the PKA signaling pathway in the regulation of Fam110c mRNA. As the LH/hCG pathway sets in motion a series of downstream events that are crucial for follicular rupture [26], we further examined the impact of progesterone, prostaglandins, and the EGF receptor signaling pathway on the regulation of Fam110c mRNA. The lack of an effect of the prostaglandin synthase 2 inhibitor NS398 or the EGF receptor tyrosine kinase inhibitor AG1478 on the hCG-induced expression of Fam110c mRNA indicates that these pathways do not contribute to the regulation of Fam110c during the periovulatory period. However, the observation that the progesterone receptor antagonist RU486 could block the induction of Fam110c mRNA expression suggests that progesterone regulates Fam110c expression. Although there is no evidence that progesterone regulates the CSPPs, previous studies have demonstrated a role for progesterone in regulating the cell cycle in the ovary. For example, progesterone inhibited in vitro proliferation of human granulosa cells obtained following an ovulatory stimulus [27, 28]. Likewise, exogenous progesterone was able to inhibit cyclin B1 expression in primate granulosa cells [29], while RU486 increased the number of proliferating bovine granulosa-lutein cells [30]. We speculate that the changes in ovarian cell proliferation observed by modulating progesterone levels may occur, in part, by progesterone action in regulating FAM110C. Such a concept is supported by the findings of Chaffin and colleagues [29], who observed that progesterone was able to regulate the timing of p27 expression, leading to the viewpoint that steroids regulate key components of the cell cycle in granulosa cells.
The regulation of Fam110c mRNA induction was explored using cultured granulosa cells. Functional analysis of the Fam110c promoter revealed that FSK plus PMA could stimulate Fam110c promoter activity. This induction was reduced when the Sp1 binding site in the Fam110c promoter was mutated, indicating that SP1 directly regulates Fam110c transcription. It has been shown that SP1 is involved in the hormone-regulated induction of a variety of ovarian genes. For example, SP1 or an intact Sp1 binding site is required for the induction of cholesterol side-chain cleavage cytochrome P450 [31], serum/glucocorticoid-inducible kinase [32], LH β-subunit [33], and the LH receptor [34]. Of particular interest is the finding that SP1 or an intact Sp1 binding site is critical for progesterone-mediated action of genes that lack a progesterone response element (PRE), such as cyclin-dependent kinase inhibitor, p21 in breast cancer [35], a Krüppel-like transcription factor 11 (KLF11) gene in uterine leiomyoma cells [36], and MMP2 in the trophoblast [37]. These findings are distinctly relevant to the present study, as the proximal Fam110c promoter lacks a PRE. Our data extend these observations of SP1-mediating expression of reproductive genes and provide the first evidence that SP1 is involved in hCG-induced expression of the Fam110c gene in rat granulosa cells.
In summary, the present findings demonstrate a transient induction of Fam110c expression in theca and granulosa cells after hCG administration. This induction requires the SP1 transcription factor and signaling through the progesterone receptor. The production of FAM110C protein may, in turn, arrest granulosa cells at the G1 phase of the cell cycle. These unique findings shed light on another potential regulator that directs the exit of granulosa cells from the cell cycle and their differentiation into luteal cells.
ACKNOWLEDGMENT
The authors would like to acknowledge the assistance of Kathy Rosewell in the overall preparation of the manuscript.
Footnotes
Supported by National Institutes of Health grants P20 RR015592 and HD057446 to T.E.C., and grant HD051727 to M.J.
REFERENCES
- Sherr CJ. G1 phase progression: cycling on cue. Cell 1994; 79: 551 555 [DOI] [PubMed] [Google Scholar]
- Robker RL, Richards JS. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol Endocrinol 1998; 12: 924 940 [DOI] [PubMed] [Google Scholar]
- Hirshfield AN. Development of follicles in the mammalian ovary. Int Rev Cytol 1991; 124: 43 101 [DOI] [PubMed] [Google Scholar]
- Richards JS. Hormonal control of gene expression in the ovary. Endocr Rev 1994; 15: 725 751 [DOI] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, Robker RL, Richards JS, McGinnis LK, Biggers JD, Eppig JJ, Bronson RT. et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 1996; 384: 470 474 [DOI] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, Kaushansky K, Roberts JM. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 1996; 85: 733 744 [DOI] [PubMed] [Google Scholar]
- Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 1996; 85: 721 732 [DOI] [PubMed] [Google Scholar]
- Li F, Liu J, Park ES, Jo M, Curry TE., Jr The B cell translocation gene (BTG) family in the rat ovary: hormonal induction, regulation, and impact on cell cycle kinetics. Endocrinology 2009; 150: 3894 3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzke S, Hauge H, Sioud M, Finne EF, Sivertsen EA, Delabie J, Stokke T, Aasheim HC. Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene 2005; 24: 1159 1173 [DOI] [PubMed] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol 2005; 15: 303 311 [DOI] [PubMed] [Google Scholar]
- Patzke S, Stokke T, Aasheim HC. CSPP. and CSPP-L associate with centrosomes and microtubules and differently affect microtubule organization. J Cell Physiol 2006; 209: 199 210 [DOI] [PubMed] [Google Scholar]
- Hauge H, Patzke S, Aasheim HC. Characterization of the FAM110 gene family. Genomics 2007; 90: 14 27 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365 386 [DOI] [PubMed] [Google Scholar]
- Jo M, Curry TE., Jr Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol 2006; 20: 2156 2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951; 193: 265 275 [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr, Ko C. Development and application of a rat ovarian gene expression database (rOGED). Endocrinology 2004; 145: 5384 5396 [DOI] [PubMed] [Google Scholar]
- Vindelov LL, Christensen IJ, Nissen NI. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 1983; 3: 323 327 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Robayna IJ, Falender AE, Ochsner S, Firestone GL, Richards JS. Follicle-Stimulating hormone (FSH) stimulates phosphorylation and activation of protein kinase B (PKB/Akt) and serum and glucocorticoid-induced kinase (Sgk): evidence for A kinase-independent signaling by FSH in granulosa cells. Mol Endocrinol 2000; 14: 1283 1300 [DOI] [PubMed] [Google Scholar]
- Salvador LM, Maizels E, Hales DB, Miyamoto E, Yamamoto H, Hunzicker-Dunn M. Acute signaling by the LH receptor is independent of protein kinase C activation. Endocrinology 2002; 143: 2986 2994 [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Ochsner S, Hsieh M, Doyle KH, Falender AE, Lo YK, Sharma SC. Novel signaling pathways that control ovarian follicular development, ovulation and luteinization. Recent Prog Horm Res 2002; 57: 195 220 [DOI] [PubMed] [Google Scholar]
- Sekar N, Veldhuis JD. Involvement of Sp1 and SREBP-1a in transcriptional activation of the LDL receptor gene by insulin and LH in cultured porcine granulosa-luteal cells. Am J Physiol Endocrinol Metab 2004; 287: E128 E135 [DOI] [PubMed] [Google Scholar]
- Sriraman V, Sharma SC, Richards JS. Transactivation of the progesterone receptor gene in granulosa cells: evidence that Sp1/Sp3 binding sites in the proximal promoter play a key role in luteinizing hormone inducibility. Mol Endocrinol 2003; 17: 436 449 [DOI] [PubMed] [Google Scholar]
- Robker RL, Richards JS. Hormonal control of the cell cycle in ovarian cells: proliferation versus differentiation. Biol Reprod 1998; 59: 476 482 [DOI] [PubMed] [Google Scholar]
- Hampl A, Pachernik J, Dvorak P. Levels and interactions of p27, cyclin D3, and CDK4 during the formation and maintenance of the corpus luteum in mice. Biol Reprod 2000; 62: 1393 1401 [DOI] [PubMed] [Google Scholar]
- Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol 2001; 187: 155 165 [DOI] [PubMed] [Google Scholar]
- Richards JS. Ovulation: new factors that prepare the oocyte for fertilization. Mol Cell Endocrinol 2005; 234: 75 79 [DOI] [PubMed] [Google Scholar]
- Chaffkin LM, Luciano AA, Peluso JJ. Progesterone as an autocrine/paracrine regulator of human granulosa cell proliferation. J Clin Endocrinol Metab 1992; 751404: 1404 1408 [DOI] [PubMed] [Google Scholar]
- Chaffkin LM, Luciano AA, Peluso JJ. The role of progesterone in regulating human granulosa cell proliferation and differentiation in vitro. J Clin Endocrinol Metab 1993; 76: 696 700 [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Schwinof KM, Stouffer RL. Gonadotropin and steroid control of granulosa cell proliferation during the periovulatory interval in rhesus monkeys. Biol Reprod 2001; 65: 755 762 [DOI] [PubMed] [Google Scholar]
- Quirk SM, Cowan RG, Harman RM. Progesterone receptor and the cell cycle modulate apoptosis in granulosa cells. Endocrinology 2004; 145: 5033 5043 [DOI] [PubMed] [Google Scholar]
- Liu Z, Simpson ER. Steroidogenic factor 1 (SF-1) and SP1 are required for regulation of bovine CYP11A gene expression in bovine luteal cells and adrenal Y1 cells. Mol Endocrinol 1997; 11: 127 137 [DOI] [PubMed] [Google Scholar]
- Alliston TN, Maiyar AC, Buse P, Firestone GL, Richards JS. Follicle stimulating hormone-regulated expression of serum/glucocorticoid-inducible kinase in rat ovarian granulosa cells: a functional role for the spl family in promoter activity. Mol Endocrinol 1997; 11: 1934 1949 [DOI] [PubMed] [Google Scholar]
- Kaiser UB, Sabbagh E, Chen MT, Chin WW, Saunders BD. Sp1 binds to the rat luteinizing hormone beta (LHbeta) gene promoter and mediates gonadotropin-releasing hormone-stimulated expression of the LHbeta subunit gene. J Biol Chem 1998; 273: 12943 12951 [DOI] [PubMed] [Google Scholar]
- Chen S, Shi H, Liu X, Segaloff DL. Multiple elements and protein factors coordinate the basal and cyclic adenosine 3′, 5′-monophosphate-induced transcription of the lutropin receptor gene in rat granulosa cells. Endocrinology 1999; 140: 2100 2109 [DOI] [PubMed] [Google Scholar]
- Richer JK, Lange CA, Manning NG, Owen G, Powell R, Horwitz KB. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J Biol Chem 1998; 273: 31317 31326 [DOI] [PubMed] [Google Scholar]
- Yin P, Lin Z, Reierstad S, Wu J, Ishikawa H, Marsh EE, Innes J, Cheng Y, Pearson K, Coon JS, Kim JJ, Chakravarti D. et al. Transcription factor KLF11 integrates progesterone receptor signaling and proliferation in uterine leiomyoma cells. Cancer Res 2010; 70: 1722 1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S, Shalev E. A proposed mechanism for progesterone regulation of trophoblast MMP2 transcription independent of classical progesterone response elements on its promoter. J Exp Clin Assist Reprod 2006; 3: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]


