ABSTRACT
Mammalian spermatogenesis is a complex process that involves spatiotemporal regulation of gene expression and meiotic recombination, both of which require the modulation of chromatin structure. Proteins important for chromatin regulation during spermatogenesis remain poorly understood. Here we addressed the role of BRG1, the catalytic subunit of the mammalian Swi/Snf-like BAF chromatin-remodeling complex, during spermatogenesis in mice. BRG1 expression is dynamically regulated in the male germline, being weakly detectable in spermatogonia, highly expressed in pachytene spermatocytes, and turned off in maturing round spermatids. This expression pattern overlaps that of Brm, the Brg1 homolog. While Brm knockout males are known to be fertile, germline-specific Brg1 deletion completely arrests spermatogenesis at the midpachytene stage, which is associated with spermatocyte apoptosis and apparently also with impaired homologous recombination and meiotic sex chromosome inactivation. However, Brg1 is dispensable for gammaH2AX formation during meiotic recombination, contrary to its reported role in DNA repair in somatic cells. Our study reveals the essential role of Brg1 in meiosis and underscores the differences in the mechanisms of DNA repair between germ cells and somatic cells.
Keywords: apoptosis, chromatin, spermatogenesis
Chromatin remodeling factor Brg1 controls development of primary spermatocytes during meiosis in mice.
INTRODUCTION
Mammalian spermatogenesis is a complex process. It begins when spermatogonia proliferate and differentiate into primary spermatocytes, which is followed by meiosis that produces haploid round spermatids. The round spermatids then undergo dramatic morphological changes to become mature sperm. Primary spermatocytes in the meiotic prophase undergo four major sequential events that culminate in DNA exchange between homologous chromosomes, namely, introduction of double strand breaks (DSBs), pairing of homologous chromosomes, crossing over of chromosomes, and resolving of Holliday junctions. These four events take place within four consecutive phases of primary spermatocytes: the leptotene, zygotene, pachytene, and diplotene stages [1, 2].
The complex nature of spermatogenesis demands precise control of transcription and recombination, which in turn involves proper modulation of chromatin structure [3, 4]. Thus, in leptotene spermatocytes, open chromatin structure at recombination hot spots is necessary for the SPO11 endonuclease to access the DNA and introduce DSBs [5]. Immediately following the generation of DSBs, the histone H2A variant H2AX in the vicinity of DSBs becomes phosphorylated at Ser-139 to form γH2AX, which serves to concentrate DNA damage response proteins at DSBs [6]. In pachytene spermatocytes, where the synapsis between homologous chromosomes is complete, γH2AX disappear from autosomes but coats sex chromosomes and represses their transcription [7–9]. Transcription repression of the genes located on sex chromosome, termed meiotic sex chromosome inactivation (MSCI), is crucial for meiosis because MSCI failure inevitably leads to pachytene apoptosis [10–12]. MSCI is mediated by extensive chromatin reorganization on sex chromosomes, including changes in histone modifications and the incorporation of histone variants such as H2AX, which culminates in the formation of repressive heterochromatin [13]. The genes controlling MSCI are not fully defined.
Two major classes of enzymes are known to regulate chromatin structure. Histone-modifying enzymes act by covalently modifying histones in nucleosomes, while ATP-dependent multiprotein chromatin-remodeling complexes use the energy of ATP-hydrolysis to physically disrupt histone-DNA contacts and move, destabilize, eject, or restructure nucleosomes [14, 15]. Multiple histone modifications have been implicated in the regulation of spermatogenesis [16–22], but little is known about the roles of chromatin remodeling complexes in spermatogenesis.
Chromatin-remodeling complexes in eukaryotic cells fall into four families (Swi/Snf, Iswi, CHD, and INO8) defined based on the ATPase/catalytic subunits they carry [14]. These complexes differ in their mechanisms of action and the target genes regulated. The prototype of the mammalian complexes is the BRG1-associated factor (BAF) complex of the Swi/Snf family; the BAF complex, also known as mammalian Swi/Snf (mSwi/Snf) complex, consists of more than 10 subunits, including the ATPase/catalytic subunit BRG1 or its homolog BRM. BRG1 and BRM are both widely expressed in somatic cells in adult mice where they play diverse, tissue-specific roles in gene regulation [23]. In addition to the well-established roles of the chromatin remodelers in transcription regulation, several members of the Swi/Snf and INO80 families also participate in the repair of DSBs caused by genotoxic agents in budding yeast and mammalian cells, and different remodelers regulate distinct steps in the repair pathway [24]. In particular, BRG1 facilitates repair of DSBs induced by irradiation in a human somatic cell line [25–27]. BRG1 is rapidly recruited to DSBs within 10 min following irradiation, where it promotes γH2AX formation. In the absence of BRG1, 1) the number and the size of γ-H2AX foci are significantly reduced as are the total cellular γH2AX level, and 2) the cells are defective in DSB repair and highly susceptible to irradiation. Whether the BAF complex also regulates DSB repair or any other events in meiosis is unclear because germline Brg1 knockout (KO) produces early embryonic lethality [28] while Brm KO mice are fertile [29]. Here we explore the role of BRG1 in spermatogenesis by deleting BRG1 specifically in the male germline.
MATERIALS AND METHODS
Mice
Brg1Flox mice on mixed genetic background have been described [30] and were mated with male mice carrying the Vasa-Cre transgene (Jackson Laboratory) to produce Brg1Flox.Flox; Cre mice. The mice were housed in the animal facility at Yale, and all the experimental procedures were approved by the Institutional Animal Care and Use Committee at Yale.
Histological Analysis and TUNEL Assay
Testes were fixed in Bouin fixative and embedded in paraffin before hematoxylin and eosin (H & E) staining. The paraffin sections were also used for immunostaining in Figure 4C (bottom two rows) and Figures 6 and 7. Immunostaining in other figures was performed on cryosections. To prepare cryosections, testes were fixed in 2% paraformaldehyde at 4°C for 3–5 h. After washing in PBS, the specimens were incubated in 20% sucrose/PBS overnight, embedded optimal-cutting temperature compound and frozen. Primary antibodies used were anti-EE2 (1:100; gift of Dr. Hiromitsu Tanaka), anti-BRG1 (1:100; 110641; Abcam), anti-BRM (1:100; 15597; Abcam), anti-H3K9me3 (1:100; 8898; Abcam), anti-phospho-S5 Pol II (1:100; IHC-00387; Bethyl Laboratories), anti-cH2AX (1:100; 05–636; Millipore), anti-RAD51 (1:100; 63801; Abcam), and anti-H1t (1:400; gift of Dr. Mary Ann Handel). Apoptosis was analyzed on cryosections using the ApopTag Fluorescein In Situ Apoptosis Detection Kit (Chemicon International). Briefly, cryosections were incubated with the TdT enzyme for 1 h at 37°C, washed, and then incubated with anti-digoxigenin conjugate. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) before final mounting for imaging.
FIG. 4. .
Brg1 KO arrested meiosis at pachytene stage IV. A–B) Lack of diplotene spermatocytes in Brg1 KO mice. Spermatocytes were stained with anti-synaptonemal complex protein 3 (SCP3) before the cells at the various stages of meiosis were enumerated and their relative abundance determined. One hundred each of WT and KO spermatocytes were scored. The data is derived from a single biological replicate. C–D) Brg1 KO blocked expression of genes marking late pachytene spermatocytes as revealed by immunofluorescence (C) and RT-PCR (D). RT-PCR was done in triplicates on RNA from testicular cells (pooled from two WT and two KO mice to avoid sampling errors), and the values (normalized to b-actin) were averaged and plotted as the ratios of the expression levels in WT to KO testes. Roman numerals within the overlaid images indicate the seminiferous epithelial stages. Original magnification ×63 (A and C).
FIG. 6. .
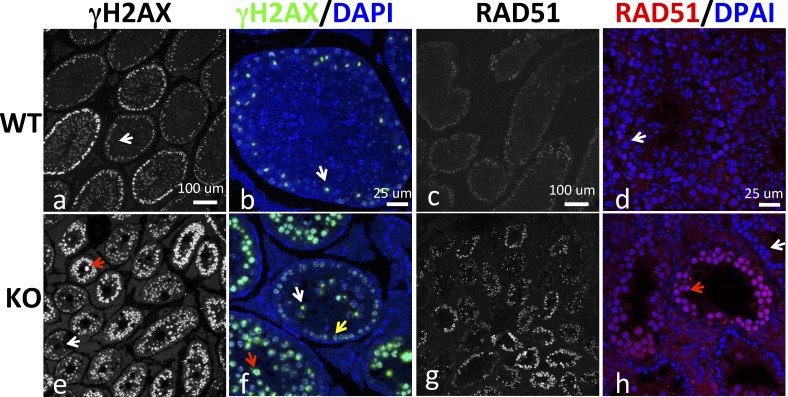
Potential defects in DSB repair in Brg1 KO testes. Testicular sections were stained for γH2AX (left two columns: a, b, e, f) and RAD51 (right two columns: c, d, g, h). The white and red arrows indicate examples of tubular sections containing pachytene spermatocytes with normal and aberrant γH2AX/RAD51 staining, respectively, while the yellow arrow in f indicates leptotene/zygotene spermatocytes. Bars = 100 μm (a, c, e, g) or 25 μm (b, d, f, h).
FIG. 7. .
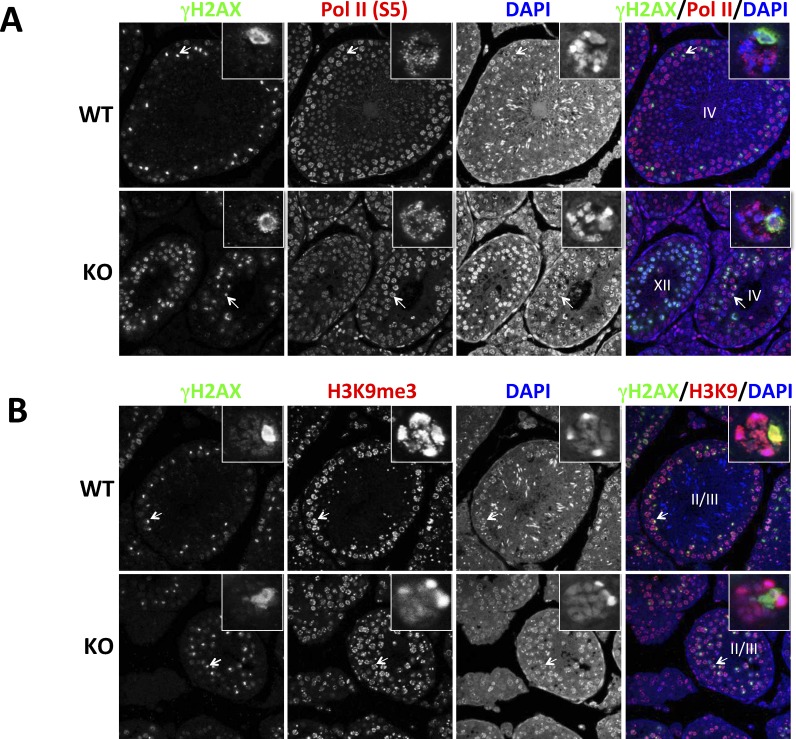
Sex body defects in Brg1 KO testes. A) Testicular sections were stained for γH2AX and Pol II (S5), the Ser-5 phosphorylated form of RNA Pol II. B) Same as A except that H3K9m3 instead of Pol II (S5) was analyzed. The insets are close-up views of the pachytene spermatocytes indicated by arrows. Roman numerals within the overlaid images indicate seminiferous epithelial stages. Original magnification ×63.
Immunofluorescence Analysis of Spermatocyte Spreads
Spermatocyte spreads were obtained as described previously [31]. Briefly, testis cells were suspended in 0.1 M sucrose supplied with protease inhibitor. The cells were applied to the slides with fixation solution (0.1% Triton X-100 in 1% paraformaldehyde). The slides were kept in a humid chamber for 3 h before air drying. The dry slides were preblocked in PBS containing 3% bovine serum albumin/10% normal goat serum/0.05% Triton X-100, incubated overnight at 4°C with primary antibodies, and followed by a secondary antibody for 1 h at room temperature. Synaptonemal complex protein 3 (SCP3) was detected using a mouse monoclonal antibody (74569; Santa Cruz Biotechnology).
RT-PCR
RT-PCR was performed using the published primers on total testicular RNA samples as described previously [32] except that the RNA was pooled from three wild-type (WT) or Brg1 KO mice to avoid sampling errors.
Fluorescence-Activated Cell Sorter Analysis
The assay was performed based on a published protocol [33] with modifications. Briefly, testes were homogenized using medicon (BD Pharmingen), and filtered through cell strainers. The cells were then pelleted, resuspended in Cytofix/Cytoperm (554714; BD Biosciences) for 10 min on ice, washed with BD Perm/Wash (554714; BD Biosciences), and resuspended in the same buffer containing 2 μg/ml propidium iodide (PI) for fluorescence-activated cell sorter (FACS) analysis.
RESULTS
Distinct Expression Patterns of Brg1 and Its Homolog Brm During Spermatogenesis
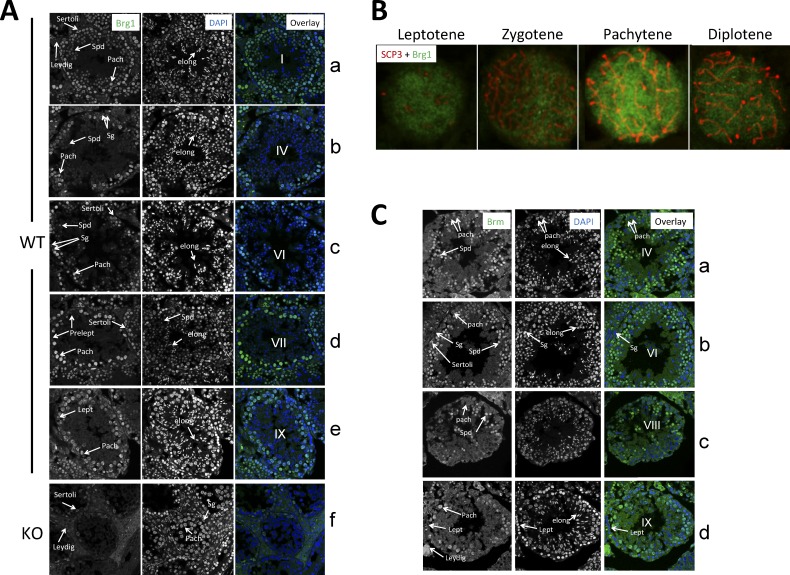
To investigate the potential role of BRG1 in spermatogenesis, we first examined BRG1 expression during spermatogenesis. Immunostaining of testicular sections revealed that BRG1 was marginally detectable in spermatogonia (Fig. 1A, rows b–c) and preleptotene spermatocytes (Fig. 1A, row d). The expression level was somewhat higher in leptotene spermatocytes (Fig. 1A, row e) and further increased in pachytene spermatocytes (see, e.g., Fig. 1A, row d). BRG1 was also present in round spermatids at stage I of the seminiferous epithelium cycle, although the expression level seemed lower than that in pachytene spermatocytes (Fig. 1A, row a). The expression became much weaker as the spermatids matured into the round spermatids seen at epithelial stage IV (Fig. 1A, row b) and completely shut off by the time they further matured into the round spermatids at stage VII (Fig. 1A, row d). In agreement with this, BRG1 was undetectable in elongating spermatids (Fig. 1A, rows a–e). Of note, the BRG1 immunofluorescence signal was specific because it was undetectable in primary spermatocytes in Brg1 KO mice (Fig. 1A, row f; see further). The lack of BRG1 staining in the germ cells in Brg1 KO mice was not an artifact because BRG1 expression was detectable in somatic (Sertoli and Leydig) cells in the same testes (Fig. 1A, row f). Also note that BRG1 was excluded from nucleoli in Sertoli cells.
FIG. 1. .
BRG1 and BRM expression in the testis. A) BRG1 expression during spermatogenesis. Shown are BRG1 immunofluorescence (left), DAPI-stained nuclei (middle), and overlay of the two images (right). B) BRG1 expression in primary spermatocytes. Cells were stained with SCP3 (red) and BRG1 (blue) antibodies. C) BRM expression during spermatogenesis. Roman numerals within the overlaid images indicate seminiferous epithelial stages. Spd, round spermatids; elong, elongating spermatids; Sg, spermatogonia; Pach, pachytene spermatocytes; Prelept, preleptotene spermatocytes. Original magnification ×63 (A–C).
To further characterize BRG1 expression in spermatocytes, we examined primary spermatocytes by staining spermatocyte spreads with anti-BRG1 and anti-SCP3. BRG1 was expressed in various subsets of spermatocytes with the expression peaking in pachytene spermatocytes (Fig. 1B). In these cells, BRG1 was rather evenly distributed in the nucleoplasm, implicating that the majority of BRG1 was not stably associated with chromatin during meiosis. Collectively, these data indicate that BRG1 expression during spermatogenesis was highly regulated: the expression began in spermatogonia, peaked in pachytene spermatocytes, down-regulated and then terminated in maturing (stage VII) round spermatids, and remained so thereafter.
We also explored a potential spermatogenic role of the Brg1 homolog Brm by determining its expression pattern. Interestingly, while the two proteins are often coexpressed in somatic tissues, BRM expression during spermatogenesis was much delayed and more temporally restricted as compared with BRG1 because BRM protein did not emerge until pachytene stage IX and did not persist beyond Stage VIII round spermatids (Fig. 1C), which has implications for the modes of action of the two proteins (see Discussion).
Brg1 Is Essential for Meiosis
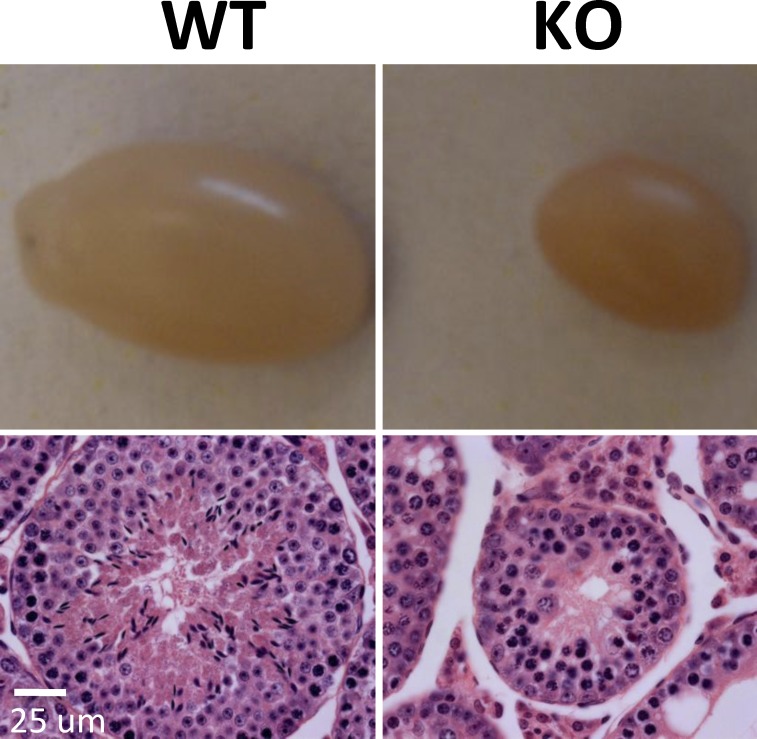
To investigate the function of BRG1 during spermatogenesis, we deleted Brg1 specifically in the male germline using a transgenic line expressing Cre from the Vasa promoter because germline Brg1 KO is embryonic lethal [28]. The Vasa-Cre transgene is known to direct Cre expression primarily in the male and female germ cells starting at Embryonic Day 15–18 [34]. As expected, Brg1Flox/Flox; Cre+/− male offspring (referred to as Brg1 KO hereafter) were generally healthy and displayed little Brg1 deletion in multiple organs examined (data not shown), consistent with the tissue specificity of Cre expression. Importantly, BRG1 protein was undetectable in the germ cells in the Brg1 KO testes as mentioned above (Fig. 1, row f), and Brg1 KO mice were sterile. Their testes were much smaller than aged-matched controls and completely devoid of sperm (Fig. 2, top, and data not shown). Histologically, the seminiferous tubules were much smaller and contained no spermatids or sperm, but spermatogonia and spermatocytes were present, suggesting Brg1 KO blocked meiosis (Fig. 2, bottom).
FIG. 2. .
Brg1 is essential for the development of male germline. Shown are gross pictures (top) and H& E stained histological sections (bottom) of WT and Brg1 KO testes from 5-mo-old mice. Bar = 25 μm.
Brg1 KO Arrests Meiosis at Pachytene Stage IV
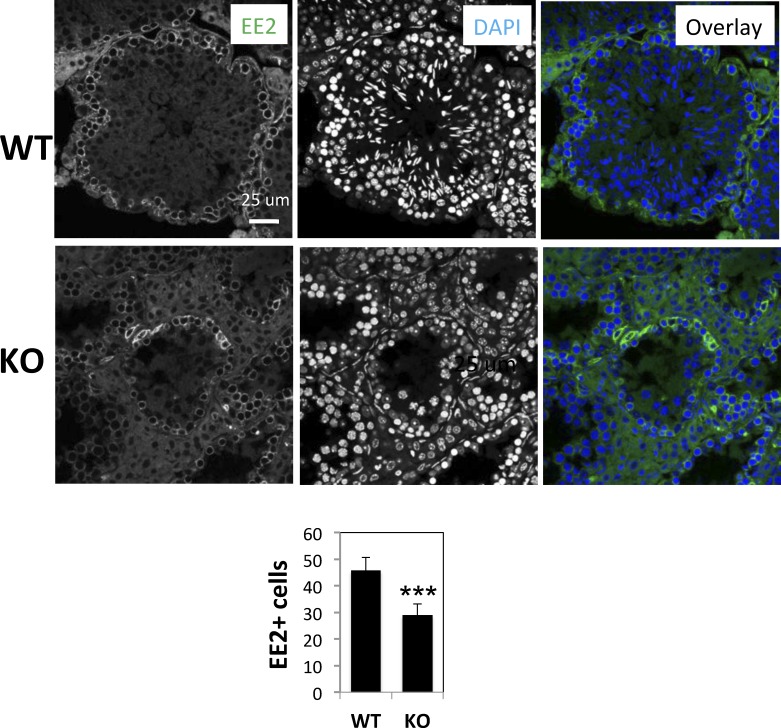
To better characterize meiotic defects in Brg1 KO testes, we analyzed germ cells at various stages of spermatogenesis. We first examined cells positive for EE2, a cytoplasmic protein that marks spermatogonia and prepachytene spermatocytes [35]. We found that the numbers of EE2+ cells per cross-section of seminiferous tubules were moderately reduced in the Brg1 KO mice (from 45.8 ± 2.3 to 29.0 ± 1.9), which is consistent with the reduction in the diameter of the tubules (Fig. 3). We next focused on primary spermatocytes because they are the arrested cell type in the mutant. We obtained spermatocyte spread and stained the cells with an antibody against SCP3, which enabled us to distinguish spermatocytes at various stages of meiosis based on chromosome morphology. While leptotene, zygotene, and pachytene spermatocytes were detected in Brg1 KO mice, diplotene cells were absent, demonstrating a defect in pachytene to diplotene transition (Fig. 4A). Furthermore, although pachytene spermatocytes were detectable in Brg1 KO mice, their abundance relative to leptotene and zygotene was significantly reduced: in WT mice, there were about 5-fold more pachytene spermatocytes than leptotene or zygotene spermatocytes, whereas in Brg1 KO mice, pachytene spermatocytes were only slightly more abundant than zygotene spermatocytes (Fig. 4B).
FIG. 3. .
Reduction of EE2+ cells in Brg1 KO testis. Testis sections were stained with anti-EE2 and counterstained with DAPI. EE2+ cells were visualized with immunofluorescence and counted, and the results displayed in the bar graph at the bottom. Six seminiferous tubular sections in WT and eight in KO testes were scored. Asterisks indicate statistical significance (P < 0.0001, two-tailed unpaired t-test). Bar = 25 μm.
The depletion of pachytene spermatocytes suggests that Brg1 KO might have arrested these cells at an early stage of pachytenema, before the pachytene spermatocytes could further mature and accumulate. To define the exact stage of arrest, we examined markers of spermatocyte development. MIWI protein, which becomes detectable in pachytene spermatocytes at epithelial stage V [36], was absent in Brg1 KO testes (Fig. 4C, top two rows) due to a dramatic (∼30-fold) reduction in the Miwi message (Fig. 4D). As expected, Brg1 KO also depleted the late pachytenema markers Calmegin [37] and Cyclin A1 [38] (∼16- and 63-fold, respectively) but not the DNA repair genes Mlh1 [39] and Dmc1 [40] expressed in leptotene/zygotene spermatocytes (Fig. 4D). In fact, Mlh1 and Dmc1 message levels were increased 2- to 3-fold in Brg1 KO testes, paralleling the enrichment of leptotene/zygotene spermatocytes in the Brg1 KO mice (Fig. 4B). These data indicate Brg1 KO arrested spermatogenesis between pachytene stages I and V. To pinpoint the stage of the arrest, we examined H1t, a testis-specific isoform of linker histone H1. H1t protein becomes detectable in stage IV pachytene spermatocytes and persists till the elongating spermatid stage [11, 41, 42]. H1t protein was present in Brg1 KO pachytene spermatocytes, which appeared apoptotic and sloughing into the lumen (Fig. 4C, bottom two rows). Interesting, H1t mRNA in total testis was down-regulated ∼5-fold (Fig. 4D), presumably because Brg1 KO blocked further development of stage IV pachytene spermatocytes, thus reducing the abundance of testicular cells expressing H1t mRNA. Taken together, these data demonstrate that Brg1 KO arrested spermatogenesis at pachytene stage IV.
Spermatogenic Arrest in Brg1 KO Testes Is Associated with Increased Apoptosis
Developmental arrest can result from or lead to apoptosis. To determine if BRG1-deficient spermatocytes were apoptotic, we performed the Tunnel assay that detects the apoptotic type of DNA fragmentation. Indeed, numerous apoptotic spermatocytes were present specifically in Brg1 KO mice (Fig. 5A). Furthermore, some of these apoptotic cells had smaller nuclei. Such cells presumably represented spermatocytes at advanced stages of apoptosis when cells undergo overt shrinkage. The data suggest that Brg1 KO led to increased death of spermatocytes.
FIG. 5. .
Spermatocyte apoptosis in Brg1 KO testis. A) Apoptotic cells were visualized using TUNEL assays detecting DNA breaks. B) FACS analysis of apoptosis based on DNA content and cell morphology. Testis single cell suspension was stained with PI to reveal haploid (1C), diploid (2C) and tetraploid (4C) cells. The contour plots at the right display side scatter (SS) versus forward scatter (FS) patterns of the 4C cells, where the blue and red circles indicate small spermatogonia and apoptotic primary spermatocytes, respectively. Of note, there were subhaploid PI peaks in both WT and KO testes, the former reflecting elongation spermatids [44, 60], while the latter presumably reflecting debris from apoptotic cells [61] and thus excluded from analysis. Sg, spermatogonia; Spt, spermatocytes. Bars = 25 μm.
To corroborate and extend these findings, we used FACS to analyze testicular cellular composition based on DNA content of the cells. Testes were homogenized and stained with PI. Wild-type testes contained haploid, diploid, and tetraploid cells as expected (Fig. 5B, top left). The diploid pool contained not only germ cells but also somatic cells and thus uninformative. The haploid cells, representing spermatids, were absent from Brg1 KO mice, consistent with the histological data (Fig. 5B, bottom left). The tetraploid cells were mostly primary spermatocytes but also contained some proliferating spermatogonia. Tetraploid cells were present in Brg1 KO testes, but the mean fluorescence intensity of PI staining was much weaker, suggesting apoptosis of these cells (Fig. 5B, bottom left); apoptotic cells are known to show reduced PI staining due to chromatin condensation [43]. To confirm that the Brg1 KO cells were apoptotic, we analyzed their forward and side scatters, which reflect cell size and granularity, respectively. Apoptotic cells at advanced stages are smaller and often also smoother. The tetraploid cells in WT mice were heterogeneous, containing both small (blue circle) and large cells representing spermatogonia and spermatocytes, respectively [44] (Fig. 5B, top right). In Brg1 KO mice, spermatogonia did not show significant defect in cell morphology, suggesting these cells were not apoptotic (Fig. 5B, bottom center and right, blue circles), although PI signal in some spermatogonia was low for unknown reasons (Fig. 5B, bottom center, blue circle). In contrast, the spermatocytes with low PI signals, but not that with normal PI signals, were enriched in small, smooth cells (Fig. 5B, bottom center, red circle). These cells apparently corresponded to the small apoptotic cells in Figure 5A. The data confirm that BRG1 KO caused death of spermatocytes. Furthermore, in Brg1 KO mice, the tetraploid spermatocytes with low and high PI signals contained 25.7% and 12.9% testicular cells, respectively (Fig. 5B, bottom left). Assuming that the two populations of cells largely represent dying and normal cells, respectively, we estimated that ∼67% of spermatocytes are apoptotic in Brg1 KO mice.
Brg1 KO Seems to Impair Homologous Recombination but Does Not Compromise γH2AX Formation
We next sought to determine the mechanism whereby Brg1 KO arrested pachytene spermatocyte development. Given the known role of BRG1 in γH2AX formation and DSB repair in somatic cells, we examined γH2AX expression. During leptonema and zygonema, γH2AX marks DSBs and shows punctate staining, whereas in pachytene spermatocytes, γH2AX is lost on autosomes (except at aberrant, unpaired regions) but becomes concentrated on the sex body, a globular chromatin domain that is the morphological correlate of the transcriptionally silenced sex chromosomes [7, 45]. We found that in WT pachytene spermatocytes, H2AX was concentrated on the sex body, as expected (Fig. 6, a and b, white arrows). In contrast, in Brg1 KO testes, such a staining pattern was seen only in ∼30% of the seminiferous tubular sections (Fig. 6, e and f, white arrows), with the pachytene spermatocytes in the majority of the sections retaining the punctate-staining pattern characteristic of leptonema and zygonema (Fig. 6, e and f, red arrows). The persistence of the punctate-staining pattern into pachytenema suggests a defect in synapsis and hence in homologous recombination, given that the loss of γH2AX staining is temporally and spatially correlated with synapsis [7], but further studies are needed to prove this. On the other hand, it is clear that BRG1 was dispensable for γH2AX formation in spermatocytes, contrary to the scenario in somatic cells.
To corroborate the above finding, we examined the expression of RAD51, an enzyme that catalyzes strand invasion during homologous recombination. RAD51 forms numerous foci during zygonema but disappears during pachynema except at unsynapsed regions [46, 47]. As expected, RAD51 expression was very low in WT pachytene spermatocytes (Fig. 6, c and d) but aberrantly elevated in Brg1 KO pachytene spermatocytes in a large fraction of seminiferous tubular sections (Fig. 6, g and h, red arrow). Thus, as in the case of H2AX, the RAD5-staining pattern at zygonema also persisted into pachytenema in many tubular sections, which reinforces the notion of the potential defects in synapsis and DSB repair.
Brg1 KO Impairs the Integrity of the Sex Body: A Potential Failure of MSCI
As mentioned above, Brg1 KO caused pachytene stage IV apoptosis. A predominant cause of such apoptosis is the failure in MSCI [11, 48]. We thus sought to determine if MSCI failure underlies pachytene stage IV apoptosis in Brg1 KO mice.
To this end, we stained the testicular sections for H2AX and Pol II (S5), the initiating form of RNA polymerase II phosphorylated at Ser-5 in the C-terminal domain; H2AX was used to mark the sex body. In WT pachytene spermatocytes, Pol II (S5) displayed punctate staining that was excluded from the sex body, as expected (Fig. 7A, top). In contrast, in the Brg1 KO pachytene spermatocytes that had developed sex bodies, Pol II (S5) was clearly detectable in the sex body, indicating a defect in sex body integrity (Fig. 7A, bottom). To explore the underlying mechanism, we examined H3K9me3, a repressive histone modification at the sex body [13]. Significantly, H3K9me3 was completely absent at the mutant sex body (Fig. 7B).
We conclude that Brg1 KO impairs the formation of repressive chromatin structure at the sex body, which allows Pol II to access the sex chromosomes. Pol II was apparently transcribing, thus causing MSCI failure and cell death, although confirmation of this model will require detection of transcripts from the sex chromosomes.
DISCUSSION
In this paper, we have demonstrated that Brg1 KO in the male germline caused pachytene stage IV apoptosis associated with potential defects in DSB repair and in MSCI, but not with any obvious defect in γH2AX formation. To our knowledge, this is the first report of an essential role of a chromatin remodeling factor in spermatogenesis.
Brg1, DSB Repair, and MSCI
In Brg1 KO testes, the majority of pachytene spermatocytes retained the γH2AX- and RAD51-staining pattern characteristic of leptotene and zygotene spermatocytes, suggesting a defect in synapsis of homologous chromosomes. However, some Brg1 KO pachytene spermatocytes were able to successfully eliminate γH2AX and RAD51 from autosomes and develop morphologically normal sex body, as in the case of WT spermatocytes. Importantly, the sex body in these so-called normal cells lacked H3K9me3 and contained transcribing Pol II, which, together with the apoptosis of pachytene spermatocytes, argues for an essential role of BRG1 in MSCI because MSCI failure is the predominant cause of pachytene stage IV apoptosis. BRG1 may act by facilitating repressive histone modifications (such as H3K9me3) on the sex chromosome, which is achievable via regulating either repressor expression or repressor access to target loci. The latter mode of action, in which the BAF complex directly represses a gene via remodeling its chromatin, appears paradoxical but is not unprecedented: the complex stimulates the binding of the transcription repressor Runx1 to the CD4 locus, thus repressing CD4 [49]. It is also possible that BRG1 may use a novel function unrelated to traditional ATP-dependent chromatin remodeling to somehow promote sex chromosome silencing [50, 51]. Despite the uncertainty in the mechanism of action, it is clear that BRG1 is dispensable for the formation of sex body but essential for its epigenetic modifications and hence functional integrity. A variety of genes (H2a, Spo11, Dnmt3l, Msh5, Dmc1) are known to control MSCI, but they all act, at least partly, by promoting sex body formation [48], and thus their modes of action are distinct from that of BRG1. Finally, it is noteworthy that the effect of BRG1 on sex chromosomes was not mediated by Suv39, the major enzyme responsible for H3K9 trimethylation, even though Suv39 KO also causes stage IV-pachytene apoptosis [52]. This is because Suv39 does not control H3K9 trimethylation in stage IV-pachytene spermatocytes [52].
The relationship between the Brg1 KO pachytene spermatocytes with aberrant versus normal γH2AX/RAD51 staining is unclear. Because the two types of cells did not coexist in the same seminiferous tubular sections, they might be at different stages of maturation. Indeed, leptotene/zygotene spermatocytes, which show punctate γH2AX/RAD51 staining and are present exclusively at advanced epithelial stages (stages IX–XII), were detectable only in conjunction with the normal pachytene spermatocytes (Fig. 6f, yellow arrow), suggesting that the normal spermatocytes were more mature than the aberrant ones. We speculate that Brg1 KO delays the synapsis of homologous chromosome, thus leading to the accumulation of the aberrant pachytene spermatocytes, but these cells can eventually form synapsis, eliminate autosomal H2AX/RAD51, and become normal spermatocytes that then die of apoptosis due to MSCI failure resulting from epigenetic defects in the sex chromosome.
A Minor Role of Brg1 in Spermatogonia?
BRG1 was only marginally detectable in spermatogonia. Consistent with this, Brg1 KO did not seem to significantly affect spermatogonia survival or differentiation, which stands in contrast to its role in other stem cells such as embryonic or neural stem cells where Brg is highly expressed and essential for their maintenance and/or differentiation [53–57]. However, spermatogonia might be partially depleted in Brg1 KO mice, based on the mild reduction in the numbers of EE2+ cells, most of which are spermatogonia. Such depletion, if exists, may reflect a role of BRG1 in the development of spermatogonia precursors because Cre is expressed in the precursors. Alternatively, the depletion may be secondary to the defects in spermatocytes and/or spermatids, which might affect spermatogonia via certain feedback loops. Finally, such depletion could be due to a cell-autonomous role of BRG1 in spermatogonia even though Brg is not highly expressed in these cells. In any case, the major role of BRG1 in spermatogenesis is in meiosis of spermatocytes, rather than maintenance or development of spermatogonia, contrary to the scenario in other stem cells.
Brg1 Versus Brm in Spermatogenesis
While Brg1 KO blocked spermatogenesis, Brm KO males are fertile [29]. This phenotypic dichotomy may reflect differences in gene functions but may also result from differences in gene expression patterns because only BRG1 is expressed in early and midpachytene spermatocytes whereas both genes are expressed in late pachytene spermatocytes and maturing round spermatids. Thus, while BRM is unable to compensate for the observed effects of BRG1 deletion, the presence of BRG1 in the BRM-expressing cells might have obscured potential phenotype in late pachytene spermatocytes and maturing round spermatids in Brm KO males. On the other hand, BRM is clearly dispensable for mature (Stage IX) round spermatids because these cells express only BRM and yet are unaffected by Brm KO [29].
It is intriguing that the expression of both BRG1 and BRM is highly regulated during spermatogenesis, given that the two proteins are in general ubiquitously, highly, and constitutively expressed in somatic cells. However, regulated expression of Brg1 has been reported for heart muscle [58]. Specifically, Brg1 is expressed in fetal heart muscle but silenced in adult heart muscle. Cardiac stress causes reactivation of Brg1 in the adult heart, leading to reexpression of fetus-specific genes and subsequent cardiac hypertrophy [58]. Future work is required to determine whether constitutive Brg1/Brm expression is detrimental to spermatogenesis.
While this paper was under review, Kim et al. [59] published similar findings except that they did not detect Brm expression in pachytene cells, contrary to our data showing transient Brm expression in late pachytene cells. Further studies are required to resolve this discrepancy.
ACKNOWLEDGMENT
We thank Mary Ann Handel for critical comments and anti-H1t, Hiromitsu Tanaka for anti-EE2, and members of the Lin and Chi laboratories for helpful discussions.
Footnotes
Supported by NIH grant 5R21AI094000-02 (T.C.), NIH grant R01HD42012 (H.L.), and the Mathers Award (H.L.).
These authors contributed equally to this work.
REFERENCES
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. (eds.) Histological and Histopathological Evaluation of the Testis. Clearwater, FL: Cache River Press, 1990. [Google Scholar]
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet 1998; 32: 619 697 [DOI] [PubMed] [Google Scholar]
- Yanowitz J. Meiosis: making a break for it. Curr Opin Cell Biol 2010; 22: 744 751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res 2011; 727: 62 71 [DOI] [PubMed] [Google Scholar]
- Brachet E, Sommermeyer V, Borde V. Interplay between modifications of chromatin and meiotic recombination hotspots. Biol Cell 2012; 104: 51 69 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003; 5: 675 679 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 2001; 27: 271 276 [DOI] [PubMed] [Google Scholar]
- Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry 2005; 44: 13202 13213 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 2003; 4: 497 508 [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development 2007; 134: 1823 1831 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Bourc'his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 2008; 182: 263 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Boyar FZ, Driscoll DJ. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proc Natl Acad Sci U S A 2004; 101: 16583 16587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 2007; 39: 251 258 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78: 273 304 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell 2002; 108: 475 487 [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450: 119 123 [DOI] [PubMed] [Google Scholar]
- Okada Y, Tateishi K, Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J Androl 2010; 31: 75 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamori N, Zhao M, Meistrich ML, Matzuk MM. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod 2011; 84: 1225 1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Liu J, An S, Nishino T, Hishikawa Y, Koji T. Immunohistochemical analysis of histone H3 modifications in germ cells during mouse spermatogenesis. Acta Histochem Cytochem 2011; 44: 183 190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou S, Liao L, Chen X, Meistrich M, Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem 2010; 285: 2758 2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota SK, Feil R. Epigenetic transitions in germ cell development and meiosis. Dev Cell 2010; 19: 675 686 [DOI] [PubMed] [Google Scholar]
- Coutinho A, Caramalho I, Seixas E, Demengeot J. Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection. Curr Top Microbiol Immunol 2005; 293: 43 71 [DOI] [PubMed] [Google Scholar]
- Chi T. A BAF-centered view of the immune system. Nat Rev Immunol 2004; 4: 965 977 [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Current Opin Genet Dev 2007; 17: 126 131 [DOI] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 2010; 29: 1434 1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J 2006; 25: 3986 3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Park E-J, Hur S-K, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009; 8: 29 39 [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A. Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 2000; 6: 1287 1295 [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J 1998; 17: 6979 6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 2003; 19: 169 182 [DOI] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 1997; 5: 66 68 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131: 839 849 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Casuriaga R, Geisinger A, Lopez-Carro B, Porro V, Wettstein R, Folle GA. Ultra-fast and optimized method for the preparation of rodent testicular cells for flow cytometric analysis. Biol Proced Online 2009; 11: 184 195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45: 413 417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshimizu U, Nishioka H, Watanabe D, Dohmae K, Nishimune Y. Characterization of a novel spermatogenic cell antigen specific for early stages of germ cells in mouse testis. Mol Reprod Dev 1995; 40: 221 227 [DOI] [PubMed] [Google Scholar]
- Deng W. Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2002; 2: 819 830 [DOI] [PubMed] [Google Scholar]
- Watanabe D, Yamada K, Nishina Y, Tajima Y, Koshimizu U, Nagata A, Nishimune Y. Molecular cloning of a novel Ca(2+)-binding protein (calmegin) specifically expressed during male meiotic germ cell development. J Biol Chem 1994; 269: 7744 7749 [PubMed] [Google Scholar]
- Sweeney C, Murphy M, Kubelka M, Ravnik SE, Hawkins CF, Wolgemuth DJ, Carrington M. A distinct cyclin A is expressed in germ cells in the mouse. Development 1996; 122: 53 64 [DOI] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 1996; 13: 336 342 [DOI] [PubMed] [Google Scholar]
- Habu T, Taki T, West A, Nishimune Y, Morita T. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res 1996; 24: 470 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res 2003; 103: 277 284 [DOI] [PubMed] [Google Scholar]
- Drabent B, Bode C, Bramlage B, Doenecke D. Expression of the mouse testicular histone gene H1t during spermatogenesis. Histochem Cell Biol 1996; 106: 247 251 [DOI] [PubMed] [Google Scholar]
- Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc 2006; 1: 1458 1461 [DOI] [PubMed] [Google Scholar]
- Malkov M, Fisher Y, Don J. Developmental schedule of the postnatal rat testis determined by flow cytometry. Biol Reprod 1998; 59: 84 92 [DOI] [PubMed] [Google Scholar]
- Hoyer-Fender S. Molecular aspects of XY body formation. Cytogenet Genome Res 2003; 103: 245 255 [DOI] [PubMed] [Google Scholar]
- Mark M, Jacobs H, Oulad-Abdelghani M, Dennefeld C, Feret B, Vernet N, Codreanu CA, Chambon P, Ghyselinck NB. STRA8-deficient spermatocytes initiate, but fail to complete, meiosis and undergo premature chromosome condensation. J Cell Sci 2008; 121: 3233 3242 [DOI] [PubMed] [Google Scholar]
- Barlow AL, Benson FE, West SC, Hulten MA. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J 1997; 16: 5207 5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Turner JM. The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 2009; 10: 207 216 [DOI] [PubMed] [Google Scholar]
- Wan M, Zhang J, Lai D, Jani A, Prestone-Hurlburt P, Zhao L, Ramachandran A, Schnitzler GR, Chi T. Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex. Eur J Immunol 2009; 39: 580 588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, Khatri R, Zhao K, Chi T. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J Exp Med 2008; 205: 2813 2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci U S A 2011; 108: 12758 12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107: 323 337 [DOI] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 2009; 27: 317 328 [DOI] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A 2009; 106: 5187 5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A 2009; 106: 5181 5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol 2006; 289: 372 383 [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007; 55: 201 215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010; 466: 62 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Fedoriw AM, Magnuson T. An essential role for a mammalian SWI/SNF chromatin-remodeling complex during male meiosis. Development 2012; 139: 1133 1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampariello F, Mauro F, Uccelli R, Spano M. Automatic analysis of flow cytometric DNA histograms from irradiated mouse male germ cells. Cytometry 1989; 10: 62 69 [DOI] [PubMed] [Google Scholar]
- Liao W, Cai M, Chen J, Huang J, Liu F, Jiang C, Gao Y. Hypobaric hypoxia causes deleterious effects on spermatogenesis in rats. Reproduction 2010; 139: 1031 1038 [DOI] [PubMed] [Google Scholar]