In neutrophils, Arf1 is activated upon GPCR stimulation. GBF1, a GEF for Arf, is primarily responsible for Arf1 activation upon GPCR stimulation and is important for chemotaxis and superoxide production. GBF1 also binds to products of PI3Kγ . The results indicate a novel mechanism that links PI3Kγ with chemotaxis and superoxide production.
Abstract
Most chemoattractants for neutrophils bind to the Gαi family of heterotrimeric G protein–coupled receptors (GPCRs) and release Gβγ subunits to activate chemotaxis and superoxide production. GIT2, a GTPase-activating protein for Arf1, forms a complex with Gβγ and is integral for directional sensing and suppression of superoxide production. Here we show that GBF1, a guanine nucleotide exchanging factor for Arf-GTPases, is primarily responsible for Arf1 activation upon GPCR stimulation and is important for neutrophil chemotaxis and superoxide production. We find that GBF1 bears a novel module, namely binding to products of phosphatidyl inositol 3-kinase (PI3K), which binds to products of PI3Kγ. Through this binding, GBF1 is translocated from the Golgi to the leading edge upon GPCR stimulation to activate Arf1 and recruit p22phox and GIT2 to the leading edge. Moreover, GBF1-mediated Arf1 activation is necessary to unify cell polarity during chemotaxis. Our results identify a novel mechanism that links PI3Kγ activity with chemotaxis and superoxide production in GPCR signaling.
INTRODUCTION
Chemotactic directional sensing and production of a number of reactive oxygen species (ROS), coupled with phagocytic activity, are essential for neutrophils for their function to kill invaders, in which most chemoattractants, including bacterial products, complement fragments, and chemokines, bind to cell surface G protein–coupled receptors (GPCRs). Gβγ subunits are released from heterotrimeric G proteins upon GPCR stimulation and constitute a directional sensing machinery by binding to p21-activating protein kinase 1 (PAK1), which simultaneously binds to αPIX, a Dbl-family guanine nucleotide exchanging factor (GEF) for Rac and Cdc42, thus forming a linear complex of Gβγ-PAK1-αPIX (Li et al., 2003). PAK1 and αPIX, as well as Rac and Cdc42, influence actin cytoskeletal remodeling, necessary for the formation of a polarized cell structure and the leading edge upon GPCR stimulation (Li et al., 2003; Niggli, 2003). On the other hand, ROS produced upon GPCR stimulation are first produced as superoxide anions by activity of the phagocyte NADPH oxidase (Cross and Segal, 2004). This enzyme is activated by recruitment and assembly of the cytoplasmic subunits at the plasma membrane (Cross and Segal, 2004; Mazaki et al., 2006). Phosphatidyl inositol 3-kinase (PI3K) consists of several subunits, including the p110 catalytic subunit, and neutrophils express the γ isoform of this subunit (Wymann and Pirola, 1998). Gβγ subunits are also known to activate p110γ by direct interaction (Wymann and Pirola, 1998), whereas disruption of the p110γ gene blocks GPCR-induced ROS production in mouse bone marrow neutrophils (Hirsch et al., 2000; Li et al., 2000; Sasaki et al., 2000). p110γ−/− neutrophils also show reduction of directional sensing and persistence during GPCR-mediated chemotaxis, in which membrane protrusions are formed rather randomly all over the cell periphery (Hannigan et al., 2002). The mechanisms by which PI3Kγ activity is involved in these properties of GPCR-mediated chemotaxis, as well as in ROS production, remain largely unclear.
Besides actin cytoskeletal remodeling, intracellular vesicle trafficking also plays crucial roles in cell migration and directional sensing (Bretscher, 1996; Mazaki et al., 2006). Arf-family small GTPases are essential for intracellular vesicle trafficking and membrane remodeling (D'souza-Schorey and Chavrier, 2006). There are six members of Arf-GTPases in mammals, Arf1–6, whereas Arf2 is missing in humans. These Arf-GTPases are subclassified into three classes, depending on their structural similarities: Arf1–3 belong to class I; Arf4, 5 belong to class II; and Arf6 belongs to class III. Class I Arfs mainly reside in the Golgi and are primarily involved in Golgi-originated trafficking by regulating the assembly of coat complexes on the budding vesicles (Roth, 1999). Class I Arfs may also localize to the plasma membrane ruffles to be involved in cell migration (Boulay et al., 2008). On the other hand, the class III Arf, namely Arf6, is localized to the cell periphery and primarily regulates recycling of plasma membrane components and several cell surface receptors (Peters et al., 1995; D'souza-Schorey and Chavrier, 2006). Cellular functions of class II Arfs are still largely unidentified, partly due to the lack of biochemical methods to precisely measure their activities in vivo.
We showed previously that GIT2, an GTPase-activating protein (GAP) for Arfs, is an integral component of the Gβγ-directional sensing machinery in bone marrow–derived neutrophils by forming a Gβγ-PAK1-αPIX-GIT2 linear complex (Mazaki et al., 2006). Biochemical assays indicated that GIT2 is engaged in the suppression of Arf1, but not Arf6, activity in neutrophils (Mazaki et al., 2006), whereas GIT2 exhibits GAP activity against different Arf isoforms in vitro (Vitale et al., 2000). Even in the absence of GIT2, neutrophils can be well polarized and form leading edges upon GPCR stimulation, and cell migration speed, as well as adhesion to fibrinogen, is not notably affected (Mazaki et al., 2006). In the absence of GIT2, however, neutrophils substantially, although not completely, lose their directional sensing, in which components of the Gβ-mediated directional sensing machinery are not well accumulated to the leading edge, and leading edges are largely misoriented against chemoattractants (Mazaki et al., 2006). In the absence of GIT2, moreover, leaky ROS production is observed without GPCR stimulation, and aberrantly high levels of ROS are produced upon GPCR stimulation (Mazaki et al., 2006). Consistently, recruitment of p22phox—a subunit of the phagocyte NADPH oxidase—to the leading edge is not notably affected by the loss of GIT2, whereas the direction of p22phox recruitment and ROS production is largely misoriented (Mazaki et al., 2006). Thus GIT2 appears to be necessary for directional sensing and also for the suppressive control of Arf1 activity and superoxide production. Such suppressive function of GIT2 is likely to be necessary not only to inhibit the leaky production of ROS, but also for moderate, well-controlled production of ROS upon GPCR stimulation.
Following our previous results, we initiated this study to identify GEF(s) primarily responsible for Arf1 activation upon GPCR stimulation in neutrophils. To further verify our previous model indicating the importance of Arf1 activity in neutrophil functions, we were also interested in whether such an ArfGEF(s) activating Arf1 is also pivotal for chemotaxis and superoxide production. Here, we identified GBF1, an ArfGEF previously shown to be localized to the Golgi (Claude et al., 1999), to be primarily responsible for the activation of Arf1, but not Arf6, upon GPCR stimulation in differentiated HL-60 cells and to be necessary for directional sensing and ROS production of neutrophils. Crucial functions of GBF1 in GPCR signaling were found to be mediated via its novel domain, which binds to products of PI3Kγ activity. Our results describe an aspect of the mechanisms by which PI3Kγ plays a crucial role in superoxide production and also in directional sensing during GPCR-stimulated chemotaxis of neutrophils.
RESULTS
Requirement for GBF1 in GPCR-mediated chemotaxis
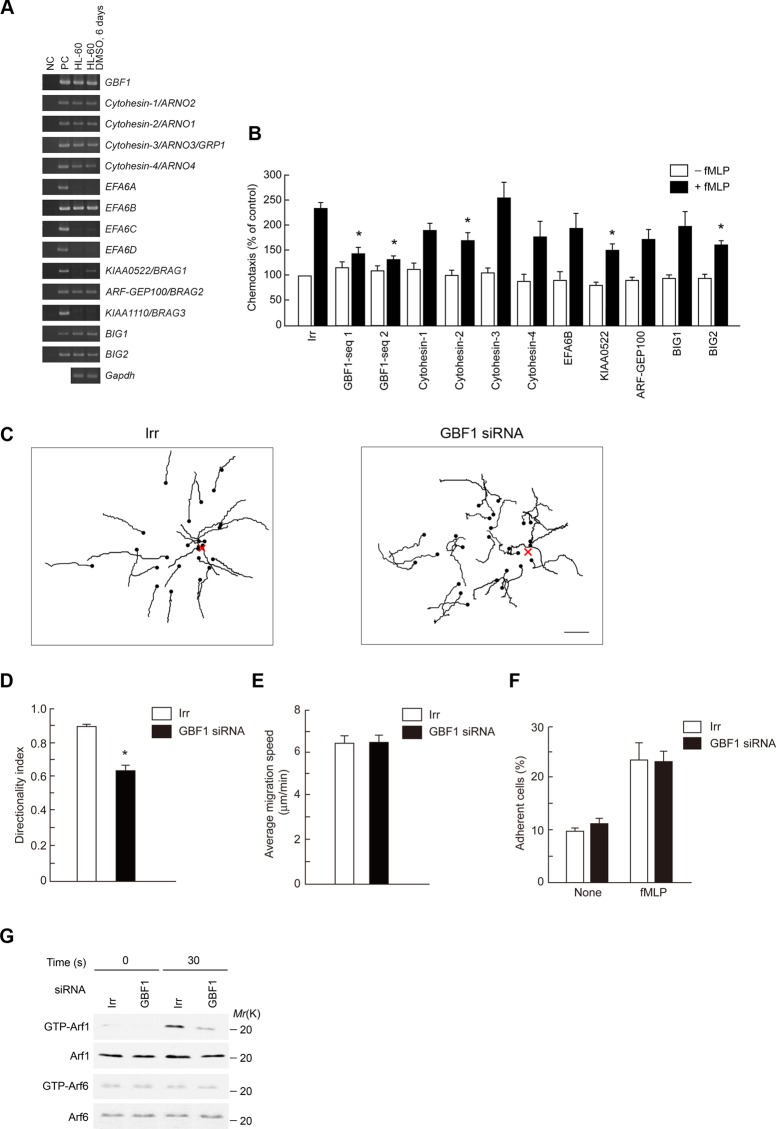
We used HL-60 cells, differentiated into a granulocytic lineage by dimethyl sulfoxide (DMSO) treatment (Matzner et al., 1987), as a model for neutrophils, since neutrophil primary cultures are difficult to maintain in vitro long enough to be manipulated such as by nucleotide transfections. ArfGEFs consist of 15 members each encoding the Sec7 domain, which is a putative ArfGEF domain (Gillingham and Munro, 2007). We found that differentiated HL-60 cells express 10 ArfGEFs (Figure 1A). In this analysis, we excluded Fbx8, which is an E3 ligase for Arf6 (Yano et al., 2008). We then suppressed the expression of each of them by the small interfering RNA (siRNA) method (Supplemental Figure S1, A and B) and measured their Transwell chemotactic migration activities toward an N-formyl-Met-Leu-Phe peptide (f MLP). We found that knockdown of GBF1, cytohesin-2/ARNO1, KIAA0522/BRAG1, and BIG2 each significantly affects f MLP-induced chemotactic activities without notably affecting the basal motile activities measured in the absence of f MLP (Figure 1B). Among these ArfGEFs, knockdown of GBF1 seemed to be most effective in inhibiting chemotaxis. Directional sensing, measured by two-dimensional chemotaxis toward a source of f MLP, was also largely impaired by GBF1 siRNA treatment (Figure 1, C and D, and Supplemental Movies S1 and S2), whereas cell migratory speeds, as well as f MLP-induced adhesion to fibrinogen (Miura et al., 2000), was not notably affected (Figure 1, E and F). These results indicate that GBF1 is pivotal for directional sensing but not for migration speed and substrate adhesion in GPCR-stimulated chemotaxis of neutrophils.
FIGURE 1:
GBF1 is crucial for f MLP-induced chemotaxis and is primarily responsible for Arf1 activation in differentiated HL-60 cells. (A) Expression of ArfGEF mRNA was analyzed by using RT-PCR, coupled with agarose gel electrophoresis. cDNAs (10 pg) corresponding to each indicated ArfGEF were used as a positive control (PC). NC, without template cDNAs. (B–G) Cells transfected with siRNA duplexes against each indicated ArfGEF or with irrelevant sequences (Irr) were subjected to Transwell chemotactic migration by placing cells on the upper chamber, which the lower chambers without or filled with f MLP, and incubating for 120 min (B); two-dimensional migration assay toward a point source of f MLP (indicated by X), being supplied by use of a micropipette, for 20 min (C–E); adhesion assay to fibrinogen in the absence or presence of f MLP for 60 min (F); and measurement of Arf1 activity after incubation with or without f MLP for 30 s (G). Also shown in the two-dimensional migration are trajectories of each cell migration, in which dots indicate positions at the end of the migration (C), directionality index (D), and average migration speed (E). (F) Percentages of adherent cells. (G) Activities of Arf1 and Arf6 were measured by GST-GGA pull-down coupled with anti-Arf1 or anti-Arf6 immunoblot, as indicated. Bottom, immunoblots of the total cell lysates (5 μg) by the indicated antibodies. Data are representative of three independent experiments. Error bars, SEM (B, D–F). *p < 0.01 against the irrelevant control (n = 6 wells [B, F]) and 25 cells [D, E]). Bar, 50 μm (C).
GBF1 is responsible for activation of Arf1 in GPCR signaling
f MLP stimulation activates Arf1, but not Arf6, in bone marrow–derived neutrophils (Mazaki et al., 2006), which is also true for differentiated HL-60 cells (Figure 1G). GBF1 activates class I and II Arfs, but not class III Arf, in vitro (Claude et al., 1999; Niu et al., 2005). We found that GBF1 siRNA treatment largely abolishes the f MLP-induced activation of Arf1 without affecting Arf6 activity (Figure 1G). Therefore GBF1 appears to be engaged in the activation of Arf1, but not Arf6, in neutrophils upon GPCR stimulation.
PI3Kγ activity is involved in activation of Arf1 and translocation of GBF1 to the leading edge in GPCR signaling
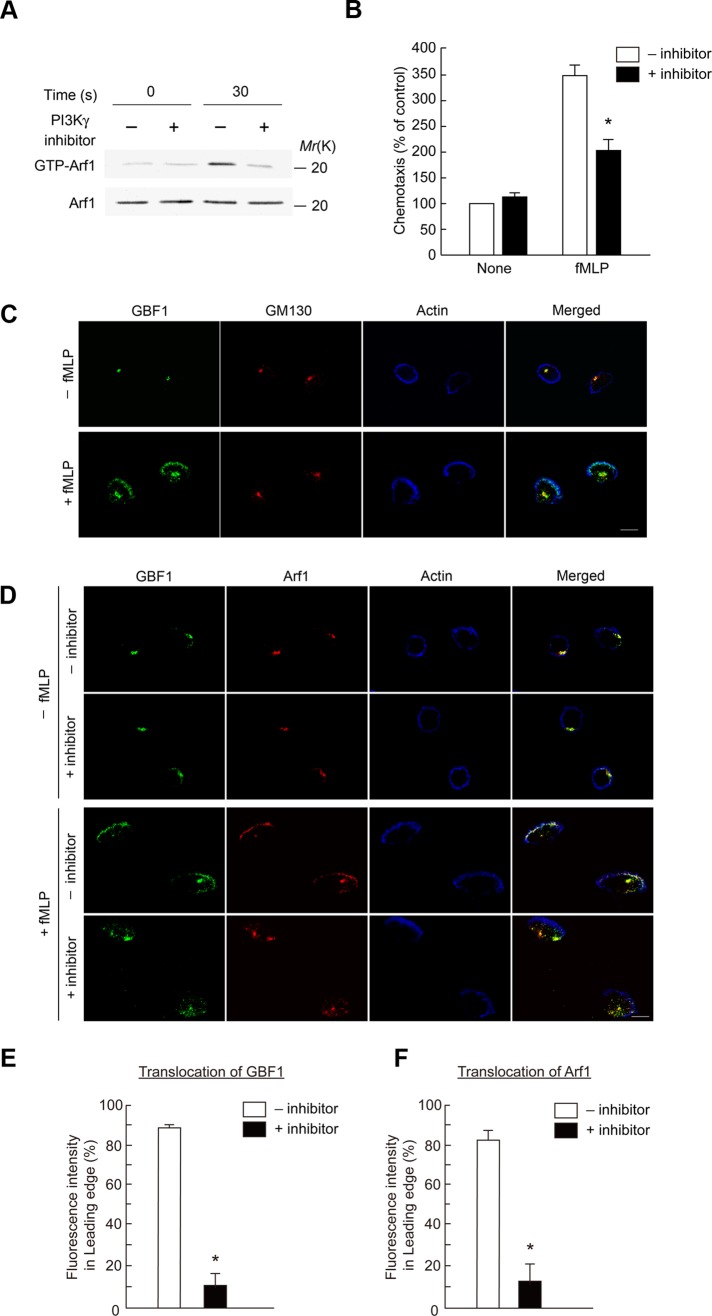
We next addressed the mechanism by which GPCR signaling uses GBF1 to activate Arf1. Gβγ subunits interact with several proteins, such as PAK1 and PI3Kγ, to activate them (Wymann and Pirola, 1998; Bokoch, 2003). We first tested whether Gβγ also binds to GBF1, by expressing a glutathione S-transferase (GST) fusion protein of GBF1 in 293T cells, together with Gβ1 and Gγ2, and found that Gβ1/γ2 is not coprecipitated with GST-GBF1 (unpublished data). PI3Kγ is activated upon GPCR stimulation, which then activates several other enzymes, such as Akt and Erk1/2 (Hirsch et al., 2000; Li et al., 2000; Sasaki et al., 2000). We then examined whether the PI3Kγ activity is involved in the function of GBF1 and first found that a specific inhibitor of PI3Kγ activity, 5-(2,2-difluoro-benzo[1,3]dioxol-5-ylmethylene)-thiazolidine-2,4-dione (AS-604850; Camps et al., 2005), clearly blocks f MLP-induced activation of Arf1 (Figure 2A). This inhibitor also blocked f MLP-induced chemotaxis (Figure 2B), as reported previously with peritoneal neutrophils (Camps et al., 2005). Furthermore, f MLP-induced activation of the Arf1 activity was blocked by siRNA-mediated knockdown of PI3Kγ (Supplemental Figure S2).
FIGURE 2:
Requirement of PI3Kγ activity in Arf1 activation and translocation of GBF1. (A, B) PI3Kγ inhibitor blocks f MLP-induced Arf1 activation (A) and chemotaxis (B) of differentiated HL-60 cells. Cells treated with or without AS-604850 for 30 min at 37°C were subjected to the measurement of Arf1 activity after incubation with or without f MLP for 30 s (A) and to the Transwell chemotactic migration toward f MLP for 120 min (B). (A, bottom) Anti-Arf1 immunoblot of the total cell lysates (5 μg). (C–F) Differentiated HL-60 cells were incubated with or without f MLP for 15 min and subjected to immunostaining analysis using antibodies as indicated. (D–F) Cells pretreated with or without AS-604850 for 30 min at 37°C (marked as +inhibitor and –inhibitor, respectively) before the addition of f MLP. F-actin was visualized by phalloidin conjugated with Alexa Fluor 647. Percentages of GBF1 and Arf1 molecules translocated to the leading edge in f MLP-stimulated cells, treated with or without AS-604850, were calculated by dividing the immunofluorescence intensity of GBF1 molecules at the leading edge that were colocalized with F-actin by that of the whole cell (E, F). In A, C, and D, data are representative of three independent experiments. Error bars, SEM (B, E, F). Asterisks represent statistical difference from control cells (p < 0.01; n = 6 wells [B] and >25 cells [E, F]). Bars, 10 μm (C, D).
GBF1 has been shown to be localized to the Golgi and hence colocalized with GM130, a Golgi marker protein, in normal rat kidney cells (Kawamoto et al., 2002). We found that GBF1 is colocalized with GM130 also in differentiated HL-60 cells (Figure 2C and Supplemental Figure S3A). We then found that a significant fraction of GBF1 molecules are translocated to the leading edge upon f MLP stimulation, which is formed upon polarization of cells (Figure 2C and Supplemental Figure S3A). Phosphatidylinositol-3,4,5-trisphosphate (PI[3,4,5]P3) is primarily observed at the leading edges in GPCR-stimulated cells, including differentiated HL-60 cells stimulated with f MLP (Sun et al., 2004; Supplemental Figure S3B). AS-604850 blocked translocation of GBF1 to the leading edge upon f MLP stimulation, whereas it did not block the cell polarization and formation of the leading edge (Figure 2, D and E). Translocation of GBF1 to the leading edge was also blocked by the PI3Kγ knockdown (Supplemental Figure S3C). These results, together with results described earlier, indicate that PI3Kγ activity is involved in translocation of GBF1 to the leading edge and also the activation of Arf1 in GPCR signaling but not the cell polarization or the formation of leading edges.
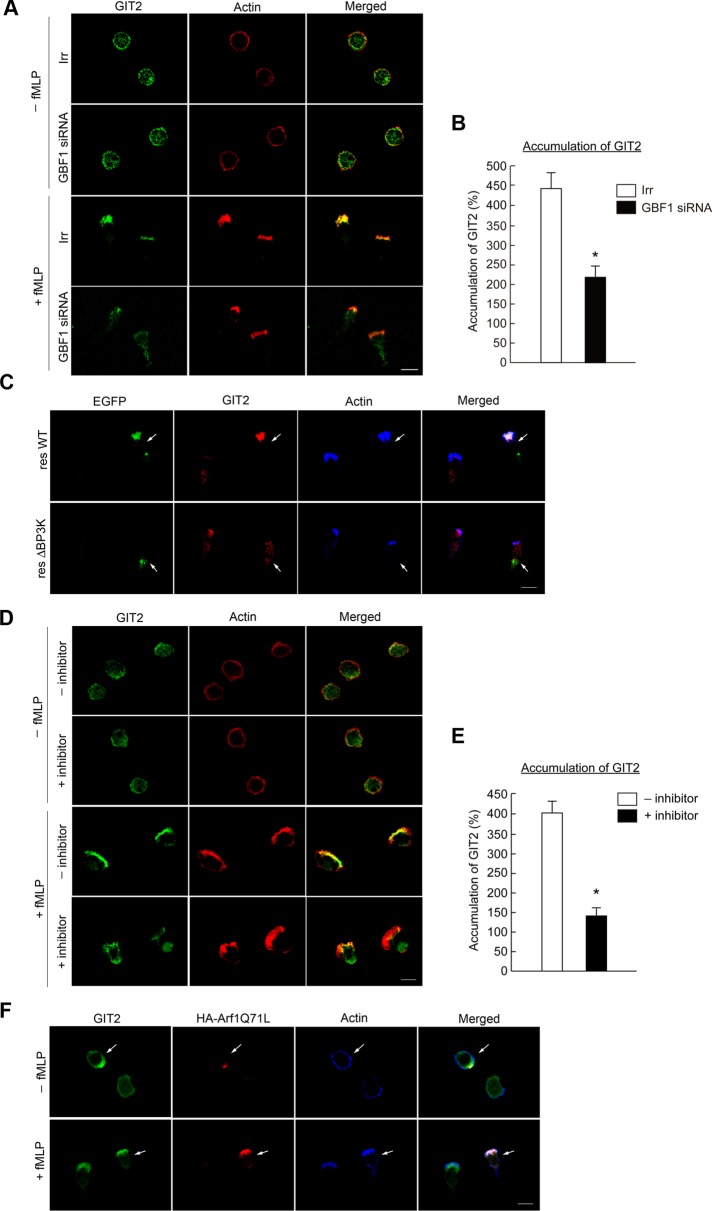
GBF1, via its novel domain, binds to products of PI3Kγ activity to translocate to the leading edge to activate Arf1 in GPCR signaling
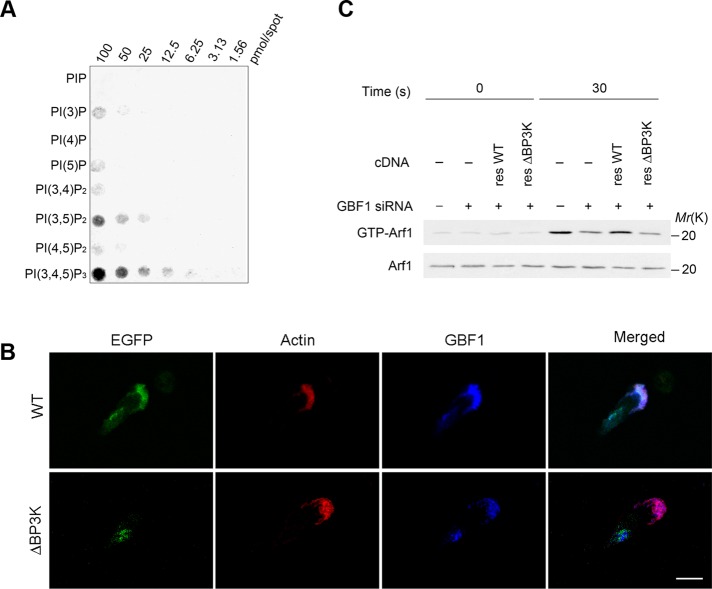
ArfGEFs of the cytohesin family, the EFA6 family, and the BRAG family, each has one pleckstrin homologue (PH) domain to the C-terminus of the Sec7 domain and thereby interact with phosphatidylinositol phosphates (Casanova, 2007). GBF1 has no canonical PH domain, but instead has homology downstream of Sec7 domain1 and domain2 (HDS1 and HDS2) in tandem to the C-terminus of the Sec7 domain, which were both predicted in silico to bear high contents of α-helices (Mouratou et al., 2005). We then examined whether the HDS1 and HDS2 domains binds to phosphatidylinositol phosphates. We found, however, that all of the GST-fusion proteins produced in Escherichia coli, each containing HDS1, HDS2, or both, do not show appreciable binding to phosphatidylinositol phosphates in a filter binding assay (unpublished data). The C-terminus of the HDS2 domain is followed by a region containing two short sequences, each recognized by the SEG program (Wootton and Federhen, 1996) to be rich in alanines and prolines (we tentatively call this the “SEG” region; see Supplemental Figure S4). We then produced GST-fusion protein containing HDS1, HDS2, and the SEG region in E. coli and found that this GST-fusion protein clearly binds to PI[3,4,5]P3 and also to other phosphoinositides, such as PI[3,5]P2, to lesser extents in a filter binding assay, in which GST itself did not exhibit any detectable binding (Figure 3A and Supplemental Figure S5A). GST-fusion protein containing only the SEG region also does not show such binding (unpublished data). Therefore it is likely that predicted HDS1 and HDS2 domains, together with the SEG region, form a single functional module, which binds to phosphatidylinositol phosphates. We named it BP3K, for binding to products of PI3K. The binding of the BP3K domain to PI[3,4,5]P3 appeared to be almost equivalent to that of the PH domain of cytohesin3 to PI[3,4,5]P3 in a filter binding assay (Supplemental Figure S5B). However, unlike the BP3K domain, the cytohesin3 PH domain did not show notable binding to PI[3,5]P2 (Supplemental Figure S5B), suggesting nonequal binding properties between these two domains. Binding of BP3K domain to PI[3,4,5]P3 was also confirmed in solution (Supplemental Figure S5C).
FIGURE 3:
Requirement of BP3K domain in Arf1 activation and translocation of GBF1. (A) In vitro binding of the GBF1 BP3K domain with phosphatidylinositol phosphates. A nitrocellulose membrane loaded with phosphoinositides, as indicated, was incubated with GST-BP3K, and after washing, bound GST proteins were visualized using an anti-GST antibody. (B) Differentiated HL-60 cells, transfected with wild-type GBF1 cDNA (WT) or GBF1ΔBP3K cDNA (ΔBP3K), each tagged with EGFP, were incubated with f MLP and subjected to anti-GBF1 immunostaining after fixation. Transfected GBF1 proteins were visualized by fluorescence from their EGFP tag. F-actin was visualized by phalloidin conjugated with Texas Red. Bar, 10 μm. (C) Requirement for the BP3K domain of GBF1 in f MLP-induced Arf1 activation of differentiated HL-60 cells. Cells treated with GBF1 siRNA (+) or an irrelevant RNA duplex (–) were transfected with a rescue construct of wild-type GBF1 cDNA (resWT) or its BP3K-deletion mutant (resΔBP3K) and analyzed for their activation of Arf1 in response to f MLP. (C, bottom) Anti-Arf1 immunoblot of the total cell lysates (5 μg). In A–C, data are representative of three independent experiments. Note that the anti-GBF1 polyclonal antibody used was not reactive against EGFP-GBF1ΔBP3K, since this antibody was raised against an amino acid sequence located within the BP3K domain (see Materials and Methods). Hence “GBF1” in B represents only the endogenous GBF1.
We showed earlier that PI[3,4,5]P3 is primarily observed at the leading edges in GPCR-stimulated cells and that PI3K activity is necessary for the translocation of GBF1 to the leading edge upon GPCR signaling. To validate in vivo the possible binding of the BP3K domain to phosphatidylinositol phosphates, we then constructed a mutant form of enhanced green fluorescent protein (EGFP)–tagged GBF1 in which the BP3K domain is deleted (EGFP-GBF1ΔBP3K). We found that this mutant is localized to the Golgi in unstimulated cells but not at all translocated to the leading edge upon their f MLP stimulation (Figure 3B). Endogenous GBF1 within the same cells, as well as EGFP-tagged wild-type GBF1, was translocated from the Golgi to the leading edge upon f MLP stimulation (Figure 3B). To further validate the role of the BP3K domain, we then expressed EGFP-GBF1ΔBP3K in GBF1 siRNA–pretreated cells and found that this mutant does not rescue the function of GBF1 in terms of Arf1 activation in response to f MLP, whereas the wild-type rescue construct does (Figure 3C). These results collectively indicate that the BP3K domain of GBF1 binds to products of the PI3Kγ activity, and this binding is necessary for the translocation of GBF1 to the leading edge and also the activation of Arf1 upon GPCR signaling.
GBF1 translocation is necessary for the recruitment of Arf1 to the leading edge
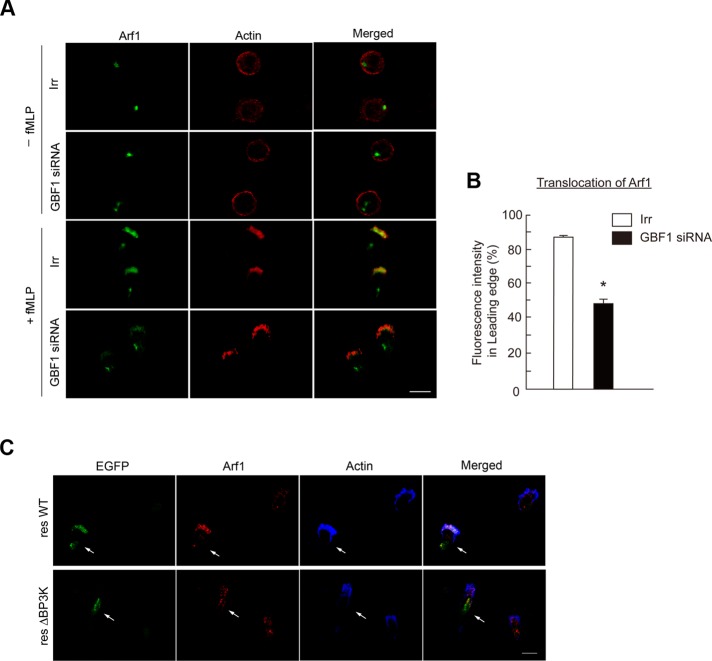
We then found that Arf1 is also translocated to the leading edge upon f MLP stimulation to be colocalized with GBF1, whereas Arf1 seemed to be also largely colocalized with GBF1 at the Golgi in unstimulated cells (Figure 2D and Supplemental Figure S6). This translocation of Arf1 was blocked by AS-604850 (Figure 2, D and F) and also by GBF1 siRNA treatment (Figure 4, A and B). Moreover, GBF1ΔBP3K was unable to rescue the translocation of Arf1 in GBF1 siRNA–treated cells (Figure 4C). On the other hand, f MLP-induced cell polarization and formation of the leading edge were not notably affected by the GBF1 siRNA treatment (Figure 4A). Therefore GBF1, as well as its PI3Kγ-mediated translocation to the leading edge, appears to be necessary for the translocation of Arf1 to the leading edge upon GPCR stimulation.
FIGURE 4:
Requirement for GBF1 in Arf1 translocation to the leading edge. (A, B) Differentiated HL-60 cells transfected with GBF1 siRNA or an irrelevant RNA duplex (Irr) were incubated with or without f MLP as indicated and subjected to anti-Arf1 immunostaining (A), and percentages of Arf1 molecules translocated to the leading edge in f MLP-stimulated cells were calculated (B). F-actin was visualized by phalloidin conjugated with Texas Red (A). (C) Differentiated HL-60 cells transfected with GBF1 siRNA together with the cDNAs of GBF1 resWT or resΔBP3K, each tagged with EGFP, were incubated with f MLP and subjected to anti-Arf1 immunostaining. Transfected GBF1 proteins were visualized by fluorescence from their EGFP tag. F-actin was visualized by phalloidin conjugated with Alexa Fluor 647. In A–C, incubation with f MLP was for 15 min, and data are representative of three independent experiments. Bars, 10 μm (A, C). Error bars, SEM (B); *p < 0.01 against the irrelevant control (n > 25 cells).
GBF1 translocation is necessary for the accumulation of GIT2 at the leading edge
GIT2 also accumulates at leading edges upon GPCR stimulation, whereas a fraction of this protein seems to already exist at the plasma membrane in unstimulated cells (Mazaki et al., 2006). We found that GBF1 siRNA treatment inhibits the efficient accumulation of GIT2 to the leading edge in f MLP-stimulated cells, whereas almost all of the GIT2 molecules accumulated at the leading edge in the control cells upon f MLP stimulation (Figure 5, A and B). Expression of GBF1ΔBP3K in GBF1 siRNA–treated cells was unable to rescue the accumulation of GIT2 at the leading edge (Figure 5C). AS-604850 also blocked the efficient accumulation of GIT2 at the leading edge upon f MLP stimulation (Figure 5, D and E). GIT2 has to bind at least transiently to GTP-Arf1 to exert its GAP activity. We hence tested whether Arf1, when activated by GBF1, has a potential to recruit GIT2, by expressing a GTP hydrolysis–deficient mutant of Arf1 (i.e., the GTP-bound form), Arf1Q71L. We found that GIT2 molecules tend to accumulate to be colocalized with Arf1Q71L in f MLP-unstimulated cells (Figure 5F), whereas GIT2 is distributed rather randomly at the plasma membrane and the cytoplasm in the unstimulated cells not expressing Arf1Q71L (Figure 5, A and F). However, not all GIT2 molecules were colocalized with Arf1Q71L in unstimulated cells, whereas most GIT2 molecules become well colocalized with Arf1Q71L upon f MLP stimulation (Figure 5F). No notable binding was observed between GBF1 and GIT2 (unpublished data). Therefore our results suggest that GBF1, as well as its translocation to the leading edge, is crucial for the efficient accumulation of GIT2 to the leading edge upon GPCR stimulation, whereas this GIT2 accumulation does not seem to be simply mediated by Arf1 upon its activation by GBF1.
FIGURE 5:
Involvement of GBF1 in GIT2 accumulation at the leading edge. (A, B) Differentiated HL-60 cells transfected with GBF1 siRNA or an irrelevant RNA duplex (Irr) were incubated with or without f MLP for 15 min and subjected to anti-GIT2 immunostaining (A), and percentages of GIT2 molecules accumulated at the leading edge in f MLP-stimulated cells were calculated (B). F-actin was visualized by phalloidin conjugated with Texas Red. (C) Differentiated HL-60 cells transfected with GBF1 siRNA together with cDNAs for GBF1 resWT or resΔBP3K, each tagged with EGFP, were incubated with f MLP and subjected to anti-GIT2 immunostaining. Transfected GBF1 proteins were visualized by fluorescence from their EGFP tag. F-actin was visualized by phalloidin conjugated with Alexa Fluor 647. (D, E) Differentiated HL-60 cells pretreated with or without AS-604850 (marked as +inhibitor and –inhibitor, respectively) for 30 min at 37°C were incubated with or without f MLP, as indicated, and subjected to anti-GIT2 immunostaining (D), and percentages of GIT2 molecules accumulated at the leading edge in f MLP-stimulated cells were calculated (E). F-actin was visualized by phalloidin conjugated with Texas Red. (F) Differentiated HL-60 cells transfected with Arf1Q71L-HA cDNA were incubated with or without f MLP and subjected to anti-GIT2 and anti-HA immunostaining. F-actin was visualized by phalloidin conjugated with Alexa Fluor 647. Arrows indicate cells expressing Arf1Q71L-HA. Error bars, SEM (B, E). *p < 0.01 against control cells (n > 25 cells). In A–F, incubation with f MLP was for 15 min. In A, C, D, and F, data are representative of three independent experiments. Bars, 10 μm.
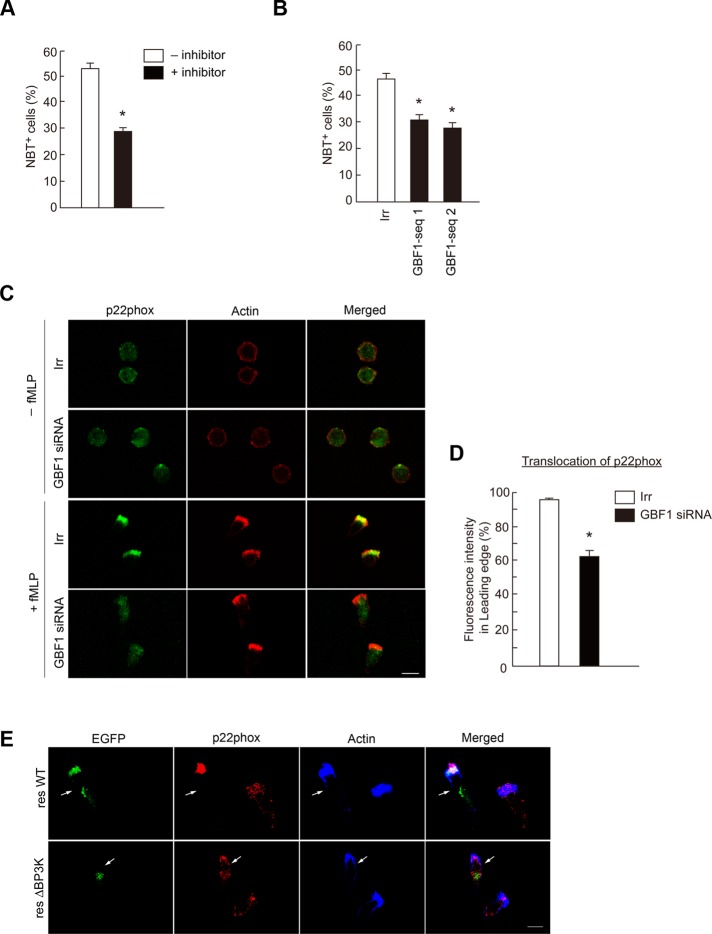
GBF1 translocation is necessary for the recruitment of p22phox to the leading edge
PI3Kγ is essential for GPCR-induced ROS production in bone marrow–derived neutrophils, as mentioned earlier. We confirmed that AS-604850 significantly blocks f MLP-induced ROS production in differentiated HL-60 cells, in which ROS production was assessed as a positive staining with nitroblue tetrazolium (NBT; Filippi et al., 2004; Figure 6A). We next examined the possible involvement of GBF1 in superoxide production and found that GBF1 siRNA treatment also significantly reduces f MLP-induced ROS production (Figure 6B). We then found that GBF1 siRNA treatment inhibits the efficient recruitment of p22phox to the leading edge upon f MLP stimulation (Figure 6, C and D). GBF1ΔBP3K was unable to rescue the translocation of p22phox in the GBF1 siRNA–treated cells (Figure 6E). Therefore our results indicate that GBF1 and its translocation to the leading edge are involved in the GPCR-induced recruitment of the phagocyte NADPH oxidase subunit(s) to the leading edge, whereas many other factors and small GTPases are also involved in this process (Cross and Segal, 2004). On the other hand, expression of a GTP-binding–deficient mutant of Arf1 (i.e., the GDP-bound form), Arf1T31N, did not block the f MLP-induced recruitment of p22phox to the leading edge (Supplemental Figure S7). Therefore activation of Arf1 by GBF1 may not be essential for this process.
FIGURE 6:
Involvement of GBF1 in ROS production. (A, B) Differentiated HL-60 cells treated with or without AS-604850 for 30 min at 37°C (A) or transfected with GBF1 siRNA (B) were incubated with f MLP in the presence of NBT, and percentages of the NBT-reactive cells were scored. Error bars, SEM; asterisks represent statistical difference from control cells (p < 0.01, n = 6 for each assay). (C–E) Differentiated HL-60 cells transfected with GBF1 siRNA or an irrelevant RNA duplex (Irr; C, D) or transfected with GBF1 siRNA together with cDNAs for GBF1 resWT or the resΔBP3K each tagged with EGFP (E) were incubated with or without f MLP and subjected to anti-p22phox immunostaining. F-actin was visualized by phalloidin conjugated with Texas Red (C) or with Alexa Fluor 647 (E). Transfected GBF1 proteins were visualized by fluorescence from their EGFP tag in E. Data are representative of three independent experiments. Bars, 10 μm. Percentages of p22phox molecules translocated to the leading edge in f MLP-stimulated cells, with or without GBF1 siRNA treatment, were calculated (D). Error bars, SEM; *p < 0.01 against the irrelevant control (n > 25 cells). In A–E, incubation with f MLP was for 15 min.
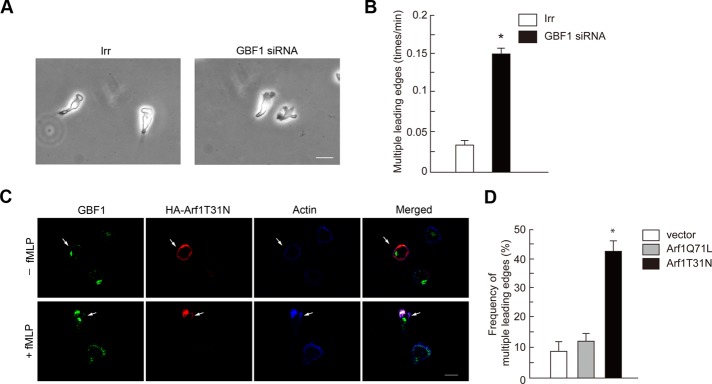
GBF1-mediated Arf1 activation is necessary for directional sensing and unified cell polarity in GPCR-mediated chemotaxis
GBF1 is primarily responsible for Arf1 activation in GPCR signaling and crucial for directional sensing during GPCR-induced chemotaxis. We sought to investigate whether the activation of Arf1 by GBF1 is involved in GPCR-induced chemotactic activities. In addition to impaired directional sensing, we noticed that GBF1 siRNA treatment frequently generated cells with multihead leading edges during f MLP-induced chemotaxis, which are seldom observed in the control irrelevant siRNA-treated cells (Figure 7, A and B, and Supplemental Movies S1 and S2). We then found that expression of Arf1T31N, but not Arf1Q71L, also frequently induces similar multihead leading edges when cells are stimulated by f MLP (Figure 7, C and D; also see Figure 5F). In the absence of f MLP, expression of Arf1T31N or Arf1Q71L did not alter the cell morphology (Figures 5F and 7C). Similar morphologies with multiple, random membrane ruffles around the cell periphery was observed in p110γ−/− neutrophils, as mentioned earlier. Moreover, GIT2−/− neutrophils do not frequently exhibit such a multihead morphology, whereas these cells also show impaired directional sensing during GPCR-induced chemotaxis (Mazaki et al., 2006). Therefore GBF1 and the activation of Arf1 appear to be important for formation of a single, uniform leading edge within a single cell upon GPCR stimulation. Impaired cell polarity may cause reduction of directional sensing during chemotaxis.
FIGURE 7:
GBF1 and Arf1 activation are necessary for unified cell polarity. (A, B) Differentiated HL-60 cells were treated with GBF1 siRNA or an irrelevant RNA duplex (Irr), as indicated, and incubated with f MLP for 15 min. (A) Morphology of cells. (B) Frequency of the appearance of cells with multiple leading edges. (C, D) Differentiated HL-60 cells were transfected with cDNAs for Arf1T31N-HA or Arf1Q71L-HA or with the HA vector alone and incubated with f MLP for 15 min. Cells were then subjected to anti-HA immunostaining after fixation, in which their F-actin was visualized by phalloidin conjugated with Alexa Fluor 647 (C). Immunocytochemical images of cells expressing Arf1Q71L-HA are shown in Figure 5F. Frequencies of the appearance of cells with multiple leading edges and positive for HA staining are shown in D. In A and C, data are representative of three independent experiments. Bars, 10 μm. In B and D, results represent the mean ± SEM, from four independent experiments, in each of which 25 cells were examined. *p < 0.01.
DISCUSSION
In this study, we identify GBF1 as crucial for GPCR-mediated chemotaxis and superoxide production. We show that GBF1 binds to products of PI3Kγ activity, such as PIP3, via a novel, previously unidentified domain, which we named BP3K. Through this binding, GBF1 is translocated from the Golgi to the leading edge. We also provide a line of evidence showing that this translocation of GBF1 is necessary to activate Arf1 at the leading edge and that the activation of Arf1 by GBF1 is necessary for directional sensing during GPCR-induced chemotaxis, especially by unifying cell polarity. GBF1 translocation is also involved in the recruitment of p22phox to the leading edge upon GPCR stimulation and is hence important for superoxide production. The translocation of GBF1 is moreover necessary to accumulate GIT2 to the leading edge, whereas this recruitment of GIT2 does not seem to be mediated only by activated Arf1. It hence can be assumed that GIT2 is not preengaged with GPCRs to be associated with Gβγ, but GBF1-mediated accumulation of GIT2 to the leading edge is necessary to enable GIT2 to be engaged with the Gβγ-mediated directional sensing machinery (also see later discussion). Therefore GBF1 appears to be involved in chemotactic directional sensing at least in two different ways: one by the activation of Arf1, and the other by the recruitment of GIT2.
Through binding of the BP3K domain to products of PI3Kγ activity, the functions of GBF1 appear to be under the control of PI3Kγ activity in GPCR signaling. Consistently, the functions of GBF1 that we examined were all blocked by a specific inhibitor of PI3Kγ activity. Moreover, the phenotypes of PI3K p110γ−/− neutrophils during GPCR-induced chemotaxis, that is, impaired ROS production and directional sensing, as well as random formation of membrane ruffles, were observed upon silencing of GBF1 in differentiated HL-60 cells stimulated with f MLP. Taking these together with our biochemical results on the properties of the BP3K domain, we hence propose that GBF1 links PI3Kγ activity with directional sensing and superoxide production in GPCR signaling of neutrophils. PI3Kγ has been recognized as an excellent molecular target for drug development against inflammation and excess-immune reactions (Camps et al., 2005), although the precise mechanism by which PI3Kγ is involved in chemotaxis and superoxide production has been largely unclear. Our identification of the linkage between PI3Kγ and GBF1 will contribute to such activities of drug development targeting PI3Kγ and its downstream signaling.
We first hypothesized that GBF1 activates Arf1 at the Golgi upon GPCR stimulation and that the activated Arf1 then translocates to the leading edge by forming budding vesicles, as been proposed as the general mode of Arf1 function (Roth, 1999). Our results show, however, that the BP3K domain of GBF1 is specially required for the activation of Arf1, as well as for the translocation of Arf1 to the leading edge. Moreover, the BP3K domain binds to PI[3,4,5]P3, and PI[3,4,5]P3 is mostly found at the leading edge of GPCR-stimulated cells. We therefore conclude that GBF1 activates Arf1 after these two proteins are translocated to the leading edge. Arf1 hence seems to be involved in unifying cell polarity after Arf1 is translocated and activated at the leading edge.
The HSD1 and HDS2 domains of GBF1 are present immediately at the C-terminus of the Sec7 domain, and this position is equivalent to that of the PH domains of other ArfGEFs. HSD1 and HDS2 domains are predicted to be rich in α-helix and moreover contain clusters of basic amino acids. Hence we first believed that HSD1 and HDS2 domains bind to phosphatidylinositol phosphates. However, we found that these HSD1 and HDS2 domains on their own do not exhibit notable binding to phosphatidylinositol phosphates, but instead we found that in combination with the C-terminal “SEG” region, HSD1 and HDS2 domains participate in binding to phosphatidylinositol phosphates. We hence hypothesized that these domains and region of GBF1 may altogether constitute a single module binding to phosphatidylinositol phosphates and named it BP3K. Moreover, we found that mutation of these basic amino acids within the HSD2 domain (R1198, R1205, R1209, and R1213) into alanines did not notably affect the BP3K binding to phosphatidylinositol phosphates (unpublished data). Therefore the BP3K binding to phosphatidylinositol phosphates may not be simply mediated by the basic amino acid clusters within the HSD1/2 domains. BP3K binding to phosphatidylinositol phosphates needs to be examined by the fine-structural analysis.
The HDS1 and HSD2 domains have been also predicted with other ArfGEFs, such as BIG1 and BIG2 (Mouratou et al., 2005). However, basic amino acid clusters found in the HDS1 and HDS2 domains of GBF1 are not well conserved in those of BIG1 and BIG2 (Mouratou et al., 2005). Moreover, the C-terminal “SEG” region, rich in alanines and prolines, found in GBF1 are not present in BIG1 and BIG2. Therefore it is not clearly predictable whether the HDS1/2 domains of BIG1 and BIG2 also bind to products of PI3K.
In conclusion, our study identified a novel module, namely BP3K, which interacts with phosphatidylinositol phosphates and hence places GBF1 under the control of PI3Kγ activity in GPCR signaling. On the basis of our results, we propose the following stepwise mechanisms. GPCR stimulation first generates Gβγ, which then activates PI3Kγ at the leading edge (Niggli, 2003). Activated PI3Kγ generates phosphatidylinositol phosphates, which then act to recruit GBF1 to the leading edge. Translocated GBF1 then recruits and activates Arf1 at the leading edge. In this step, GBF1 is simultaneously involved in the accumulation of GIT2 to the leading edge, which is necessary to engage GIT2 in the suppressive control of Arf1 activity and suppressive control of the superoxide production. This accumulation of GIT2 may also be necessary for GIT2 to be a component of the Gβγ-mediated directional sensing machinery. According to this model, the PI3Kγ activity is primarily necessary for the recruitment of GBF1 to the leading edge, and inhibition of PI3Kγ activity thereby blocks both chemotaxis and superoxide production in GPCR-stimulated neutrophils. Questions to be solved include the detailed mechanisms of how GBF1 is involved in the recruitment of Arf1, GIT2, and p22phox to the leading edge upon GPCR stimulation, since these processes do not seem to be simply mediated by the activated Arf1. GBF1 may possibly activate another class I Arf, namely Arf3, and also class II Arfs in human neutrophils. Activation of these Arfs by GBF1 and their involvement in chemotaxis and superoxide production should be clarified in the future. Moreover, the mechanisms by which the Arf1 activated by GBF1 plays a role in unifying the cell polarity to form a single leading edge in chemotaxis, as well as what roles GIT2 plays in terms of directional sensing after being engaged with the Gβγ complex, also deserve further investigation.
MATERIALS AND METHODS
Cells
HL-60 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (Gibco, Carlsbad, CA) and 2 mM l-glutamine. To be differentiated into a granulocytic lineage, cells were cultured in the presence of 1.25% DMSO for 6 d, as described previously (Matzner et al., 1987). AS-604850 (Merck, Darmstadt, Germany) was applied to cells at a final concentration of 5 μM.
Antibodies and chemicals
Mouse polyclonal antibodies against cytohesin-4, EFA6B, and KIAA0522 and rabbit polyclonal antibody against GEP100 were generated as described previously (Morishige et al., 2008). Rabbit polyclonal antibody against GIT2 was as described previously (Mazaki et al., 2001). BIG2 antibody was from K. Nakayama (Kyoto University, Kyoto, Japan; Shin et al., 2004). Other antibodies were purchased from commercial sources: rabbit polyclonal antibodies against hemagglutinin (HA) tag (MBL, Nagoya, Japan) and BIG1 (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit monoclonal antibody against p110γ (Cell Signaling Technology, Beverly, MA); mouse monoclonal antibodies against HA tag (Jackson ImmunoResearch Laboratories, West Grove, PA), GST tag (Millipore, Billerica, MA), GBF1 (BD Biosciences), Arf6 (Santa Cruz Biotechnology), Arf1 (Abcam, Cambridge, MA), PI[3,4,5]P3 (Echelon Biosciences, Salt Lake City, UT), cytohesin-1, -2, and -3 (Sigma-Aldrich, St. Louis, MO); and goat polyclonal antibodies against p22phox and GBF1 (Santa Cruz Biotechnology). Donkey antibody against mouse and rabbit immunoglobulin Gs (IgGs), conjugated with horseradish peroxidase, were from Jackson ImmunoResearch Laboratories. Goat antibodies against rabbit and mouse IgGs, conjugated with Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647, donkey antibodies against goat IgG, conjugated with Alexa Fluor 488 or Alexa Fluor 555, and phalloidins, conjugated with Texas Red or Alexa Fluor 647, were from Invitrogen (Carlsbad, CA). All other chemical reagents were purchased from Sigma-Aldrich or Nacalai Tesque (Kyoto, Japan), unless otherwise described.
Reverse transcription-PCR
Total RNA was extracted from HL-60 cells using TRIzol (Invitrogen) and reverse transcribed by SuperScript II Reverse Transcriptase (Invitrogen) using random primers at 42°C for 60 min. Detection of mRNA by reverse transcription (RT)-PCR was performed as described previously (Miyata et al., 2008; Morishige et al., 2008).
cDNAs
A cDNA encoding human GBF1 was a gift from T. Nagase (Kazusa DNA Research Institute Kisarazu, Chiba, Japan); pGEX6P-1/cytohesin-3 PH plasmid was from T. Takenawa (Kobe University, Kobe, Japan); and pcDNA3/Arf1T31N-HA and pcDNA3/Arf1-Q71L-HA were from K. Nakayama. For construction of pEGFP/GBF1 and pEGFP/GBF1ΔBP3K (amino acids 886–1370 deletion of GBF1), a NotI liker was ligated into the BamHI site of the pEGFP-C1 vector (Clontech, Mountain View, CA). GBF1 and GBF1ΔBP3K cDNAs were then ligated into the SalI–NotI sites of this vector to be ligated to the carboxyl terminal of EGFP and expressed in cultured cells. A rescue cDNA for GBF1 was constructed by substituting the nucleotides within the siRNA target to 5′-GCACGACCTAGTTGCGAAATC-3′ without changing the coding amino acids. For the expression of GST-BP3K in E. coli, the GBF1 cDNA fragment encoding the BP3K domain (amino acids 886–1370) was ligated into the SalI–NotI site of the pGEX-4T3 vector (GE Healthcare, Piscataway, NJ) to be ligated with the carboxyl terminal of the GST protein. Bacterial expression of GST proteins and their purification using glutathione beads (GE Healthcare) were performed according to the manufacturer's instructions.
Transfections
HL-60 cells incubated with DMSO for 5 d were subjected to nucleotide transfections. For cDNA transfection, 2 × 106 cells were transfected with 1 μg of pEGFP-C1/GBF1 or pEGFP-C1/GBF1ΔBP3K. For siRNA transfection, 2 × 106 cells were transfected with 3 μg of siRNAs each specific to ArfGEFs, p110γ, or an irrelevant RNA duplex (siCONTROL, RISC-free siRNA1; Dharmacon, Lafayette, CO). Transfections were performed using Nucleofector (Program T19; Lonza, Basel, Switzerland). Cells were then cultured for another 2 d in the absence of DMSO before being subjected to analyses. GBF1 siRNA targeting sequences were 5′-GCCAGACCAAGCTGTGAGATA-3′ (sequence #1) and 5′-AGATGCTGCTTTCTGCCTAGA-3′ (sequence #2). siRNA targeting sequences against other ArfGEFs were as described previously (Morishige et al., 2008). p110γ siRNA was purchase form Invitrogen.
Chemotaxis
Transwell chemotaxis assays were carried out by using 24-well Transwell chambers (5.0-μm pore size; Corning Life Sciences, Tewksbury, MA), as described previously (Mazaki et al., 2006). Briefly, after washing twice with Hank's balanced salt solution (HBSS) containing 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.2) and 1% bovine serum albumin (BSA), 5 × 105 cells were placed on the upper side of the top chambers; the bottom chambers were filled with HBSS containing 20 mM HEPES (pH 7.2), 1% BSA, and 10 nM f MLP. After incubation for 120 min at 37°C, nonmigrated cells remaining on the upper side of the chamber membranes were removed by use of cotton swabs, and cells that migrated to the lower side of the chamber membrane were stained with Diff-Quick (Sysmex, Kobe, Japan) and counted.
For two-dimensional chemotaxis, live cells migrating toward a point source of 10 nM f MLP, supplied from a micropipette at a pressure of 50 hPa (Eppendorf, Hamburg, Germany), were recorded by collecting images every 15 s for 20 min, by use of an inverted microscope (Axiovert 135; Carl Zeiss, Jena, Germany) equipped with a digital camera (AxioCam and AxioVision software; Carl Zeiss). Stacks of images were then analyzed by ImageJ software (National Institutes of Health, Bethesda, MD) with the chemotaxis and migration tool (Ibidi, Munich, Germany) plugged in for analysis of the migration track, directional index, and average migration speed of each moving cell. Frequency of the appearance of multiple leading edges during chemotaxis was calculated by counting cells bearing clefts (length, >1 μm; depth, >2 μm) within their leading edges and dividing this number by the total cell number observed within each microscopic field. Cells that migrated more than 20 μm in their tracks during 20 min of incubation were used for analysis.
Cell adhesion
Cell adhesion assays were performed as described (Mazaki et al., 2006). Briefly, differentiated HL-60 cells were washed and then placed in flat-bottomed, 96-well plates coated with fibrinogen (20 μg/ml) and incubated for 60 min at 37°C. After washing to remove nonadherent cells, the percentages of cells that remained were determined by use of the acid phosphatase assay.
Immunofluorescence microscopy
Differentiated HL-60 cells were washed and then allowed to attach to coverslips by incubating at 37°C for 10 min in HBSS containing 20 mM HEPES (pH 7.2) and 0.1% BSA. Coverslips were then placed on Dunn chambers (Hawksley, Lancing, United Kingdom), in which the outer well of the chambers was filled with the same solution with or without 10 nM f MLP, and incubated for 15 min at 37°C before fixation with 4% paraformaldehyde in phosphate-buffered saline (PBS). Fixed cells were permeabilized for 10 min with 0.2% saponin in PBS. Immunostaining of cells and acquisition of confocal images using a laser scanning microscope (FV500; Olympus, Tokyo, Japan) were performed as previously described (Mazaki et al., 2001). Each experiment was performed three times, in each of which >50 cells were analyzed, and representative pictures are shown in each figure. Numerical analyses were performed by using ImageJ software, in which the leading edge was identified as being stained with phalloidin.
Arf activities
For measurement of Arf activities, 1 × 106 cells were washed, preincubated in HBSS for 5 min at 37°C, and then stimulated with 100 nM f MLP or left untreated for the indicated times in the same solution at 37°C. Cells were then solubilized, and GTP-bound Arf1 and Arf6 were pulled down using 50 μg of GST-GGA31–226 and detected by immunoblotting coupled with gel electrophoresis, using their specific antibodies as described previously (Luton et al., 2004).
Lipid overlay assay
Lipid overlay assay was performed using PIP Array (Echelon), according to the manufacturer's instructions. Briefly, membranes were blocked with 1% skim milk in TBST (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween-20) at room temperature for 1 h and then incubated with 1 μg/ml GST proteins in the same solution at 4°C overnight. Membranes were then washed three times in TBST and incubated with mouse monoclonal anti-GST antibody at room temperature for 60 min, followed by incubation with anti–mouse IgG conjugated with horseradish peroxidase, to visualize GST proteins.
Protein-lipid bead–binding assay
Protein-lipid bead–binding assay was performed using PI[3,4,5]P3 beads (Echelon), according to the manufacturer's instructions. Briefly, 4 μg of GST-fusion protein was incubated with 25 μl of PI[3,4,5]P3 beads in binding buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.25% Nonidet P-40, 1% protease inhibitor cocktail [Nacalai Tesque]) for 3 h at room temperature under rotary agitation. After being washed three times with binding buffer, samples were boiled in Laemmli SDS sample buffer, separated by SDS–PAGE, and analyzed by silver staining.
Statistical analysis
For all experiments, the difference between groups was calculated with Student's t test.
Supplementary Material
Acknowledgments
We thank A. Hirano for technical assistance, E. Hayashi for secretarial work, T. Nagase, K. Nakayama, and T. Takenawa for plasmids and antibodies, N. Sakaguchi and H. Nakanishi (Kumamoto University, Kumamoto, Japan) for useful discussions, and H. A. Popiel for critical reading of the manuscript. This work was supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan and grants from the Novartis Foundation for the Promotion of Science and the Takeda Foundation for the Promotion of Natural Science. Y.M. is supported by Special Coordination Funds for Promoting Science and Technology from the Japan Science and Technology Agency.
Abbreviations used:
- BP3K
binding to products of PI3K
- DMSO
dimethyl sulfoxide
- f MLP
N-formyl-Met-Leu-Phe peptide
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchanging factor
- GPCR
G protein–coupled receptor
- GST
glutathione S-transferase
- PAK1
p21-activating protein kinase 1
- PBS
phosphate-buffered saline
- PH
pleckstrin homologue
- PI3K
phosphatidyl inositol 3-kinase
- ROS
reactive oxygen species
- siRNA
small interfering RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0062) on May 9, 2012.
REFERENCES
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Boulay PL, Cotton M, Melancon P, Claing A. ADP-ribosylation factor 1 controls the activation of the phosphatidylinositol 3-kinase pathway to regulate epidermal growth factor-dependent growth and migration of breast cancer cells. J Biol Chem. 2008;283:36425–36434. doi: 10.1074/jbc.M803603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher MS. Getting membrane flow and the cytoskeleton to cooperate in moving cells. Cell. 1996;87:601–606. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- Camps M, et al. Blockade of PI3Kγ suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melancon P. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- Cross AR, Segal AW. The NADPH oxidase of professional phagocytes—prototype of the NOX electron transport chain systems. Biochim Biophys Acta. 2004;1657:1–22. doi: 10.1016/j.bbabio.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat Immunol. 2004;5:744–751. doi: 10.1038/ni1081. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Hannigan M, Zhan L, Li Z, Ai Y, Wu D, Huang CK. Neutrophils lacking phosphoinositide 3-kinase γ show loss of directionality during N-formyl-Met-Leu-Phe-induced chemotaxis. Proc Natl Acad Sci USA. 2002;99:3603–3608. doi: 10.1073/pnas.052010699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase γ in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- Kawamoto K, Yoshida Y, Tamaki H, Torii S, Shinotsuka C, Yamashina S, Nakayama K. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic. 2002;3:483–495. doi: 10.1034/j.1600-0854.2002.30705.x. [DOI] [PubMed] [Google Scholar]
- Li Z, et al. Directional sensing requires Gβγ-mediated PAK1 and PIXα-dependent activation of Cdc42. Cell. 2003;114:215–227. doi: 10.1016/s0092-8674(03)00559-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-β2 and -β3 and PI3Kγ in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- Luton F, Klein S, Chauvin JP, Le Bivic A, Bourgoin S, Franco M, Chardin P. EFA6, exchange factor for ARF6, regulates the actin cytoskeleton and associated tight junction in response to E-cadherin engagement. Mol Biol Cell. 2004;15:1134–1145. doi: 10.1091/mbc.E03-10-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzner Y, Gavison R, Rachmilewitz EA, Fibach E. Expression of granulocytic functions by leukemic promyelocytic HL-60 cells: differential induction by dimethylsulfoxide and retinoic acid. Cell Differ. 1987;21:261–269. doi: 10.1016/0045-6039(87)90481-7. [DOI] [PubMed] [Google Scholar]
- Mazaki Y, et al. An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol Biol Cell. 2001;12:645–662. doi: 10.1091/mbc.12.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaki Y, et al. Neutrophil direction sensing and superoxide production linked by the GTPase-activating protein GIT2. Nat Immunol. 2006;7:724–731. doi: 10.1038/ni1349. [DOI] [PubMed] [Google Scholar]
- Miura Y, Tohyama Y, Hishita T, Lala A, De Nardin E, Yoshida Y, Yamamura H, Uchiyama T, Tohyama K. Pyk2 and Syk participate in functional activation of granulocytic HL-60 cells in a different manner. Blood. 2000;96:1733–1739. [PubMed] [Google Scholar]
- Miyata M, Raven JF, Baltzis D, Koromilas AE, Sabe H. IRES-mediated translational control of AMAP1 expression during differentiation of monocyte U937 cells. Cell Cycle. 2008;7:3273–3281. doi: 10.4161/cc.7.20.6883. [DOI] [PubMed] [Google Scholar]
- Morishige M, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat Cell Biol. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jurgens G, Melancon P, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V. Signaling to migration in neutrophils: importance of localized pathways. Int J Biochem Cell Biol. 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
- Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, Klausner RD. Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments. J Cell Biol. 1995;128:1003–1017. doi: 10.1083/jcb.128.6.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MG. Snapshots of ARF1: implications for mechanisms of activation and inactivation. Cell. 1999;97:149–152. doi: 10.1016/s0092-8674(00)80723-0. [DOI] [PubMed] [Google Scholar]
- Sasaki T, et al. Function of PI3Kγ in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- Shin HW, Morinaga N, Noda M, Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CX, Downey GP, Zhu F, Koh AL, Thang H, Glogauer M. Rac1 is the small GTPase responsible for regulating the neutrophil chemotaxis compass. Blood. 2004;104:3758–3765. doi: 10.1182/blood-2004-03-0781. [DOI] [PubMed] [Google Scholar]
- Vitale N, Patton WA, Moss J, Vaughan M, Lefkowitz RJ, Premont RT. GIT proteins, A novel family of phosphatidylinositol 3,4, 5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J Biol Chem. 2000;275:13901–13906. doi: 10.1074/jbc.275.18.13901. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Wymann MP, Pirola L. Structure and function of phosphoinositide 3-kinases. Biochim Biophys Acta. 1998;1436:127–150. doi: 10.1016/s0005-2760(98)00139-8. [DOI] [PubMed] [Google Scholar]
- Yano H, Kobayashi I, Onodera Y, Luton F, Franco M, Mazaki Y, Hashimoto S, Iwai K, Ronai Z, Sabe H. Fbx8 makes Arf6 refractory to function via ubiquitination. Mol Biol Cell. 2008;19:822–832. doi: 10.1091/mbc.E07-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.