ABSTRACT
The mechanisms by which the region-specific expression patterns of clustered genes evolve are poorly understood. The epididymis is an ideal organ to examine this, as it is a highly segmented tissue that differs significantly in structure between closely related species. Here we examined this issue through analysis of the rapidly evolving X-linked reproductive homeobox (Rhox) gene cluster, the largest known homeobox gene cluster in metazoans. In the mouse, we found that most Rhox genes are expressed primarily in the caput region of the epididymis, a site where sperm mature and begin acquiring forward motility. This region-specific expression pattern depends, in part, on the founding member of the Rhox cluster—Rhox5—as targeted mutation of Rhox5 greatly diminishes the expression of several other family members in the caput region. In the rat, Rhox5 expression switches from the caput to the site of sperm storage: the cauda. All Rhox genes under the control of Rhox5 in the mouse epididymis display a concomitant change in their regional expression in the rat epididymis. Our results lead us to propose that widespread changes in the region-specific expression pattern of genes over evolutionary time can be the result of alterations of one or only a few master regulatory genes.
Keywords: epididymis, evolution, gene expression, transcriptional regulation, X chromosome
The Rhox homeobox gene cluster has a dramatically different region-specific expression pattern in the mouse versus the rat epididymis, and the shift in expression is dictated by one of its family members.
INTRODUCTION
The epididymis is a highly specialized tissue that functions in maturation, transport, and storage of sperm within morphologically and molecularly distinct segments that display region-specific gene expression [1, 2]. Classically, the epididymis is divided into three regions—the caput, corpus, and cauda—that extend from the proximal to the distal end. Evidence suggests that these regions have specific functional roles. For example, the caput appears to restrict lumicrine signaling triggered by testis-derived growth factors to the proximal end of the epididymis [3] and load epididymis-derived proteins into the sperm membrane [1]. Caput segments are also rich in acid-base regulators (both enzymes and ion transporters) that contribute to acidification of the lumen and elicit water retention essential for sperm compaction and metabolic senescence [4]. Sperm maturation continues as the cells pass through the corpus segments, a site where evidence suggests that their flagellar machinery is activated [5]. Final maturation occurs in the cauda, where spermatozoa are also stored until ejaculation [6].
To date, the mechanisms by which the unique microenvironments in the epididymis are established and maintained are poorly understood [7]. While several transcription factors have been shown to be expressed in the developing epididymis [8, 9], it remains unclear which, if any of these, establish the segment-specific gene expression patterns in the epididymis. One appealing candidate is the transcription factor, androgen receptor (AR), which clearly has a role in the development of the epididymis [6, 10], but it remains to be determined whether it controls any segment-specific developmental events. Other promising candidates to have a role in this are the homeobox transcription factors HOXA10 and HOXA11 since their loss in mice causes region-specific epididymal defects, including “homeotic transformation events,” in which one segment acquires the appearance of another segment or organ [11]. A confounding factor in the identification of other candidates has been that knockout of some transcription factors leads to disruption or loss of the entire Wolffian duct, precluding analysis of their role, if any, in the formation of the epididymis [9]. Likewise, the transcription factors that dictate the segment-specific functions of the mature epididymis are only beginning to be explored. ETS family members have been implicated in initial segment functions, based on electroporation experiments performed with a dominant-negative ETS factor in the epididymis in vivo [12]. Targeted disruption of the helix-loop-helix transcription factor, ID3, which is primarily expressed in principal cells of the cauda region [13], did not disrupt epididymal segmentation, but did result in the misexpression of several genes specifically in the cauda [14].
Given that we know very little about the transcription factors that drive the development and function of the epididymis, it is not surprising that the regulatory mechanisms responsible for the evolutionary diversification of the epididymis are completely unknown. While the basic functions of the epididymis are the same in all mammalian species, these functions appear to be accomplished in different ways in different species. For example, species differ in the sequential protein milieu that sperm are exposed to as they travel through distinct morphological segments [6], and there are also functional differences between equivalent regions of the epididymis in different species [15]. In this article, we provide evidence that the reproductive homeobox (Rhox) genes are candidates to influence the development, function, and evolution of the epididymis. Like all homeobox genes, Rhox genes encode transcription factors harboring a homeodomain, a 60 amino-acid DNA-binding motif with three alpha helices, one of which makes base-specific contacts with DNA [16]. The Rhox genes are clustered together at a single syntenic site on the X chromosome in a wide variety of mammals, including mice, rats, dogs, marmosets, hamsters, and primates, including humans [16–19] (see also NCBI's online resources). The mouse Rhox genes have been studied in more detail than those from other species. Intriguingly, most mouse Rhox genes are expressed in the epididymis [16, 20, 21], raising the possibility that they have a functional role in this organ. Indeed, targeted mutation of the founding member of the Rhox gene cluster—Rhox5 (previously called Pem)—compromises male fertility in mice and reduces both the number and the proportion of sperm with normal forward motility [16]. While it is not known how Rhox5 promotes the maturation of normally motile sperm, Rhox5 is highly expressed in the caput region of the epididymis [21], the site that several lines of evidence indicate is where sperm motility capability acquisition occurs [15, 22, 23]. Thus, Rhox5 may regulate target genes in somatic cells in the caput epididymis that, in turn, create a local niche in this epididymal region that promotes the acquisition of forward motility potential [17]. Rhox5 and other Rhox family members are also promising candidates to have a role in the rapid evolutionary changes characteristic of the epididymis [24]. This follows from the fact that the Rhox genes themselves are rapidly evolving both in sequence and in copy number [25–27]. In this communication, we demonstrate that the Rhox gene cluster has also undergone a dramatic shift in its region-specific expression in the epididymis over evolutionary time. We provide evidence that this shift is driven by Rhox5, which we find dictates the expression of most other Rhox cluster genes in the mouse epididymis. Our results lead us to propose that Rhox genes encode homeobox factors that function in a transcription network that elicits changes in region-specific gene expression in the epididymis over evolutionary time.
MATERIALS AND METHODS
Animals and Experimental Design
All animals were handled according to National Institutes of Health guidelines and in compliance with institutional approved animal protocols. The Rhox5-null mice were described previously [16, 28]. Adult male C57BL/6 or mice of the indicated postnatal age were euthanized by CO2 asphyxiation and their testes and epididymides extirpated. Fat was trimmed away from each epididymis, and one was fixed in 4% paraformaldehyde and the other processed further by cutting first between the fifth and sixth (as defined in Johnston et al. [29]) connective tissue septa, then between the seventh and eighth connective tissue septa, to generate three pieces corresponding to caput, corpus, and caudal epididymis. These latter segments were immediately ground in 500 μl Trizol reagent (Invitrogen) and frozen until RNA was extracted according to the manufacturer's protocol.
Adult male Sprague-Dawley rats were obtained from the University of Virginia Vivarium sources and maintained on a 12L:12D cycle with food and water ad libitum. The rats were anesthetized by i.p. injection of urethane (1 g/kg body weight). The epididymis was microdissected from the efferent ducts and vas deferens and for convenience of dissection was bisected at the narrowest point of the corpus region. Both epididymal halves were immediately placed in separate Petri dishes that contained ice-cold saline and were defatted with sharp dissection. This and all subsequent manipulations were performed with the use of a dissecting microscope while maintaining the dissection dish and medium on ice at all times. One operator proceeded to microdissect individual segments from the caput and proximal corpus regions, while another investigator microdissected individual segments from the distal corpus and cauda regions, as described previously [3, 30, 31]. The pooled tissues from both epididymides comprised one sample for subsequent RNA extraction. When all the segments making up one sample were collected, the samples were immediately stored at −80°C. This procedure was repeated until five to eight samples of each segment were collected for RNA extraction.
Quantitative Real-Time RT-PCR Analysis
Total RNA was isolated from mouse and rat epididymides using the Trizol reagent (Invitrogen) according to manufacturer's recommendations. The quantity and quality of total RNA were determined by spectrometry and denaturing agarose gel electrophoresis, respectively. The cDNA was synthesized from total RNA (2 μg) using the iScript Select cDNA synthesis Kit (BioRad). Real-time quantitative RT-PCR (qPCR) analysis of mRNA expression was performed using a MyiQ Single-Color Real-Time PCR Detection System (BioRad) with iQ SYBR Green supermix (BioRad) as the detector according to the manufacturer's recommendations. Primers shown in Supplemental Table S1 (all Supplemental Data are available online at www.biolreprod.org) were designed to amplify cDNAs of around 200 base pairs, and all exhibited similar amplification efficiency (97 ± 3%) as assessed by amplification of cDNA dilution series. PCR cycle parameters were 95°C for 15 sec and 60°C for 1 min for 40 cycles. The threshold line was set in the linear region of the plots above the baseline noise, and threshold cycle (CT) values were determined as the cycle number at which the threshold line crossed the amplification curve. PCR without template or template substituted with total RNA was used as a negative control to verify experimental results. After amplification, the specificity of the PCR was determined by both melt-curve analysis and gel electrophoresis to verify that only a single product of the correct size was present. Data were normalized against Rpl19 and are shown as the average fold increase ± SEM.
In Situ Hybridization
In situ hybridization analysis in mouse epididymides was conducted using methods described previously [32]. Briefly, deparaffinized, rehydrated, and deproteinated cross sections (5 μm) of the epididymis from each mouse were hybridized with radiolabeled sense or antisense cRNA probes generated from linearized plasmid DNA templates using in vitro transcription with [35S-α] UTP. For Rhox8, we used a 500-nt cDNA probe (corresponding to 681-1181 nt downstream of the transcription start site) to exclude the large GAA-repeat region so as to preclude potential cross-reactivity with trinucleotide repeat genes; all other Rhox cDNA probes were full length [16]. After hybridization, washing, and ribonuclease A digestion, slides were dipped in NTB liquid photographic emulsion (Kodak), stored at 4°C for 4–30 days, and developed in Kodak D-19 developer. Slides were then counterstained with Gill modified hematoxylin (Stat Lab), dehydrated through a graded series of alcohol to xylene (Fisher), and protected with a coverslip.
Statistical Analyses
All qPCR data were subjected to one-way analysis of variance, and differences between individual means were tested by a Tukey multiple-range test using Prism 4.0 (GraphPad); qPCR data were corrected for differences in sample loading using the Rpl19 data as a covariate. Tests of significance were performed using the appropriate error terms according to the expectation of the mean squares for error. A P-value of 0.05 or less was considered significant. Data are presented as least-square means with SEM.
RESULTS
Region-Specific Expression of Rhox Genes in the Adult Mouse Epididymis
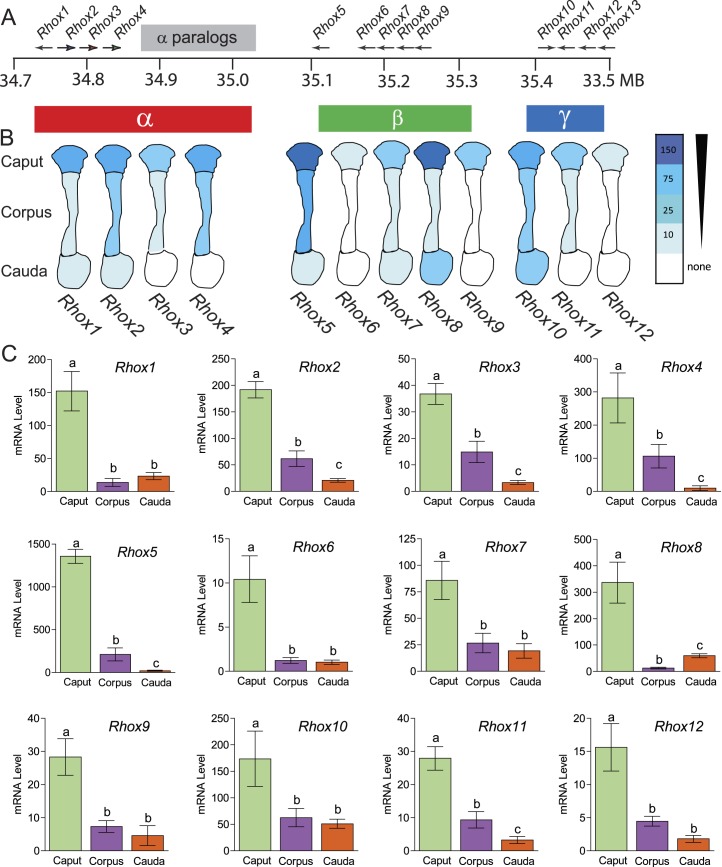
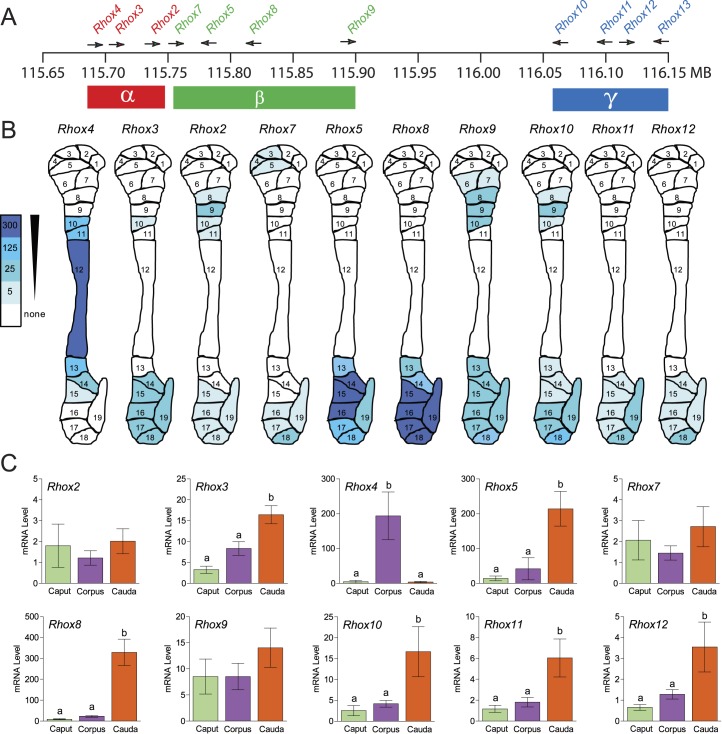
The mouse Rhox gene cluster contains single copies of 10 genes (Rhox1 and Rhox5–Rhox13) [16, 33] and several almost identical copies (paralogs) of three genes (Rhox2, Rhox3, and Rhox4) that were probably derived by tandem duplication of a trimer unit containing these three genes [17] (Fig. 1A). To determine the expression of these Rhox genes in the mouse epididymis, we used qPCR analysis with primer pairs specific for each of the single-copy genes and “pan paralog-specific” primers possessing 100% complementarity with all members of the Rhox2, Rhox3, or Rhox4 paralog families. This analysis revealed that most Rhox genes are expressed in the epididymis (Fig. 1B). The only exception—Rhox13—failed to give a detectable signal with several independent epididymis samples (data not shown), so we excluded this gene from our subsequent analysis. All the other Rhox genes are expressed mainly in the caput region of the epididymis (Fig. 1, B and C; >60% of the signal was in the caput, relative to corpus and cauda). We divided these Rhox genes into three classes based on their expression pattern. Class I, which consists of Rhox2, Rhox3, Rhox4, Rhox5, and Rhox11, exhibits a gradient of expression that taper down from caput to cauda. Class II, which is comprised of three Rhox genes that are all highly expressed in placenta—Rhox6, Rhox9, and Rhox12 [16]—displays only modest expression in the epididymis and is barely detectably expressed in the corpus and cauda. Class III, which has only a lone member—Rhox8—has the unique feature of being expressed higher in the cauda than the corpus region. These results are consistent with published data for Rhox5, Rhox6, and Rhox9, available in the Mammalian Reproductive Genetics (Database mrg.genetics.washington.edu [31]).
FIG. 1. .
Regional expression of mouse Rhox cluster family members in the epididymis. A) Schematic of the mouse Rhox gene cluster, which contains 13 distinct primary genes and a seven-unit tandem duplication of Rhox2, Rhox3, and Rhox4, indicated as “α-paralogs” for simplicity. B) Expression pattern of the Rhox genes in the three main regions of the mouse epididymis as determined from the expression analysis in C. C) qPCR analysis of total cellular RNA from epididymides obtained from six adult animals that were dissected into caput, corpus, and caudal sections. Values were normalized against ribosomal L19 (Rpl19) mRNA and are expressed as fold above background (±SEM), which was arbitrarily given a value of 1. Equivalent results were obtained when cyclophilin mRNA was used as the internal control (data not shown). Letters denote means that were significantly different (P < 0.05).
Rhox5-Null Mice Have Dramatically Altered Expression of Most Other Rhox Genes
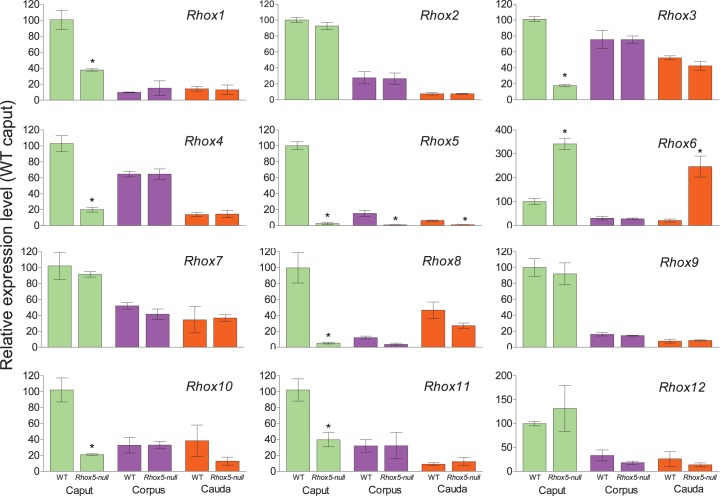
We previously demonstrated that RHOX5 protein is expressed in segments 2–4 of the caput epididymis [21] and that Rhox5-null male mice sperm have motility defects [16], which together suggests that Rhox5 may regulate genes in the caput epididymis that promote sperm maturation. Given this candidate role for Rhox5 and our finding that most other Rhox genes are also selectively expressed in the caput (Fig. 1), we wondered if Rhox5 serves to activate the expression of other Rhox genes in the caput epididymis. To test this, we determined the expression levels of other Rhox genes in the epididymis of Rhox5-null mice. We found that Rhox5-null mice had depressed expression of Rhox1, Rhox3, Rhox4, Rhox8, Rhox10, and Rhox11 in the caput region of the epididymis (Fig. 2), implying that Rhox5 activates the expression of these Rhox genes. The depressed expression of other Rhox genes in Rhox5-null mice occurred selectively in the caput region, not in the corpus or cauda, which is consistent with our finding that Rhox5 is expressed predominantly in the caput (Fig. 1) [21]. We also noted that Rhox8 and Rhox10 exhibited a trend toward reduced expression in other regions of the epididymis, but this reduction was not statistically significant (P > 0.05; Fig. 2). We also identified a Rhox gene—Rhox6—that appeared to be negatively regulated by Rhox5, as its expression was elevated (>5-fold increase) in the caput epididymis of Rhox5-null mice (Fig. 2). This suggests that Rhox5 normally serves to extinguish Rhox6 expression, as Rhox6 is barely expressed in the epididymis of wild-type mice (near the limits of detection). Rhox6 expression was also elevated in the cauda region of Rhox5-null mice (Fig. 2), suggesting that Rhox5 also negatively regulates Rhox6 expression in this region (note that, while not apparent from Fig. 2C, Rhox5 mRNA is clearly expressed in the cauda region, as we found its level is 12.5 Ct units above that of the L19 internal control; background was 17 Ct units above this internal control). Deregulation of the aforementioned Rhox genes in response to loss of Rhox5 was not a general feature of the entire Rhox cluster, but a specific effect, as Rhox2, Rhox7, Rhox9, and Rhox12, did not display a significant change in expression in response to the absence of Rhox5 (Fig. 2).
FIG. 2. .
Caput-specific regulation of Rhox gene expression by RHOX5 in the adult epididymis. Epididymides from six adult animals (either wild-type [WT] or Rhox5-null) were dissected and analyzed as in Figure 1. However, for comparative purposes, rather than fold above background signal, expression data are shown as fold relative to the normalized WT caput qPCR signal, which was arbitrarily given a value of 100. *Segments exhibiting significantly different relative expression between WT and Rhox5-null animals (P < 0.001).
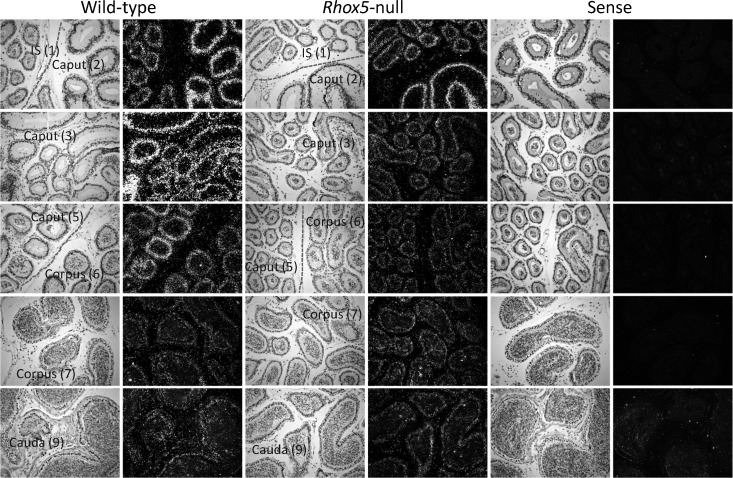
To verify the ability of Rhox5 to regulate the expression of other Rhox genes in a region-specific manner, we examined the expression pattern of the Rhox gene most highly expressed in the epididymis—Rhox8—by in situ hybridization. Using an antisense Rhox8 probe, we observed a strong hybridization signal in the caput region and lower signals in the other two regions of the epididymis (Fig. 3), in agreement with our qPCR results (Fig. 1). The Rhox8 in situ hybridization signal in the corpus was heavily concentrated in a segment near the boundary with the caput. Expression in the cauda was more diffuse than in the caput or corpus, probably because of the lower cell number relative to lumen size in cross sections in the corpus. In Rhox5-null mice, there was a strong reduction in the Rhox8 in situ hybridization signal in the caput region and little or no reduction in the corpus and cauda (Fig. 3), which agrees with what we observed by qPCR analysis (Fig. 2). Of note, we observed variable expression and downregulation of Rhox5 and Rhox8, respectively, in segment 2. However, downregulation of Rhox8 in segments 3–4, where RHOX5 is maximally expressed [21], was always pronounced. In contrast, the Rhox8 signal was not significantly reduced in the initial segment subregion of the caput epididymis in Rhox5-null mice, which is consistent with our prior finding that RHOX5 protein is not detectable in the initial segment of wild-type mice [21]. As a negative control, we used a Rhox8 sense probe, which we found did generate a significant hybridization signal in any of the epididymal tissue regions (Fig. 3). Similar in situ hybridization analyses were performed for Rhox4, Rhox7, and Rhox11, but we failed to observe a consistent signal (data not shown), probably because these genes are expressed at lower levels than Rhox8 in the epididymis (Fig. 1).
FIG. 3. .
In situ hybridization analysis of Rhox8 expression in the adult epididymis. WT and Rhox5-null epididymal samples (n = 6) were prepared from littermates that were coprocessed and developed for the same lengths of time. Rhox8 mRNA was detected by antisense probe in the initial segment (IS), caput, corpus, and cauda (numbers correspond to previously characterized segments of the mouse epididymis [31]). No significant signal was observed with the sense probe in any region. Original magnification ×200.
Developmental Expression and Regulation of the Rhox Genes in the Mouse Epididymis
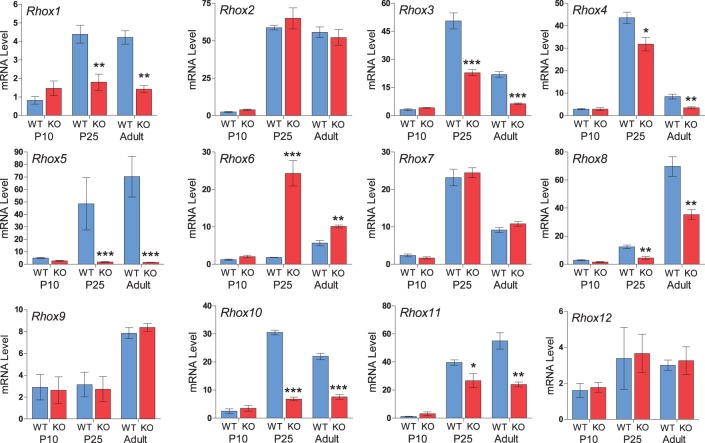
We next examined whether Rhox5-mediated regulation of other Rhox genes occurs during postnatal development of the epididymis. All the Rhox genes were detectably expressed at Postnatal Day 10 (P10), but there was no difference in their expression between Rhox5-null and control mice (Fig. 4). By P25, when Rhox5 is strongly upregulated, several Rhox genes displayed altered expression in Rhox5-null mice as compared to control mice. The particular Rhox genes that exhibited altered expression at P25—Rhox1, Rhox3, Rhox4, Rhox6, Rhox8, Rhox10, and Rhox11—were the same genes that displayed altered expression at the adult stage (Fig. 4). In contrast, the expression levels of Rhox2, Rhox7, Rhox9, and Rhox12 were not altered in Rhox5-null mice, confirming the specificity and significance of RHOX5 cross regulation of neighboring Rhox genes.
FIG. 4. .
Temporal expression and regulation of Rhox mRNA during postnatal epididymal development. WT and Rhox5-null (KO) epididymal samples (n = 6) were prepared from animals of the indicated postnatal age and the relative expression of each Rhox gene was determined by qPCR as described in Figure 1. Data are presented as fold relative to their specific background value established by the signal observed in WT epididymis on P7. P7 expression levels were similar to no-RT control reactions (data not shown) and arbitrarily given a value of 1. Asterisks indicate segments exhibiting significantly different relative expression between WT and Rhox5-null animals (*P < 0.05, **P < 0.01, ***P < 0.001).
Evolution of Rhox Gene Cluster Expression
The Rhox gene cluster is rapidly evolving, as described in the Introduction. Based on the most recent build of the rat genome, the rat Rhox gene cluster possesses fewer genes than the mouse Rhox cluster, as it lacks orthologs for Rhox1, Rhox6, and Rhox13, and it does not have multiple copies of the Rhox2/3/4 trimer unit. Despite these differences, we found that, like their mouse orthologs [16], all the rat Rhox genes are expressed selectively in reproductive-associated tissues (Fig. 5). Analysis of 18 adult tissues by qPCR analysis showed that rat Rhox cluster genes are detectably expressed only in the epididymis, testes, ovary, and placenta, consistent with the notion that the Rhox cluster has a conserved role in male and female reproduction.
FIG. 5. .
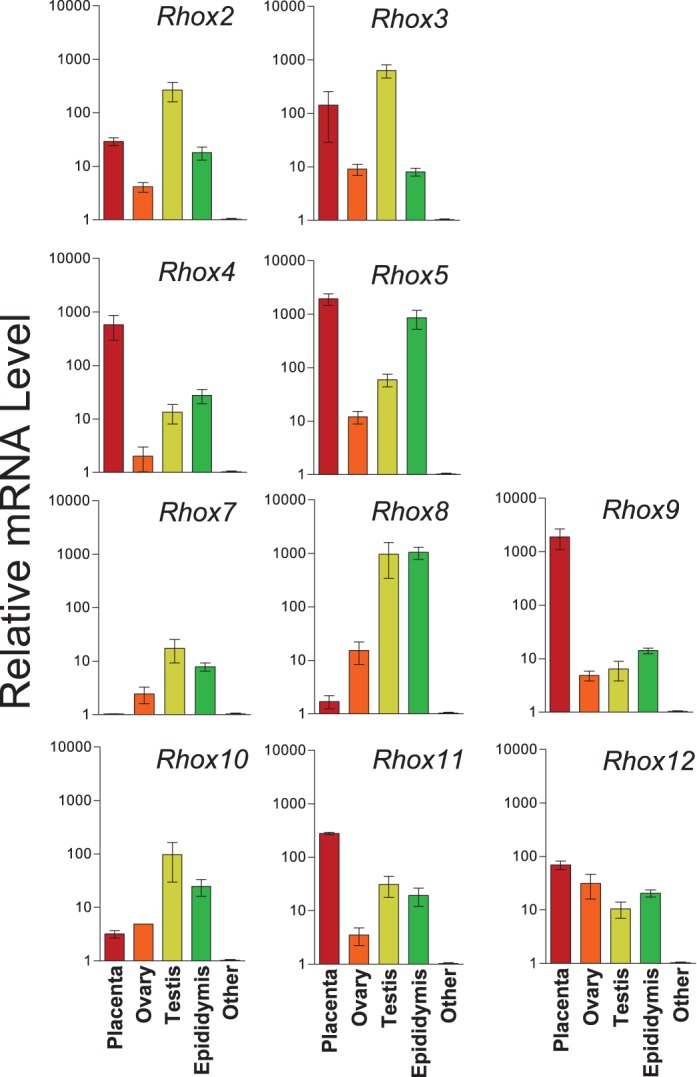
Preferential expression of the rat Rhox genes in reproductive-associated tissues. qPCR analysis was used to assess the level of Rhox mRNA, performed as in Figure 1. Each bar represents the average level of expression relative to background for at least three separate RT reactions assayed in duplicate (note that values are on a log scale). The only tissues shown are those that detectably expressed Rhox genes. The adult tissues that we tested that did not have detectable expression were brain, skin, heart, lung, spleen, tongue, kidney, liver, seminal vesicle, prostate, stomach, thymus, and intestine, which are grouped together under “other.”
We previously reported that the rat Rhox5 gene is expressed mainly in the cauda region of the epididymis [34]. This in striking contrast to what we found for the mouse Rhox5 gene, which we found was expressed mainly in the caput region (Fig. 1). Given our evidence that several Rhox family members depend on Rhox5 for expression in mice (Fig. 2), this predicts that the switch of Rhox5 expression to the cauda in the rat led to a concomitant switch in the expression of other Rhox family members to the cauda region. To test this prediction, we isolated each of the 19 rat epididymides segments that have been defined in past studies [29, 35]. These segments are defined by boundaries that are established by inherent connective tissue septa that have been proposed to establish functional and gene expression “zones” [3]. For our analysis, we dissected each of the 19 segments from three rat epididymides and pooled each segment before RNA isolation; qPCR analysis of the 19 segments confirmed our earlier finding [34] that Rhox5 has a cauda-specific expression pattern, as we found that Rhox5 mRNA was confined to segments 13–19, all of which are all in the cauda region (Fig. 6). Supporting our prediction that other Rhox family members “acquire” the expression pattern of Rhox5 in the rat, we found that most other rat Rhox genes were also preferentially expressed in the cauda region (Fig. 6 and Supplemental Table S2). This included Rhox3, Rhox8, Rhox10, and Rhox11, whose mouse orthologs depend on Rhox5 for normal expression in the mouse epididymis (Fig. 2). In contrast, the rat caput region expressed only low levels of most Rhox genes. No rat Rhox genes were detectably expressed in the most proximal region of the caput; only Rhox7 exhibited detectable expression in the mid-caput region (albeit very low expression), and only Rhox2, Rhox9, and Rhox10 were detectably expressed in the distal caput region (Fig. 6). While we have no direct evidence for a causal role of Rhox5 regulating other Rhox family members in the rat, these data are consistent with a caput-to-cauda switch in Rhox gene cluster expression elicited by Rhox5 (Fig. 7; see also Discussion).
FIG. 6. .
RHOX genes are preferentially expressed in the cauda region of the rat epididymis. A) Schematic of the rat Rhox gene cluster, which contains 11 gene orthologous to those in the mouse Rhox gene cluster displayed in Figure 1. B) Expression pattern of the Rhox genes in the 19 segments of the rat epididymis as determined from the expression analysis in Supplemental Table S2. RNA was prepared from four rats and the average expression in each segment was determined by qPCR as described in Figure 1. C) Histogram presentation of the data in B, determined by electronically pooling segments 1–9, 10–14, and 15–19, which correspond to the caput, corpus, and caudal regions of the epididymis, respectively. Shown is the mean expression ± SEM for each Rhox gene, in each epididymal segment, expressed as fold relative to background signal, which was arbitrarily given a value of 1. RNA levels were normalized against Rpl19 mRNA and weighted by the proportion of RNA contained in each segment to account for differences in relative size of each region.
FIG. 7. .
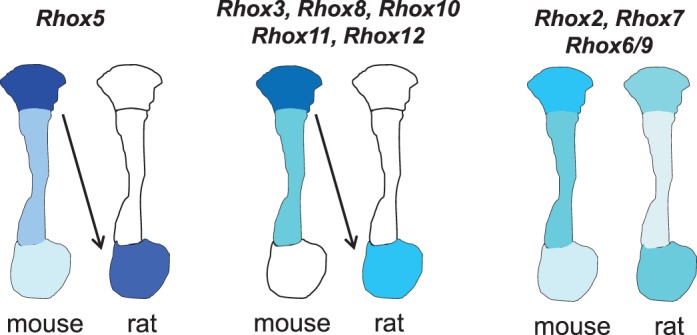
Model: evolutionary shift in the region-specific expression pattern of Rhox gene expression during evolution. In mice, Rhox5 and the Rhox genes under its control (Rhox3, Rhox8, Rhox10, Rhox11, and Rhox12) are expressed predominantly in the caput region (left and middle). In rats, these Rhox genes are all expressed primarily in the cauda region, leading us to posit that a caput-to-caudal switch in Rhox5 expression is responsible for the corresponding switch in the region-specific expression of the Rhox genes under its control. In contrast, those genes that are not RHOX5-regulated (Rhox2, Rhox6/9, and Rhox7) possess similar expression patterns in mice and rats (right).
DISCUSSION
One of the primary goals of evolutionary biologists is to determine how genetic variation arises, how it is maintained, and ultimately how it drives biological diversity. In contrast, developmental biologists concern themselves with how organs take shape and integrate form into function. Reproductive biologists are ideally suited to integrate both of these interests because reproductive organs and the associated cells tend to display major differences between different species. This may be partly the result of selection pressure associated with speciation. It may also derive from the fact that the ultimate goal of the reproductive tract—the generation of gametes—occurs after embryonic development in many species and thus is likely to be less subject to evolutionary constraints. Thus, even though the gonads are established during embryonic development, pubertal development occurs postnatally, and the proliferation and differentiation of germ cells continues throughout the reproductive life of higher organisms. Regardless of the precise mechanisms responsible, it is clear that the most of the genes involved in reproduction undergo rapid evolution [36, 37]. Even genes encoding proteins absolutely essential for reproduction, such as SRY, the male-determining transcription factor, are undergoing extremely high rates of evolution driven by positive selection [38].
Here, we used the epididymis as a model system to begin to examine the mechanisms of evolutionary diversification. Long discounted as being merely a storage site for sperm, accumulating evidence suggests that the epididymis actually performs many other functions, including promotion of sperm maturation and protection of sperm from damage (see the Introduction). Furthermore, these functions appear to occur in specific regions of the epididymis, making this organ an excellent model system for studying evolutionary development [9, 39]. In support of the notion that each segment provides specific functions, unique gene expression profiles have been demonstrated by microarray analysis for each epididymal segment [30, 31, 35, 40–42]. In addition, proteomic analyses have begun to define the secretome of individual epididymal regions [5, 43, 44].
In this study, we examined the expression and regulation of the rapidly evolving Rhox gene cluster in the epididymis. We first confirmed the previously reported caput-specific expression of the founding member of this cluster—Rhox5—and then demonstrated that most of the other Rhox genes exhibited the same or similar region-specific expression pattern (Fig. 1, B and C). Using Rhox5-null mice, we then demonstrated that this coordinate expression of Rhox cluster genes was largely the result of Rhox5 activating the expression of the other Rhox genes in the caput region of the mouse epididymis (Fig. 2). We then examined the rat epididymis and made the surprising discovery that most Rhox genes, including Rhox5, displayed predominant expression in the cauda region rather than the caput region (Fig. 6). This indicated that during the expansion and diversification of rodents, there was a major switch in the region-specific expression pattern of nearly the entire Rhox gene cluster. While we do not know the mechanism behind this switch, a likely possibility is that it is due to a change in the expression pattern of Rhox5, which, in turn, shifted the pattern of expression of downstream Rhox genes. To test this directly, it would be necessary to disrupt the Rhox5 gene in rats, which would be a technically challenging task. Regardless of the mechanism responsible for this evolutionary switch, our findings suggest the existence of a Rhox gene network in the mouse epididymis in which RHOX5 serves as a master regulator of subordinate Rhox genes.
While most Rhox cluster genes coexpressed with Rhox5 in the caput region in mice were coexpressed with Rhox5 in the cauda region in rats, there was one exception: Rhox4. Why might this be? One possibility is that the rat Rhox4 gene promoter has lost responsiveness to RHOX5. In support of this, the 900-nt promoter region previously defined for mouse Rhox4b [45] has only 81% sequence identity with the corresponding region of rat Rhox4 (RGSC v3.4). Notably, a candidate-binding site for RHOX5 (a homeobox consensus-binding site in the mouse Rhox4b promoter [45]) is disrupted in the rat Rhox4 promoter. Rhox4 is also positioned differently in the rat and mouse Rhox clusters. In rats, it is the most 5′ Rhox gene, whereas in mice, it exists as several paralogs in the middle of the Rhox cluster, between Rhox1 and Rhox5, in the opposite orientation compared to in the rat (according to rat and mouse genome builds 3.4 and 37.2, respectively). If Rhox genes are regulated by global mechanisms that involve, for example, long-range enhancers and/or insulators, this shift in Rhox4 gene position could explain why Rhox5 does not regulate Rhox4 expression in the rat.
The Rhox genes are not the only class of homeobox genes that have been implicated in regulating genes that control the development and function of the epididymis. Lbx2, Lhx1, Tlx2, Lim2, Emx2, Pax2, and Pax8 are all expressed in the epididymis when it is forming during embryogenesis, suggesting that these homeobox genes have roles in the development and/or function of this organ [8, 9, 46, 47]. Studies with knockout mice have shown that some members of the best-characterized homeobox subfamily—the Hox genes—dictate the segment-specific development of parts of the mouse epididymis [11]. Many members of the Hox subfamily are also expressed in the mature epididymis, where some exhibit region-specific expression [46, 48]. However, it is not known whether the region-specific expression of Hox genes differs between species, which would be required for them to drive evolutionary shifts in the region-specific expression of their subordinate genes.
A major distinguishing feature of Hox genes is that they are highly conserved in sequence, even in species as distant in the phylogenetic scale as flies and mammals [49]. This suggests that the functions of Hox genes are relatively constrained, limiting their ability to have roles in the evolutionary diversification of the epididymis. In contrast, the Rhox genes are evolving rapidly and thus have more flexibility in terms of driving evolutionary change. This was first demonstrated for Rhox5, which has undergone extremely rapid evolution in rodents [27]. A high proportion of the nucleotide changes result in nonsynonymous amino-acid substitutions, indicating that Rhox5 has undergone strong positive selection for alterations in many of its amino acids. These amino-acid changes have occurred not only in the RHOX5 N-terminal region, whose function is unknown, but also in portions of the homeodomain region that are known to confer protein-protein interactions in other homeobox proteins [49, 50]. A recent study provided evidence that the RHOXF2 gene, which is present in higher mammals, has also undergone positive selection for amino-acid alterations, including in the homeodomain region [18]. The Rhox gene cluster is also undergoing rapid change in copy number over recent evolutionary time. For example, the mouse Rhox gene cluster has 33 genes, while the rat Rhox gene cluster has only 11 genes [17]. Humans have 3 RHOX genes, and other primates have between 1 and ≥ 6 RHOX genes [18]. It is not known what evolutionary forces are responsible for eliciting these dramatic gains and/or losses of Rhox/RHOX genes over relatively short periods of time. Despite its rapid evolution, the Rhox gene cluster appears to have maintained its selective expression in reproductive-associated tissues. All Rhox cluster genes are expressed in the testis, epididymis, ovary, and/or placenta in both mice and rats (Fig. 5 and MacLean et al. [16]). Few Rhox genes are expressed in any other organs besides these reproductive organs in mice and rats (Fig. 5 and MacLean et al. [16]). This suggests that the Rhox gene cluster encodes a large set of transcription factors that are devoted to driving and supporting fertility [17].
Ablation of Rhox5 in mice results in a reduction in the proportion of sperm with forward motility and male subfertility, suggesting that RHOX5's presence in the caput epididymis is important for the final stages of sperm maturation [16]. Consistent with this, evidence suggests that the caput region of the epididymis directs the acquisition of sperm motility [6]. Why does Rhox5 activate the expression of so many other Rhox genes in the caput? One possibility is that these different caput-expressed Rhox genes encode RHOX transcription factors that have distinct target genes. Thus, by regulating subordinate Rhox genes, Rhox5 greatly expands the number of target “structural” genes it can regulate in the epididymis. In support of this, the key amino acids that determine base-specific contacts with DNA are among the most variable between RHOX proteins [16, 25]. In addition, RHOX proteins differ considerably in regions of the homeodomain known to mediate protein-protein interactions as well as in the amino-terminal region, which varies both in length and sequence [16]. This raises the possibility that RHOX proteins differ in the cofactors that they attract, leading to different transcriptional outcomes.
We have begun to define RHOX5-regulated genes in the testes [51, 52], but nothing is yet known about RHOX5 targets in the epididymis apart from other Rhox genes, as reported in this article (Figs. 2–4). Obvious candidates are genes encoding proteins that promote spermatozoa progressive motility and protect spermatozoa from damage [5]. Particularly intriguing is genes that have undergone a switch in expression from the mouse caput region to the rat cauda region, as most members of the Rhox cluster have undergone this shift. Genes that have been identified in this category include B4galnt1 (also known as Galgt1), Fah, Fads1, Elovl6, Acox1, and Acox3 [35]. One of these genes—B4galnt1—is essential for spermatogenesis based on the finding B4galnt1-knockout mice fail to complete male meiosis due to improper lipid interactions [53]. Another class of genes likely to be regulated by RHOX5 in the epididymis is secondary androgen-responsive genes; that is, genes that are regulated by androgen/AR indirectly through androgen/AR-regulated transcription factors. This follows from the recent finding that Rhox5 is an androgen- and AR-inducible gene (via DNA demethylation) in epididymal cells [54] and the evidence that RHOX5 regulates secondary androgen-responsive genes in the testis [51, 55].
RHOX5 is expressed in a region of the epididymis containing cells that are known to actively secrete many proteins into the epididymal lumen [44]. Many of these proteins are absorbed by the differentiating spermatozoa in transit in the caput region, thereby potentially providing protection from environmental toxicants or other functions [5]. Since some homeobox proteins are known to have the ability to be both secreted and absorbed by cells [56, 57], it is not unreasonable to suppose that RHOX5 and other RHOX proteins are secreted by epididymal cells and taken up by spermatozoa. Indeed, evidence suggests that at least one HOX protein—HOXB2 (also known as HOXBES2)—is secreted by epididymal cells and absorbed by spermatozoa [58]. Given that spermatozoa are almost completely transcriptionally silent, it is unlikely that HOX or RHOX proteins regulate transcription in these cells; a more likely possibility is that they are sequestered in spermatozoa until after fertilization, at which point they regulate gene expression in the developing embryo. In support of this, RHOX5 protein is known to be present in the early developing mouse embryo [17, 59]. If indeed RHOX proteins are transmitted to spermatozoa in the epididymis, the divergence of RHOX proteins in both sequence and number could present a unique “tag” for each species that blocks cross-species fertilization and hence plays a role in speciation.
Why are most Rhox cluster genes preferentially expressed in a different region of the epididymis in mice versus rats? One possibility is that mouse and rat Rhox genes exhibit different region-specific expression patterns because they have evolved to regulate different genes. If so, it will be important to determine which came first: changes in target genes followed by selection for altered Rhox region-specific expression pattern or vice versa? The converse possibility is that each gene in the Rhox gene cluster has retained the ability to regulate the same set of target genes despite being expressed in a different region of the epididymis in mice and rats. If this is the case, the Rhox gene cluster could be responsible for dictating species-specific differences in the epididymis. It is well known that the sequential protein milieu that sperm are exposed to as they travel through distinct morphological segments differs between species [6]. While the most prevalent secreted proteins produced in the epididymis are largely conserved across species, they vary greatly in their concentration and region specificity [5]. For example, clusterin exhibits differences in region specificity, as it is predominantly in the caput in swine and horses, in the corpus in sheep, and at similar concentration across the entire human epididymis [5]. Glutathione peroxidase is a major component of the epididymal secretome in cattle, swine, sheep, and horses, where it is thought to play a crucial protective role against DNA damage, particularly in aging animals, but it is completely absent in the human epididymis [60]. There are also functional differences between equivalent regions of the epididymis in different species [15]. For example, sperm acquire near maximal competency for progressive motility in the third and seventh segment of the human and porcine epididymides, respectively [5]. This is associated with the loading of sialoglycoproteins on the sperm surface that occurs in these regions, but whether these have a causal role is not known. The human epididymis displays characteristics that differ from rodent epididymides; the segments are less well defined histologically, and most epididymal genes do not display regional specificity in their expression [40, 42, 61]. This also appears to be the case for RHOXF1 and RHOXF2, which, unlike the mice and rat Rhox genes, are expressed at similar levels in all three regions of the human epididymis (Supplemental Fig. S1).
In mice, a large proportion of the sperm from the corpus region of epididymides exhibit progressive motility, whereas few sperm from the equivalent region of rat epididymides have this ability [15]. Similarly, the corpus region of mice epididymides has a much higher proportion of sperm that can bind or fertilize eggs than the equivalent region in rat epididymides [15]. Given that the caput region in neither of these species has progressively motile sperm that bind or fertilize eggs, this implies that these qualities are acquired either late in the caput region and/or during transit through the corpus region. We suggest that this more proximal acquisition of progressive motility and sperm binding/fertilization ability in the mouse versus the rat could be the result of the more proximal expression of Rhox genes in the mouse versus rat epididymis. However, it is important to bear in mind that the order and/or timing at which events occur in the epididymis may not always be fundamentally important for generating functional spermatozoa; instead, it may only be important that each critical event occurs at some point during the path of spermatozoa prior to their storage [15]. This idea is supported by partial epididymectomy and tubule ligation experiments that showed that functional spermatozoa are generated despite portions of the epididymis being mechanically bypassed [3]. This brings up the possibility that some species may differ with respect to region-specific events in the epididymis as a result of selective pressures other than those directly influencing spermatozoa quality.
Given that so many Rhox genes depend on Rhox5 for proper expression in the epididymis, why does loss of Rhox5 result in only modest defects in sperm maturation and fertility [16]? One explanation is that Rhox genes expressed independently of Rhox5 provide compensation. Candidates to mediate this role are Rhox2 and Rhox7, both of which are expressed at high levels in the epididymis regardless of the presence or absence of Rhox5 (Figs. 2 and 4). Another candidate is Rhox6, which we found was dramatically upregulated in Rhox5-null epididymides (Figs. 2 and 4), indicating that Rhox6's expression in the epididymis is normally repressed by Rhox5. Given that Rhox6 is directly adjacent to Rhox5, it is tempting to speculate that it is uniquely subject to negative regulation because of “local” regulatory control, which, when circumvented by loss of Rhox5, leads to a compensatory response. Future studies will be required to test this. With the exception of Rhox5, little is known about the regulation of Rhox gene cluster family members [17, 59].
It is well established that the epididymis is important for the development of fertile sperm and that this process depends on both hormones and secretory proteins from the testis and epididymis. New advances in proteomic and genomic analysis in the epididymis have identified several factors likely to be important for acquisition of sperm motility and gamete interaction. However, little is known about the interplay between these factors and how each is regulated. A common core genetic program may drive the maturation of sperm in all mammalian species, but different species may implement this program in unique ways. Some mammalian species may also possess some of their own unique factors for sperm maturation and preservation. In this article, we provided evidence that the RHOX transcription factors are good candidates to regulate the luminal environment necessary for sperm development in the epididymis. Our results also suggest that RHOX transcription factors drive some of the evolutionary events responsible for species-specific differences in the epididymis. Further study of their function and regulation may lead to insights into human infertility and the generation of novel contraception approaches.
ACKNOWLEDGMENT
The authors would like to thank Daniel Johnston for his helpful correspondence and Anjana Bhardwaj and Miriam Buttigieg for their help with mouse epididymal tissue collection.
Footnotes
Supported by the National Institutes of Health HD53808, HD45595, HD65584, and HD55268.
REFERENCES
- Hinton BT, Palladino MA. Epididymal epithelium: its contribution to the formation of a luminal fluid microenvironment. Microsc Res Tech 1995; 30: 67 81 [DOI] [PubMed] [Google Scholar]
- Hinton BT, Palladino MA, Rudolph D, Lan ZJ, Labus JC. The role of the epididymis in the protection of spermatozoa. Curr Top Dev Biol 1996; 33: 61 102 [DOI] [PubMed] [Google Scholar]
- Turner TT, Johnston DS, Jelinsky SA, Tomsig JL, Finger JN. Segment boundaries of the adult rat epididymis limit interstitial signaling by potential paracrine factors and segments lose differential gene expression after efferent duct ligation. Asian J Androl 2007; 9: 565 573 [DOI] [PubMed] [Google Scholar]
- Pastor-Soler N, Pietrement C, Breton S. Role of acid/base transporters in the male reproductive tract and potential consequences of their malfunction. Physiology (Bethesda) 2005; 20: 417 428 [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belleannee C, Jones R, Labas V, Belghazi M, Guyonnet B, Druart X, Gatti JL, Dacheux F. Mammalian epididymal proteome. Mol Cell Endocrinol 2009; 306: 45 50 [DOI] [PubMed] [Google Scholar]
- Robaire B, Hinton BT, Orgebin-Crist MC. (eds.) The Epididymis. New York: Elsevier; 2006. [Google Scholar]
- Cornwall GA, Hann SR. Specialized gene expression in the epididymis. J Androl 1995; 16: 379 383 [PubMed] [Google Scholar]
- Rodriguez CM, Kirby JL, Hinton BT. Regulation of gene transcription in the epididymis. Reproduction 2001; 122: 41 48 [DOI] [PubMed] [Google Scholar]
- Joseph A, Yao H, Hinton BT. Development and morphogenesis of the Wolffian/epididymal duct, more twists and turns. Dev Biol 2009; 325: 6 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutskikh A, De Gendt K, Sharp V, Verhoeven G, Poutanen M, Huhtaniemi I. Targeted inactivation of the androgen receptor gene in murine proximal epididymis causes epithelial hypotrophy and obstructive azoospermia. Endocrinology 2011; 152: 689 696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Wilkinson MF. Homeobox genes and the male reproductive : Robaire B, Hinton BT. (eds.), The Epididymis: From Molecules to Clinical Practice. New York: Kluwer Academic/Plenum Publishers; 2002: 269 283. [Google Scholar]
- Yang L, Fox SA, Kirby JL, Troan BV, Hinton BT. Putative regulation of expression of members of the Ets variant 4 transcription factor family and their downstream targets in the rat epididymis. Biol Reprod 2006; 74: 714 720 [DOI] [PubMed] [Google Scholar]
- Carroll M, Hamzeh M, Robaire B. Expression, localization, and regulation of inhibitor of DNA binding (Id) proteins in the rat epididymis. J Androl 2006; 27: 212 224 [DOI] [PubMed] [Google Scholar]
- Carroll M, Luu T, Robaire B. Null mutation of the transcription factor inhibitor of DNA binding 3 (id3) affects spermatozoal motility parameters and epididymal gene expression in mice. Biol Reprod 2011; 84: 765 774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TT. De Graaf's thread: the human epididymis. J Androl 2008; 29: 237 250 [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, MacLeod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell 2005; 120: 369 382 [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Wilkinson MF. The Rhox genes. Reproduction 2010; 140: 195 213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu AL, Wang YQ, Zhang H, Liao CH, Wang JK, Zhang R, Che J, Su B. Rapid evolution and copy number variation of primate RHOXF2, an X-linked homeobox gene involved in male reproduction and possibly brain function. BMC Evol Biol 2011; 11: 298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svingen T, Koopman P. Involvement of homeobox genes in mammalian sexual development. Sex Dev 2007; 1: 12 23 [DOI] [PubMed] [Google Scholar]
- Lindsey JS, Wilkinson MF. Pem: a testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Dev Biol 1996; 179: 471 484 [DOI] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Wilkinson MF. Pem homeobox gene regulatory sequences that direct androgen-dependent developmentally regulated gene expression in different subregions of the epididymis. J Biol Chem 2002; 277: 48771 48778 [DOI] [PubMed] [Google Scholar]
- Hinton BT. The testicular and epididymal luminal amino acid microenvironment in the rat. J Androl 1990; 11: 498 505 [PubMed] [Google Scholar]
- Orgebin-Crist MC. Sperm maturation in rabbit epididymis. Nature 1967; 216: 816 818 [DOI] [PubMed] [Google Scholar]
- Dean MD, Good JM, Nachman MW. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Mol Biol Evol 2008; 25: 383 392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean JA, II, Lorenzetti D, Hu Z, Salerno WJ, Miller J, Wilkinson MF. Rhox homeobox gene cluster: recent duplication of three family members. Genesis 2006; 44: 122 129 [DOI] [PubMed] [Google Scholar]
- Sutton KA, Wilkinson MF. The rapidly evolving Pem homeobox gene and Agtr2, Ant2, and Lamp2 are closely linked in the proximal region of the mouse X chromosome. Genomics 1997; 45: 447 450 [DOI] [PubMed] [Google Scholar]
- Sutton KA, Wilkinson MF. Rapid evolution of a homeodomain: evidence for positive selection. J Mol Evol 1997; 45: 579 588 [DOI] [PubMed] [Google Scholar]
- Pitman JL, Lin TP, Kleeman JE, Erickson GF, MacLeod CL. Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Dev Biol 1998; 202: 196 214 [DOI] [PubMed] [Google Scholar]
- Johnston DS, Turner TT, Finger JN, Owtscharuk TL, Kopf GS, Jelinsky SA. Identification of epididymis-specific transcripts in the mouse and rat by transcriptional profiling. Asian J Androl 2007; 9: 522 527 [DOI] [PubMed] [Google Scholar]
- Turner TT, Johnston DS, Finger JN, Jelinsky SA. Differential gene expression among the proximal segments of the rat epididymis is lost after efferent duct ligation. Biol Reprod 2007; 77: 165 171 [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 2005; 73: 404 413 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshioka S, Reardon SN, Rucker EB, III, Spencer TE, DeMayo FJ, Lydon JP, MacLean JA., II WNTs in the neonatal mouse uterus: potential regulation of endometrial gland development. Biol Reprod 2011; 84: 308 319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 2008; 423: 194 200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JS, Wilkinson MF. An androgen-regulated homeobox gene expressed in rat testis and epididymis. Biol Reprod 1996; 55: 975 983 [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod 2007; 76: 561 570 [DOI] [PubMed] [Google Scholar]
- Findlay GD, Swanson WJ. Proteomics enhances evolutionary and functional analysis of reproductive proteins. Bioessays 2010; 32: 26 36 [DOI] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ. Selection in the rapid evolution of gamete recognition proteins in marine invertebrates. Cold Spring Harb Perspect Biol 2010; 3: a002931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW, Brennan FE, Hampikian GK, Goodfellow PN, Sinclair AH, Lovell-Badge R, Selwood L, Renfree MB, Cooper DW, Graves JA. Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 1992; 359: 531 533 [DOI] [PubMed] [Google Scholar]
- Jones RC. Evolution of the vertebrate epididymis. J Reprod Fertil Suppl 1998; 53: 163 181 [PubMed] [Google Scholar]
- Dube E, Chan PT, Hermo L, Cyr DG. Gene expression profiling and its relevance to the blood-epididymal barrier in the human epididymis. Biol Reprod 2007; 76: 1034 1044 [DOI] [PubMed] [Google Scholar]
- Guyonnet B, Marot G, Dacheux JL, Mercat MJ, Schwob S, Jaffrezic F, Gatti JL. The adult boar testicular and epididymal transcriptomes. BMC Genomics 2009; 10: 369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimon V, Koukoui O, Calvo E, Sullivan R. Region-specific gene expression profiling along the human epididymis. Mol Hum Reprod 2007; 13: 691 704 [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belghazi M, Lanson Y, Dacheux F. Human epididymal secretome and proteome. Mol Cell Endocrinol 2006; 250: 36 42 [DOI] [PubMed] [Google Scholar]
- Guyonnet B, Dacheux F, Dacheux JL, Gatti JL. The epididymal transcriptome and proteome provide some insights into new epididymal regulations. J Androl 2011; 32: 651 664 [DOI] [PubMed] [Google Scholar]
- Lee WK, Kim YM, Malik N, Ma C, Westphal H. Cloning and characterization of the 5′-flanking region of the Ehox gene. Biochem Biophys Res Commun 2006; 341: 225 231 [DOI] [PubMed] [Google Scholar]
- Snyder EM, Small CL, Bomgardner D, Xu B, Evanoff R, Griswold MD, Hinton BT. Gene expression in the efferent ducts, epididymis, and vas deferens during embryonic development of the mouse. Dev Dyn 2010; 239: 2479 2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan V, Bomgardner D, Tremblay JJ. Expression of the Ladybird-like homeobox 2 transcription factor in the developing mouse testis and epididymis. BMC Dev Biol 2008; 8: 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomgardner D, Hinton BT, Turner TT. 5′ hox genes and meis 1, a hox-DNA binding cofactor, are expressed in the adult mouse epididymis. Biol Reprod 2003; 68: 644 650 [DOI] [PubMed] [Google Scholar]
- Duboule D. The Guidebook to the Homeobox Genes. New York: Oxford University Press; 1994. [Google Scholar]
- Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity 2006; 97: 88 96 [DOI] [PubMed] [Google Scholar]
- Hu Z, MacLean JA, Bhardwaj A, Wilkinson MF. Regulation and function of the Rhox5 homeobox gene. Ann N Y Acad Sci 2007; 1120: 72 83 [DOI] [PubMed] [Google Scholar]
- Hu Z, Shanker S, MacLean JA, II, Ackerman SL, Wilkinson MF. The RHOX5 homeodomain protein mediates transcriptional repression of the netrin-1 receptor gene Unc5c. J Biol Chem 2008; 283: 3866 3876 [DOI] [PubMed] [Google Scholar]
- Rabionet M, van der Spoel AC, Chuang CC, von Tumpling-Radosta B, Litjens M, Bouwmeester D, Hellbusch CC, Korner C, Wiegandt H, Gorgas K, Platt FM, Grone HJ. et al. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J Biol Chem 2008; 283: 13357 13369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Song HW, Beildeck M, Kerkhofs S, Castoro R, Shanker S, De Gendt K, Suzuki K, Claessens F, Issa JP, Orgebin-Crist MC, Wilkinson MF. DNA demethylation-dependent AR recruitment and GATA factors drive Rhox5 homeobox gene transcription in the epididymis. Mol Endocrinol 2012; 26: 538 549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Dandekar D, O'Shaughnessy PJ, De Gendt K, Verhoeven G, Wilkinson MF. Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Mol Endocrinol 2010; 24: 60 75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layalle S, Volovitch M, Mugat B, Bonneaud N, Parmentier ML, Prochiantz A, Joliot A, Maschat F. Engrailed homeoprotein acts as a signaling molecule in the developing fly. Development 2011; 138: 2315 2323 [DOI] [PubMed] [Google Scholar]
- Maizel A, Tassetto M, Filhol O, Cochet C, Prochiantz A, Joliot A. Engrailed homeoprotein secretion is a regulated process. Development 2002; 129: 3545 3553 [DOI] [PubMed] [Google Scholar]
- Prabagaran E, Bandivdekar AH, Dighe V, Raghavan VP. HOXBES2: a novel epididymal HOXB2 homeoprotein and its domain-specific association with spermatozoa. Biol Reprod 2007; 76: 314 326 [DOI] [PubMed] [Google Scholar]
- MacLean JA, Bettegowda A, Kim BJ, Lou CH, Yang SM, Bhardwaj A, Shanker S, Hu Z, Fan Y, Eckardt S, McLaughlin KJ, Skoultchi AI, et al. The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol Cell Biol 2011; 31: 1275 1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabory E, Damon C, Lenoir A, Henry-Berger J, Vernet P, Cadet R, Saez F, Drevet JR. Mammalian glutathione peroxidases control acquisition and maintenance of spermatozoa integrity. J Anim Sci 2009; 88: 1321 1331 [DOI] [PubMed] [Google Scholar]
- Thimon V, Calvo E, Koukoui O, Legare C, Sullivan R. Effects of vasectomy on gene expression profiling along the human epididymis. Biol Reprod 2008; 79: 262 273 [DOI] [PubMed] [Google Scholar]