The immune system has adopted a remarkable series of genetic tricks to cope with pathogens, extant and possible. For example, diverse sets of Ig heavy- and light-chain genes are generated by specific DNA recombinations in the developing B lymphocytes present in bone marrow. This process of V(D)J recombination can generate >107 distinct antibody V regions from relatively few (≈102) genetic building blocks and is driven by the punctuated expression of the recombination activating genes Rag1 and Rag2 during B cell genesis (1). Maturing B cells leave the bone marrow and travel to peripheral lymphoid follicles, where they become competent to respond to an antigen. The antigen activates B cells by binding to membrane Ig, inducing migration from follicles into adjacent zones of T lymphocytes and a concerted up-regulation of surface molecules that mediate T and B cell collaboration (2). In the T cell zone, activated antigen-specific B cells provide and receive survival and proliferation signals; the progeny of these cells subsequently specialize to become antibody-secreting plasmacytes or return to the follicle and initiate the germinal center (GC) reaction (2, 3). GCs support another trick for generating Ig diversity: V(D)J hypermutation. B cells in GCs accumulate high frequencies of point mutations (and, less commonly, deletions and insertions); the rate of mutation in the Ig V region is thought to be ≈1 mutation/103 bp/cell division, a rate 106-fold above that for other gene loci. Ig hypermutation usually is restricted to GC B cells and exhibits a distinctive pattern of nucleotide misincorporations by favoring transition mutations and strand polarity (1, 4). The small, GC B cell population undergoes repeated rounds of hypermutation, selection, and proliferation. In this darwinian microcosm, mutations that better the ability of the B cell to bind antigen are selected; mutations that diminish binding are thought to result in programmed cell death (1, 4).
Despite intensive searches, genes that direct V(D)J hypermutation in the way that Rag1 and Rag2 drive V(D)J recombination have not been found. This failure has lead to the hunch that normally expressed gene products have been coopted into the hypermutation mechanism. Most recently, the error-prone DNA polymerase β and components of several DNA repair pathways have fallen under suspicion. In this issue of Proceedings, Winter et al. (5) examine V(D)J hypermutation in mice deficient in the nucleotide excision repair gene Xpa or the mismatch repair (MMR) factor PMS2. Both nucleotide excision repair and MMR have been proposed as components of the hypermutation mechanism, but Winter and his colleagues find only slight reductions in the frequency of V(D)J mutations in Xpa−/− and PMS2−/− mice. Normally, these negative findings might not attract notice, but a recent report in Science by Cascalho et al. (6) claimed PMS2 to be integral to V(D)J hypermutation. Cascalho et al. observed 6- to 22-fold reductions in V(D)J mutation frequencies whereas Winter and colleagues find only a 2-fold suppression. Other laboratories (ref. 7; U. Storb, personal communication) also have observed little or no reduction of V(D)J mutations in mice deficient for MMR. Remarkably, Winter, Cascalho, and Storb used the same PMS2−/− stock (8) although important experimental and analytical differences exist between the various groups. Discrepant findings between groups of very competent workers suggests caution is in order in the interpretation of PMS2’s role in hypermutation.
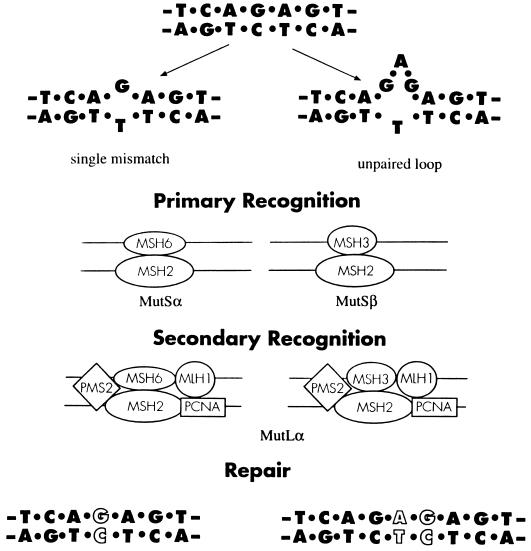
Mammalian MMR proceeds via two recognition steps: a primary identification and binding to mismatched DNA by MutS homologues and the subsequent recruitment of an adapter complex, the MutL homologue, to direct repair (Fig. 1). Mice deficient in MSH2 or PMS2 exhibit impaired MMR as evidenced by the instability of microsatellite DNA and are to lesser and greater degrees cancer-prone. However, PMS2−/− male mice are sterile, in contrast to the fully fertile MSH2−/− animals (8, 9), which suggests that the components of MMR do have additional and independent functions. Cascalho and her colleagues (6) have suggested that PMS2 may reverse its usual correction function in GC B cells, perhaps by losing the ability to discriminate the newly synthesized DNA strand from its parental template. Mutations introduced during DNA replication might then be inserted, and fixed, into the parental strand. This proposal is both clever and attractive because it explains the paradoxical reduction of V(D)J mutations in the absence of MMR and provides a plausible mechanism for hypermutation.
Figure 1.
A general outline for mammalian MMR. MMR begins with the binding of MutSα or MutSβ heterodimers to the distorted segment of mispaired DNA. MutSα and MutSβ are composed of MSH2 and MSH6 or MSH3, respectively. Primary recognition by the MutSα complex is best for 1–2 base mismatches whereas unpaired loops of 3–4 bases preferentially recruit MutSβ. The MutLα dimer of PMS2 and MLH1 then joins the MutS-DNA complex for secondary recognition and eventual repair and resolution. In the absence of PMS2, MMR is therefore generally deficient. The proliferating cell nuclear antigen (PCNA) associated with MLH1 may catalyze MMR by stimulating nuclease activity and facilitating removal of the mismatch. (Adapted from ref. 9.)
However, alternative explanations, including diminished cellular viability in response to generalized failure to repair DNA mismatches, have not been excluded rigorously. For example, should replication slippage (10) occur during B cell proliferation in GCs, only insertions that maintain reading frame and do not debilitate the Ig receptor could be tolerated in the absence of PMS2. A recent study (11) emphasizes the presence of short, in-frame insertions or deletions in the mutated Ig genes of human GC and memory B cells. Many of these involved local sequence motifs that could form loop intermediates and nucleate replication slippage (10, 11). Thus, efficient MMR may allow B cells to persist in GCs and thereby accumulate higher frequencies of point mutations.
Alternative explanations might also be found in the distinctive animals studied by Cascalho et al. (6). She and her colleagues in the laboratory of Matthias Wabl (University of California, San Francisco) have created a mouse line that largely is prohibited from using V(D)J recombination to generate antibody diversity. These “quasimonoclonal” (QM) mice carry a single functional Ig heavy chain gene, disrupted κ light-chain alleles, and wild-type λ light-chain loci (17.2.25/H−, κ−/κ−, λ+/λ+) (12). Not surprisingly, these impoverished mice rely on V(D)J hypermutation (and the normally rare mechanism of V gene replacement) to create a diverse repertoire of antibody specificities. Indeed, selection for additional diversity appears to be so strong that mutated Ig genes, rare in normal mice, are recovered readily from the blood of unimmunized QM mice (12). It may be that diversification creates niches for clonal expansion within these animals that are occupied normally by B cells diversified by V(D)J rearrangement. The QM and PMS2−/− lines were crossed to generate mice deficient in both V(D)J diversity and MMR. For mutational analysis, blood cells expressing the B220 B-cell antigen or the transgenic Ig were purified by flow cytometry. In contrast, all other studies relied on the recovery of GC B cells from the spleens of immunized PMS2−/− mice or Peyer’s patches (lymph node-like structures that contain GCs induced by gut flora).
V(D)J mutation most often is studied in GC and memory B cells because the great majority of peripheral B lymphocytes have not been activated by an antigen and have not entered the GC reaction. In GCs, B cells acquire a characteristic phenotype, including binding of peanut agglutinin, a plant lectin (4). Thus, Winter et al. (5) analyzed peanut agglutinin-binding GC B cells from the spleens of immunized PMS2−/− and Xpa−/− mice. How do these cells compare with those studied by Cascalho et al (6)? Why hasn’t the Wabl group chosen to immunize QM or QM/PMS2−/− mice? These are very real experimental differences that merit hasty resolution. It is possible that V(D)J hypermutation in unimmunized QM mice uses a mechanism unlike that operating in GCs. D. Chaplin and collaborators at Washington University have demonstrated Ig mutation and cellular selection in lymphotoxin α-deficient mice that do not support GC formation (14).
At least two processes obscure the mechanism of V(D)J hypermutation. Cellular selection may restrict the frequencies of debilitating mutations such as missense insertions or deletions. This limitation has been removed by the construction of unselectable Ig transgenes capable of hypermutation (13). A second veil of DNA repair is now being pulled aside in studies like those of Winter (5) and Cascalho (6). Even if these repair machineries are not responsible for Ig hypermutation, the mutator now can be seen more clearly.
ABBREVIATIONS
- GC
germinal center
- MMR
mismatch repair
- QM
quasimonoclonal
Footnotes
The companion to this commentary is published on pages 6953–6958.
References
- 1.Rajewsky K. Nature (London) 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.MacLennan I C M, Gulbranson-Judge A, Toellner K-M, Casamayor-Palleja M, Chan E, Sze D M-Y, Luther S A, Orbea H A. Immunol Rev. 1997;156:53–66. doi: 10.1111/j.1600-065x.1997.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 3.Jacob J, Kelsoe G. J Exp Med. 1992;176:679–688. doi: 10.1084/jem.176.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelsoe G. Immunity. 1996;4:107–111. doi: 10.1016/s1074-7613(00)80675-5. [DOI] [PubMed] [Google Scholar]
- 5.Winter D B, Phung Q H, Umar A, Baker S M, Tarone R E, Tanaka K, Liskay R M, Kunkel T A, Bohr V A, Gearhart P J. Proc Natl Acad Sci USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascalho M, Wong J, Steinberg C, Wabel M. Science. 1998;272:1207–1210. doi: 10.1126/science.279.5354.1207. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs, H., Fukita, Y., vander Horst, G. T., de Boer, J., Weeda, G., Esser, J., de Wind, N., Engleward, B. P., Samson, L., Verbeek, S., et al. (1998) J. Exp. Med., in press. [DOI] [PMC free article] [PubMed]
- 8.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliot E A, Yu J, Ashley T, Arnheim N, et al. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl T, Karran P, Wood R D. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 10.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Wilson P C, deBouteiller O, Liuy Y-J, Potter K, Banchereau J, Capra J D, Pascual V. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascalho M, Ma A, Lee S, Masat L, Wabl M. Science. 1996;272:1649–1652. doi: 10.1126/science.272.5268.1649. [DOI] [PubMed] [Google Scholar]
- 13.Wagner S D, Neuberger M S. Immunol Rev. 1996;14:441–457. doi: 10.1146/annurev.immunol.14.1.441. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto M, Lo S F, Carruthers C J L, Min J, Mariathasan S, Huang G, Plas D R, Martin S M, Geha R S, Nahm M H, et al. Nature (London) 1996;382:462–466. doi: 10.1038/382462a0. [DOI] [PubMed] [Google Scholar]