Circular dorsal ruffles (CDRs) are highly dynamic F-actin–based membrane structures involved in bulk endocytosis of membrane receptors and macropinocytosis. A key role of ARAP1, an Arf GAP protein, is found to be in the control of the ring size of CDRs.
Abstract
Small guanosine triphosphatase (GTPase) ADP-ribosylation factors (Arfs) regulate membrane traffic and actin reorganization under the strict control of GTPase-activating proteins (GAPs). ARAP1 (Arf GAP with Rho GAP domain, ankyrin repeat, and PH domain 1) is an Arf GAP molecule with multiple PH domains that recognize phosphatidylinositol 3,4,5-trisphosphate. We found that growth factor stimulation induced localization of ARAP1 to an area of the plasma membrane inside the ring structure of circular dorsal ruffles (CDRs). Moreover, expression of ARAP1 increased the size of the CDR filamentous-actin ring in an Arf GAP activity–dependent manner, whereas smaller CDRs were formed by ARAP1 knockdown. In addition, expression of a dominant-negative mutant of Arf1 and Arf5, the substrates of ARAP1, expanded the size of CDRs, suggesting that the two Arf isoforms regulate ring structure downstream of ARAP1. Therefore our results reveal a novel molecular mechanism of CDR ring size control through the ARAP1–Arf1/5 pathway.
INTRODUCTION
The ADP-ribosylation factor (Arf) family of small GTPases are members of the Ras superfamily of GTP-binding proteins and regulate membrane traffic by recruiting coat proteins to membrane surfaces and modulating membrane lipid composition (Donaldson and Jackson, 2011). The six mammalian Arfs (five in humans) are grouped into three classes based on their primary structure and gene organization: class I, Arf1, 2, and 3; class II, Arf4 and 5; class III, Arf6 (D'souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007; Donaldson and Jackson, 2011). In particular, Arf1 and Arf6 are the best-characterized Arfs. Arf6 is localized at the plasma membrane and regulates both cortical actin cytoskeleton reorganization and endosomal membrane trafficking (Donaldson, 2003). In contrast, Arf1 controls the assembly and disassembly of vesicle coat machinery in the early cis-Golgi and also promotes actin polymerization at the Golgi membrane by modulating lipid-metabolizing enzymes, including phosphatidylinositol phosphate 5-kinase and phospholipase D (Donaldson, 2008; Donaldson and Jackson, 2011). Arf1 is also involved in the endocytosis of glycosylphosphatidylinositol-anchored proteins from the plasma membrane by regulation of another small GTPase, Cdc42 (Kumari and Mayor, 2008). In addition, Arf1 is activated at the phagocytic cup (Beemiller et al., 2006) and is essential for phagocytosis (Braun et al., 2007), implying that Arf1 functions not only in the Golgi membrane but also at the plasma membrane.
Arf GTPases have slow nucleotide exchange rates and undetectable intrinsic GTPase activity and must be regulated by specific guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), which catalyze Arf GTPase activation and deactivation, respectively (Casanova, 2007; Inoue and Randazzo, 2007; Spang et al., 2010). Arf GAPs regulate cytoskeletal structures, including focal adhesions, circular dorsal ruffles (CDRs), and invadopodia (Sabe et al., 2006; Yoon et al., 2006; Inoue and Randazzo, 2007; Ha et al., 2008). CDRs are highly dynamic and ring-shaped structures rich in filamentous actin (F-actin), which are induced at the dorsal membrane surface within minutes after growth factor stimulation with platelet-derived growth factor (PDGF) or epidermal growth factor (Buccione et al., 2004). One of the physiological roles of CDRs may be to promote the bulk internalization of certain transmembrane proteins, including growth factor receptors (Orth et al., 2006) and integrins (Gu et al., 2011).
ARAP1 is a member of the AZAP family of Arf GAPs (ASAPs, ACAPs, AGAPs, and ARAPs), which contains an Arf GAP domain, Rho GAP domain, sterile α motif (SAM), ankyrin repeat, five PH domains, and Ras association (RA) domain (Figure 1A). The Arf GAP activity of ARAP1 is stimulated in vitro by the binding of phosphoinositides, particularly phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3; Miura et al., 2002; Campa et al., 2009). Furthermore, overexpression of ARAP1 induces reorganization of the actin cytoskeleton (Miura et al., 2002; Campa et al., 2009). However, although ARAP1 is recruited to the plasma membrane in response to growth factor stimulation and may regulate the trafficking of growth factor receptors (Daniele et al., 2008; Yoon et al., 2008; Kang et al., 2010), its role in the formation of membrane ruffles, including CDRs, is unclear.
FIGURE 1:
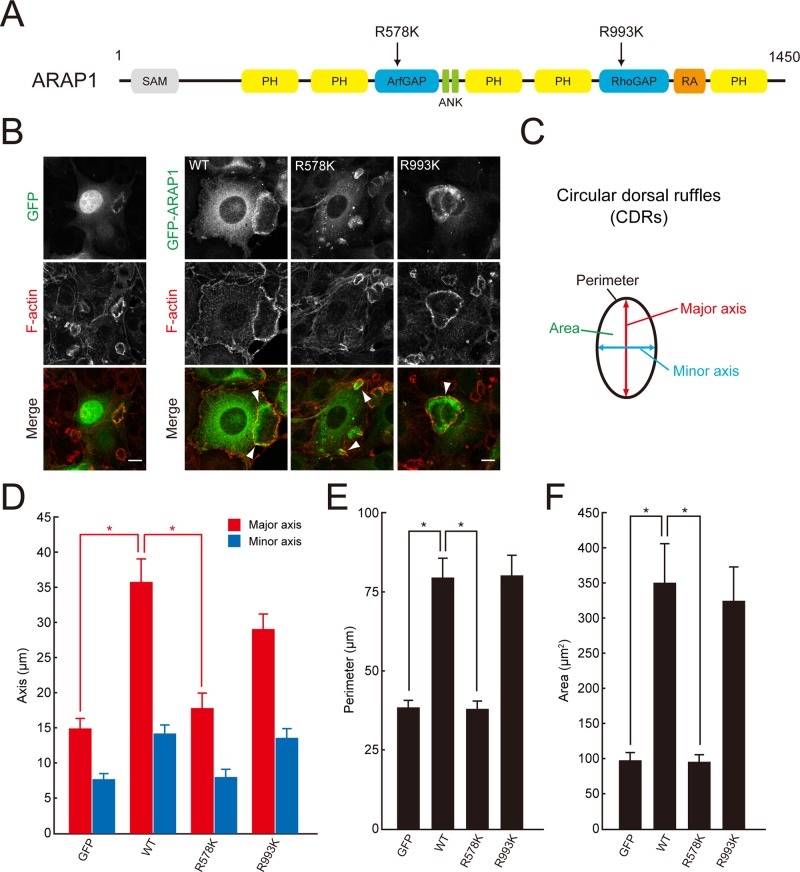
ARAP1 regulates the ring size of CDRs. (A) Schematic diagram of human ARAP1 and point mutants used in this study. ANK, ankyrin repeat; ArfGAP, Arf GTPase-activating protein; PH, pleckstrin-homology domain; RA, Ras-associating domain; Rho GAP, Rho GTPase–activating protein; SAM, sterile α-motif. (B) NIH 3T3 cells expressing GFP or GFP-ARAP1(WT, R578K, and R993K) were stimulated with 20 ng/ml PDGF for 5 min and stained with rhodamine–phalloidin. Arrowheads indicate CDR structures. Bars, 10 μm. (C) The four parameters measured in this study: the length of the major axis (red), minor axis (blue), the ring perimeter (black), and the area enclosed by the CDRs (green). (D–F) Quantification of the major and minor axes (D), the perimeter (E), and the area (F) of CDRs formed in B. Results represent the mean (SEM) of 15 cells. *p < 0.01.
In the present study, we describe a regulatory mechanism of CDR formation by ARAP1. We show that ARAP1 localizes to the inner area of the F-actin ring structure of CDRs at the plasma membrane and regulates the size of CDRs in an Arf GAP activity–dependent manner. Furthermore, Arf1 and Arf5, the substrates of ARAP1, are also involved in the modulation of the F-actin ring size. Our findings suggest a novel mechanism for the control of CDR formation through Arfs and its associated GAP molecules.
RESULTS
ARAP1 regulates the ring size of CDRs
To determine the role of ARAP1 in CDR formation, green fluorescent protein (GFP)–tagged ARAP1 (GFP-ARAP1) was expressed in NIH 3T3 cells, which form CDRs in response to PDGF stimulation (Buccione et al., 2004; Hasegawa et al., 2011). As shown in Figure 1B, GFP-ARAP1 specifically localized to an area of the plasma membrane inside the F-actin ring. The observed localization of ARAP1 was consistent with the hypothesis that the protein expanded the ring structure from the inside, thereby increasing the size of CDRs (Figure 1B; GFP-expressing vs. GFP-ARAP1–expressing cells). To evaluate the effects of ARAP1 expression on the size of CDRs, we measured four parameters of the ring structure: the lengths of the major and minor axes, the ring perimeter, and the enclosed area (Figure 1C; Zeng et al., 2011). As a result, GFP-ARAP1 increased the lengths of both major and minor axes as well as the perimeter of CDRs by approximately twofold and also expanded the inner area of CDRs by ∼3.5-fold compared with those formed in cells expressing GFP alone (Figure 1, B and D–F). Although GFP-ARAP1(R578K), an inactive mutant of the Arf GAP domain (Miura et al., 2002), also localized inside the F-actin ring, it had no effect on the size of CDRs (Figure 1, B and D–F). In contrast, GFP-ARAP1(R993K), which lacks Rho GAP activity, had a similar effect to the wild type (WT; Figure 1, B and D–F), suggesting that the formation of large CDRs is dependent on Arf GAP but not Rho GAP activity. To our knowledge, this is the first report to show that the ring size of CDRs is specifically regulated.
Dynamic localization of ARAP1 inside the ring of CDRs
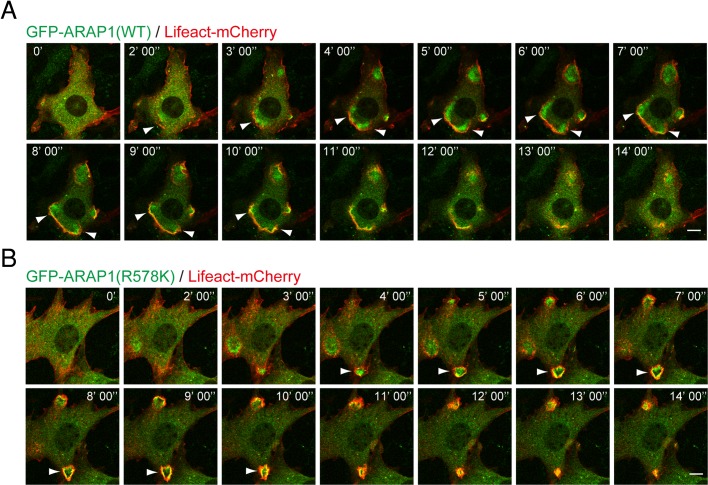
To observe the dynamic localization of ARAP1 in CDR formation, we performed time-lapse imaging of PDGF-stimulated NIH 3T3 cells that expressed GFP-ARAP1 and Lifeact-mCherry, a probe for F-actin (Riedl et al., 2008). Similar to the result in Figure 1, GFP-ARAP1(WT) localized to the inner area of CDRs (Figure 2A and Supplemental Movie S1). Subsequently, GFP-ARAP1 formed a membrane edge containing dense actin filaments, which eventually disappeared without constricting into the typical circular structure. Similar localization was observed for GFP-ARAP1(R578K), although it did not affect the size of CDRs (Figures 1 and 2B and Supplemental Movie S2). Whereas suppression of Gα12 and Gα13 trimeric G proteins has been reported to reduce the rate of CDR breakdown (Wang et al., 2006), the disassembly rate of CDRs appeared normal in cells expressing GFP-ARAP1(WT) compared with the control (unpublished data) or GFP-ARAP1(R578K)-expressing cells (Figure 2, A and B). Thus the dynamics and the size of CDRs are independently regulated, and ARAP1 is specifically involved in controlling the size of the ring structure.
FIGURE 2:
Dynamic localization of ARAP1 inside the ring of CDRs. (A, B) NIH 3T3 cells transiently transfected with GFP-ARAP1(WT) and Lifeact-mCherry (A) or GFP-ARAP1(R578K) and Lifeact-mCherry (B) were serum starved, and live imaging was performed after adding PDGF (20 ng/ml). Arrowheads indicate CDR structures. Bars, 10 μm.
Multiple domains, including PH domains, are required for the precise localization of ARAP1
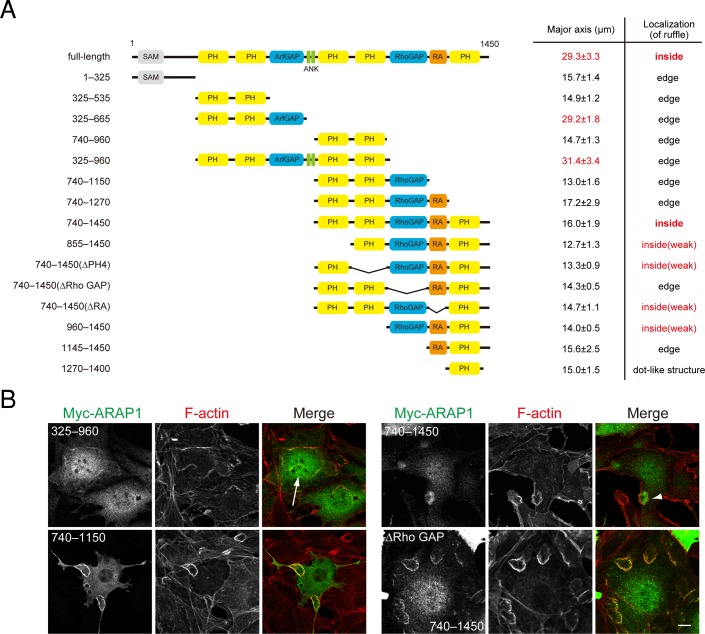
Next we tried to determine the structural basis for ARAP1 localization inside CDRs by constructing a series of deletion mutants. Although the PH domain binds to phosphoinositides to mediate membrane recruitment (Behnia and Munro, 2005), none of the individual PH domains of ARAP1 (325–535, 740–960, or 1270–1400) displayed a similar localization pattern to that of the full-length ARAP1 (Figure 3, A and B, and Supplemental Figure S1). An ARAP1 construct (960–1450) containing Rho GAP, RA, and the fifth PH domain localized inside of the F-actin ring with a lower efficiency, with ∼20% of cells forming CDRs (Figure 3A and unpublished data). Similar results (the weak and inefficient localization inside of the dorsal ring) were obtained by further additions of the third (740–1450(ΔPH4)) or fourth (855–1450) PH domain, respectively (Figure 3A and unpublished data). However, simultaneous addition of both PH domains (740–1450) resulted in more efficient localization to the inner area of the F-actin ring (Figure 3, A and B, and Supplemental Figure S1). Neither the 740–1450(∆RA) nor the 740–1450(∆Rho GAP) construct showed an efficient localization inside the CDR. These results collectively indicate that three PH domains, a Rho GAP domain, and an RA domain are required. Consistent with the foregoing data that Arf GAP activity is essential for large CDR formation, ARAP1 constructs containing the Arf GAP domain (full-length, 325–665, or 325–960) were able to induce larger CDRs (Figure 3, A and B, and Supplemental Figure S1).
FIGURE 3:
Multiple domains, including PH domains, are required for the precise localization of ARAP1. (A) Localization of ARAP1-deletion mutants and their ability to form large CDRs. Measured results (the length of the major axis) represent the mean (SEM) of five cells. (B) NIH 3T3 cells transfected with Myc-ARAP1 constructs were serum starved, stimulated with 20 ng/ml PDGF for 5 min, fixed, and stained with anti-Myc antibodies and rhodamine–phalloidin. Arrows show larger CDRs. Arrowheads show localization of ARAP1 constructs inside CDRs. Bars, 10 μm.
Each PH domain of ARAP1 may bind to distinct phosphoinositides (Daniele et al., 2008). Therefore we examined the lipid-binding specificity of the full-length ARAP1 protein to investigate which signaling pathway regulates the Arf GAP molecule. Recombinant protein of FLAG-tagged ARAP1 (FLAG-ARAP1) was purified and subjected to a cosedimentation assay using liposomes comprising various phosphoinositides. As a result, the full-length ARAP1 bound most strongly to PI(3,4,5)P3 (Supplemental Figure S2). However, it is of note that Btk-PH, a specific probe for PI(3,4,5)P3, behaves differently to ARAP1 (Hasegawa et al., 2011). Thus other factors that bind to PH, Rho GAP, or RA domains may also play a role in the precise localization of ARAP1.
ARAP1 is necessary for CDR formation
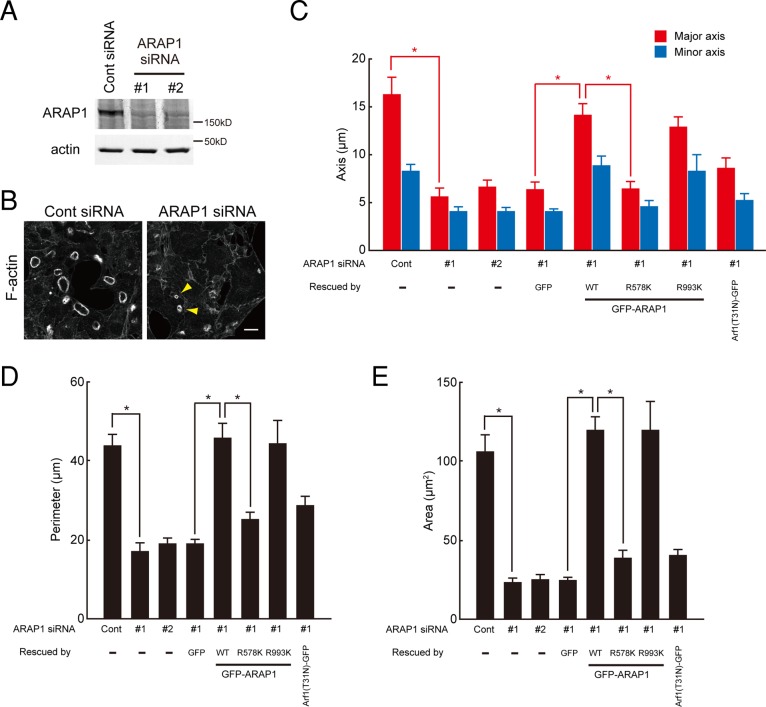
To assess whether ARAP1 is required for CDR formation, we used RNA interference to knock down ARAP1 in NIH 3T3 cells. Two distinct small interfering RNAs (siRNAs) both independently achieved significant knockdown of ARAP1 levels, as determined by immunoblotting (Figure 4A). Whereas CDR formation was induced by PDGF in ∼60% of control siRNA–treated cells, it was markedly inhibited in ARAP1 siRNA–treated cells (Figure 4B and Supplemental Figure S3). In addition, the sizes of the few CDRs that formed in ARAP1-depleted cells were much smaller than those in control cells (Figure 4, B–E, see arrowheads). This is in agreement with the results shown in Figure 1, which indicated that larger CDRs were formed in ARAP1-expressing cells. Suppression of the number and size of CDRs was efficiently rescued by exogenous expression of wild-type or R993K ARAP1 but not by R578K (Arf GAP inactive) mutant (Figure 4, C–E, and Supplemental Figure S3), further supporting the hypothesis that Arf GAP activity is necessary for precise control of the size of CDRs.
FIGURE 4:
ARAP1 is necessary for CDR formation. (A) Levels of ARAP1 protein 72 h after transfection of NIH 3T3 cells with control or ARAP1-specific siRNA, as detected by immunoblotting. (B) Phalloidin staining of control or ARAP1-depleted NIH 3T3 cells treated with PDGF for 5 min. Arrowheads indicate small CDRs. Bar, 10 μm. (C–E) Quantification of the major and minor axes (C), the perimeter (D), and the area (E) of CDRs formed in ARAP1-depleted NIH 3T3 cells. Rescue experiments were performed by transfecting GFP-ARAP1 constructs to restore normal CDR formation. Results represent the mean (SEM) of 15 cells. *p < 0.01.
Arf1/5, downstream targets of ARAP1, regulate CDR formation
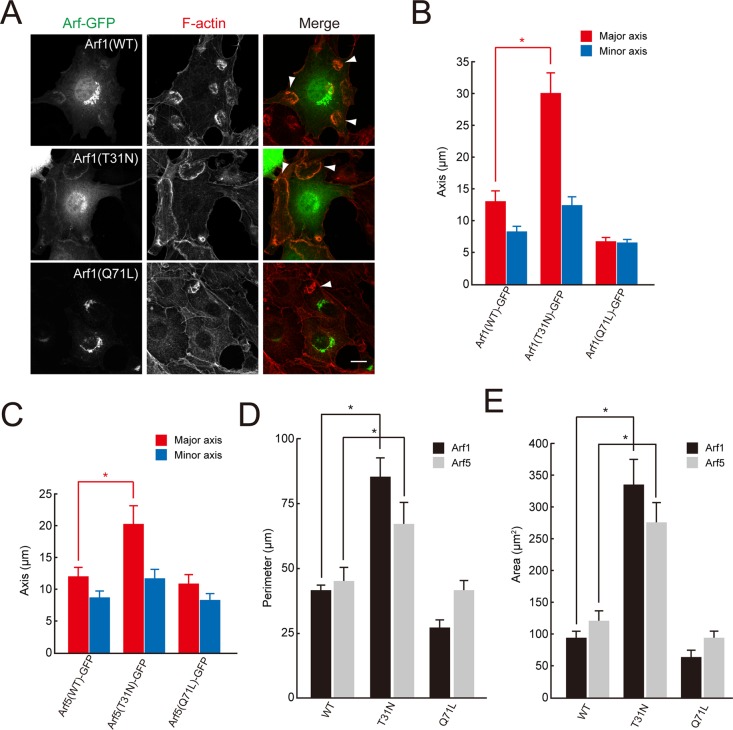
Given that the Arf GAP activity of ARAP1 was essential for CDR formation, we investigated its downstream targets by expressing in NIH 3T3 cells GFP-tagged forms of Arf (Arf1-GFP, Arf5-GFP, or Arf6-GFP), which are candidate substrates for ARAP1. Expression of Arf1(T31N)-GFP, a dominant-negative form of Arf1 defective in GTP loading, induced larger CDRs (Figure 5, A, B, D, and E). In contrast, expression of Arf1(Q71L)-GFP, a constitutively active mutant defective in GTP hydrolysis, reduced the size of CDRs and also impaired their formation (Figure 5, A, B, D, and E). Thus the GTP-bound form of Arf1 modulates the size of the F-actin ring. However, expression of Arf1(T31N)-GFP did not adequately rescue the size and number of CDRs in ARAP1-depleted cells (Figure 4, C–E, and Supplemental Figure S3). Therefore it is conceivable that ARAP1 functions via downstream targets other than Arf1 or acts as a structural component (e.g., scaffolding protein) indispensable for the ring size control of CDR.
FIGURE 5:
Arf1/5, which are downstream targets of ARAP1, regulate CDR formation. (A) NIH 3T3 cells were transiently transfected with Arf1(WT, T31N, and Q71L)-GFP, stimulated with PDGF for 5 min, fixed, and stained with rhodamine–phalloidin. Arrowheads indicate CDR structures. Bar, 10 μm. (B–E) Quantification of the major and minor axes (B, C), the perimeter (D), and the area (E) of CDRs formed in NIH 3T3 cells expressing Arf1(WT, T31N, and Q71L)–GFP or Arf5(WT, T31N, and Q71L)–GFP. Results represent the mean (SEM) of 15 cells. *p < 0.01.
We also found that expression of a dominant-negative form of Arf5 (Arf5(T31N)-GFP) resulted in the enlargement of CDRs, albeit to a lesser extent than Arf1(T31N)-GFP (Figure 5, C–E, and Supplemental Figure S4A). Because Arf5 may also be an in vivo substrate of ARAP1 (Campa et al., 2009), Arf5 could act downstream of ARAP1 to regulate the size of CDRs. Arf6 also appeared to be involved in the regulation of CDR formation, as evidenced by the reduced numbers of CDRs in cells expressing wild-type or dominant-active forms (Q67L) of Arf6 (Supplemental Figure S4, A and B).
ARAP1 and Arf1 are required for macropinocytosis
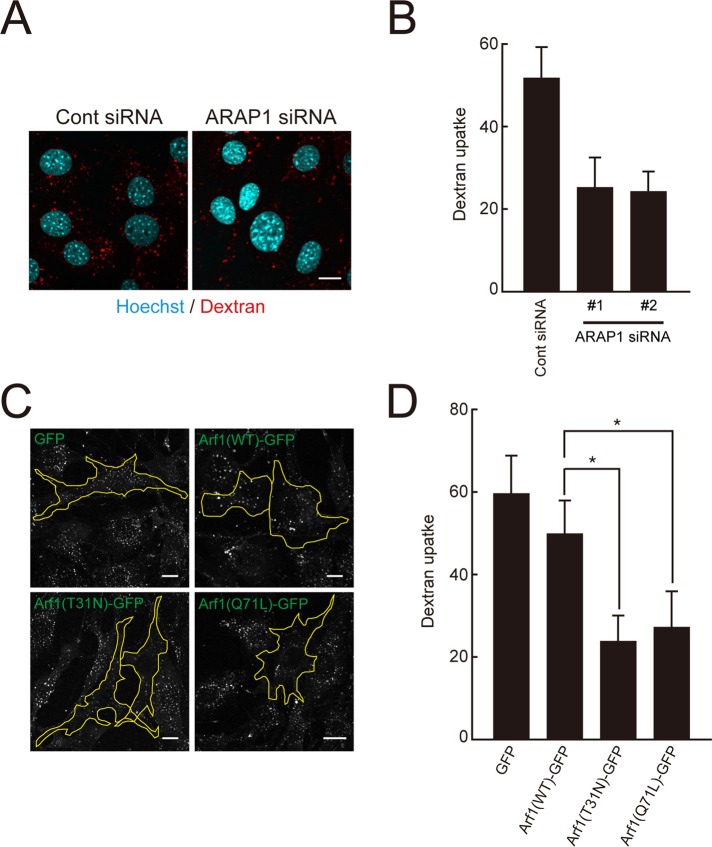
Given that CDRs may play a role in macropinocytosis, we investigated the effect of ARAP1 depletion on fluid-phase endocytosis with fluorescently labeled dextran. As shown in Figure 6, A and B, PDGF-dependent dextran uptake was significantly inhibited in ARAP1-knockdown cells. Therefore ARAP1 may be required for macropinocytosis through precise control of ring expansion and constriction.
FIGURE 6:
ARAP1 and Arf1 are required for macropinocytosis. (A) Control or ARAP1-depleted NIH 3T3 cells that internalized the fluid-phase marker rhodamine–dextran (0.2 mg/ml; shown in red) for 10 min were stained with Hoechst (cyan). Bar, 10 μm. (B) Quantification of dextran uptake in A by counting the number of dextran spots/cell. Results represent the mean (SD) of 20 cells. *p < 0.01. (C) NIH 3T3 cells expressing GFP or Arf1(WT, T31N, and Q71L)–GFP were allowed to internalize rhodamine–dextran (0.2 mg/ml) for 10 min. Cells expressed with the indicated constructs are outlined. Bars, 10 μm. (D) Quantification of dextran uptake in C by counting the number of dextran spots/cell. Results represent the mean (SD) of 30 cells. *p < 0.01.
The role of Arf1 in macropinocytosis was also examined by expressing Arf1(WT, T31N, or Q71L)-GFP in NIH 3T3 cells. The PDGF-dependent dextran internalization was suppressed in cells expressing Arf1(Q71L)-GFP compared with GFP or Arf1(WT)-GFP (Figure 6, C and D), reflecting the smaller size of CDR induced by the active form of Arf1. Furthermore, expression of Arf1(T31N)-GFP also caused a significant decrease in dextran uptake (Figure 6, C and D). Time-lapse imaging of Arf1(T31N)-GFP–expressing cells showed that a large CDR was disassembled at its initial position without F-actin ring closure (Supplemental Figure S5), supporting an inefficient engulfment of outer materials by macropinocytosis.
DISCUSSION
In this study, we demonstrate that not only exogenous expression of ARAP1, but also its knockdown inhibits CDR formation, as well as macropinocytosis. Previous studies described negative roles of some Arf GAP members in CDR formation by overexpression experiments (Jackson et al., 2000; Randazzo et al., 2000). Our data, obtained by moderate expression levels of ARAP1 in NIH 3T3 cells, strongly suggest that such inhibitory effects are caused by their ability to expand the diameter of CDRs. Consistently, knockdown of ARAP1 induced smaller CDRs, thus inhibiting their proper functions in macropinocytosis (Figure 4).
Arf5, as well as Arf1, is mainly localized at the Golgi apparatus and involved in the Golgi morphology (Donaldson and Jackson, 2011). However, the precise role of Arf5 has remained elusive. In this study, we found that Arf5 modulates the size of CDRs, albeit less effectively than Arf1 (Figure 5). Therefore Arf5 may also be regulated by ARAP1 and act cooperatively with Arf1 (Volpicelli-Daley et al., 2005) to control the size of CDRs. Of interest, whereas Arf1(Q71L) induced smaller CDRs by an enhanced constriction of the ring structure, Arf5(Q71L) had little effects on the size of CDRs (Figure 5). These findings indicate that both Arf1 and Arf5 induce larger CDRs, but the constriction of the CDR ring is mainly dependent on Arf1.
We found that the full-length ARAP1 protein bound most strongly to PI(3,4,5)P3 (Supplemental Figure S2), suggesting that it functions at the plasma membrane where D3-phosphorylated phosphoinositide is produced. Consistently, the Arf GAP activity of ARAP1 was more strongly stimulated in vitro by PI(3,4,5)P3 than with PI(3,4)P2 (Miura et al., 2002; Campa et al., 2009). We recently demonstrated that conversion from PI(3,4,5)P3 to phosphatidylinositol 3,4,-biphosphate (PI(3,4)P2) by the action of the phosphoinositide 5-phosphatase SHIP2 was necessary for CDR formation and that PI(3,4)P2 specifically accumulated at the membrane edges of the CDR ring (Hasegawa et al., 2011). Therefore, upon PDGF stimulation, ARAP1 is first activated by the increase of PI(3,4,5)P3 and then becomes deactivated in response to the dephosphorylation of PI(3,4,5)P3 by SHIP2. As a result, this phosphoinositide conversion leads to the transition of the nucleotide-binding state of Arf1/5 from a GDP-bound (ring expansion) to a GTP-bound form (ring closure; Figure 7). Indeed, our findings that the GDP-bound form of Arf1 and Arf5 induces expansion of CDR, whereas GTP-bound Arf1 promotes its constriction (Figure 5 and Supplemental Figure S4A), are consistent with this model. Further analyses are required to characterize the mechanism by which Arf1 (or Arf5) governs sequential changes in the ring size of CDR via its downstream targets.
FIGURE 7:
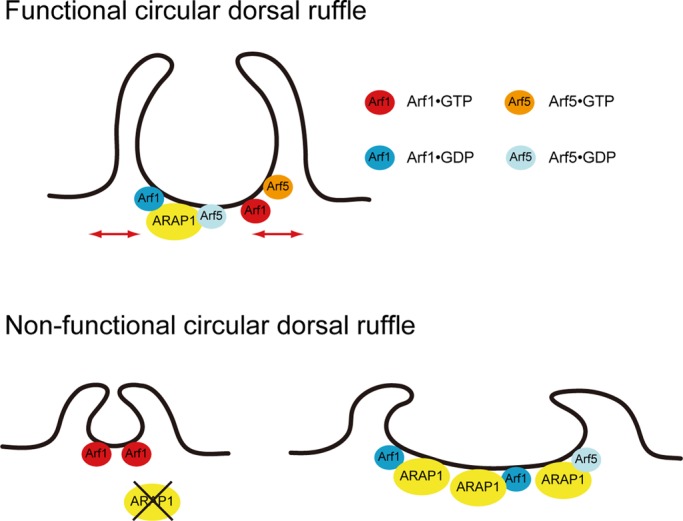
A model for CDR formation by ARAP1 and Arf1/5. Functional CDR, maintains a normal-sized CDR; nonfunctional CDR, depletion of ARAP1 (or increasing Arf1•GTP) leads to small CDR, whereas overexpression of ARAP1 (or increasing Arf1•GDP, Arf5•GDP) leads to large CDR. See Discussion for details.
ARAP3, another ARAP family protein, has also been reported to be involved in the formation of lamellipodia and CDR through its GAP activity for Arf6 (Krugmann et al., 2006). It was shown that ARAP3-deficient cells displayed a strong CDR formation by PDGF stimulation because of an increase in GTP-bound form of Arf6. In this study, we show that ARAP1 depletion results in an inhibition of CDR formation and a decrease in the size of CDRs (Figure 4 and Supplemental Figure S2). These discrepancies may be caused by the difference in substrate specificity of the two Arf GAP isoforms; ARAP3 acts only on Arf6 (Krugmann et al., 2002), whereas ARAP1 prefers Arf1/5 (weakly on Arf6) as its substrates (Miura et al., 2002; Campa et al., 2009). The molecular basis of these opposite actions by different Arf molecules on CDR formation an important subject of future studies.
One of the important physiological functions of CDRs proposed is macropinocytosis. Orth et al. (2006). found that CDRs play a major role in the bulk internalization of growth factor receptors. Recently, it was also reported that the integrin molecules are internalized and sorted to the newly formed focal adhesions via CDRs (Gu et al., 2011). These studies suggest that CDRs might function as an important membranous scaffolding platform for the internalization of transmembrane proteins. The significance of CDR formation is also evidenced by the fact that a variety of viruses and bacteria can enter host cells through macropinocytosis (Mercer and Helenius, 2009). Further studies are needed to determine the molecular mechanisms underlying CDR formation and dynamics for the definition of infectious route of pathogenic microbes.
Podosomes are an additional type of circular membrane structure found in motile cells (Buccione et al., 2004). Several common properties of CDRs and podosomes include the following: 1) formation by activated tyrosine kinases, 2) dependence on PI3 kinase activity, and 3) dependence on PI(3,4)P2 synthesis (Buccione et al., 2004; Oikawa et al., 2008; Hasegawa et al., 2011). Thus podosome formation in osteoclasts or invasive cancer cells may also be regulated by the ARAP1-Arf1/5 pathway to strictly control the ring size of the actin-based circular membrane structure.
MATERIALS AND METHODS
Reagents and antibodies
Rabbit anti-Myc polyclonal antibody was purchased from Cell Signaling Technology (Beverly, MA). Rabbit anti-CENTD2/ARAP1 polyclonal antibody was from Novus Biologicals (Littleton, CO). Mouse anti-actin monoclonal antibody was from Chemicon (Temecula, CA). All fluorescent reagents (rhodamine–phalloidin and Alexa Fluor 488, 568–conjugated goat anti-rabbit or anti-mouse secondary antibodies) were purchased from Invitrogen (Carlsbad, CA). Phosphatidic acid was from Sigma-Aldrich (St. Louis, MO). Purified phospholipids (phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine [PE], and phosphatidylcholine [PC]) were from Avanti Polar Lipids (Alabaster, AL). All phosphorylated phosphoinositides (PI(3)P, PI(4)P, PI(5)P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, and PI(3,4,5)P3) were from Cell Signaling Technology. Recombinant human PDGF-BB was from Wako Chemical.
Plasmids
Coding sequences of the human ARAP1 was amplified from HeLa cDNA by PCR with specific primers (5′-gcggatccaccatggcagaggctggggatg-3′ and 5′-gcgtcgacgacgttgcgcagaagagacaga-3′) and subcloned into pCMV-Tag3B (Agilent Technologies, Santa Clara, CA), pEGFP-C1 (Takara Bio, Otsu, Japan), and pEF-BOS-FLAG vectors. ARAP1 deletion and point mutants were amplified by PCR with specific primers and ligated into each vector. Arf1(WT, T31N, Q71L), Arf5(WT, T31N, Q71L), and Arf6(WT, T27N, Q67L) cDNA were subcloned into pEGFP-N3 (Takara Bio) vector. The Lifeact-mCherry construct was prepared as described previously (Hasegawa et al., 2011).
Protein expression and purification
Recombinant FLAG-tagged ARAP1 protein was expressed in FreeStyle 293-F cells (Invitrogen) using FreeStyle MAX reagent (Invitrogen) and then was purified with ANTI-FLAG M2-agarose (Sigma-Aldrich) according to the manufacturer's instruction.
Cell culture and transfection
NIH 3T3 cells were maintained in DMEM supplemented with 10% calf serum (CS). Plasmid transfection was performed using Lipofectamine LTX (Invitrogen). Experiments were carried out 24 h after transfection.
RNA interference
For knockdown of ARAP1, validated siRNAs were purchased from Invitrogen. siRNAs at 50 nM were transfected into NIH 3T3 cells with Lipofectamine RNAiMAX (Invitrogen), and the expression levels were assessed after 72 h by immunoblots.
Endocytosis assay
For dextran uptake, cells were starved in serum-free medium for 16 h, incubated with 0.2 mg/ml tetramethylrhodamine-labeled, lysine-fixable, 10-kDa dextran (Invitrogen) in DMEM plus 20 ng/ml PDGF for 10 min at 37°C, and then fixed and stained with Hoechst 34580 (Invitrogen) for immunofluorescence.
Immunofluorescence microscopy
Cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature (RT) and then were permeabilized with PBS containing 0.2% Triton X-100 for 5 min at RT, washed three times with PBS, and blocked with Image-iT FX signal enhancer (Invitrogen) for 30 min at RT. Cells were incubated with primary antibodies in PBS for 90 min. After three washes with PBS, cells were incubated with the appropriate secondary antibodies in PBS for 1 h. After a brief wash with PBS, coverslips were mounted onto slides using PermaFluor Mounting Medium (Thermo Scientific, Waltham, MA) and observed under a FluoView 1000-D confocal microscope (IX81; Olympus, Tokyo, Japan) equipped with 473-, 568-, and 633-nm diode lasers (Olympus) through an objective lens (60× oil immersion objective, numerical aperture 1.35; Olympus) and with FluoView software (Olympus). The lengths of the major, the minor axis, the perimeter, and the area enclosed by CDRs in the images were calculated using FluoView software. Acquired images were processed with Photoshop (Adobe, San Jose, CA).
Live-cell imaging
NIH 3T3 cells transfected with the indicated constructs were grown on glass-base dishes (Iwaki; Asahi Techno Glass, Tokyo, Japan). Cells were starved in serum-free DMEM for 16 h and then imaged in the same medium before and after the addition of PDGF (final, 20 ng/ml). The images were acquired for 10–15 min at 15-s intervals by using a FluoView 1000-D confocal microscope through an objective lens (60× oil immersion objective, numerical aperture 1.35) at RT.
Liposome cosedimentation assay
Mixtures of PE (70%), PC (20%), and 5% of various acidic phospholipids were dried under nitrogen gas and then suspended in 50 μl of buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 100 mM NaCl, 0.5 mM EDTA) for 1 h at 37°C to allow formation of liposomes. Before mixing with the liposomes, proteins were subjected to centrifugation at 150,000 × g for 15 min at 4°C to remove aggregated portions. Proteins that came to the supernatant (5 μg) were incubated with the liposomes (25 μg) for 15 min at RT and centrifuged at 150,000 × g for 20 min at 20°C. Proteins that sedimented with liposomes in the pellet and unbound proteins in the supernatant were separated and then subjected to SDS–PAGE followed by Coomassie brilliant blue staining.
Statistical analysis
Statistically significant differences were determined using Student's t test. Differences were considered significant if p < 0.01.
Supplementary Material
Acknowledgments
We are grateful to S. Kurisu for plasmids. This study was supported in part by a Grant-in-Aid for Creative Scientific Research from the Japan Society for the Promotion of Science (JSPS) to T.T. and T.I.; a Grant-in-Aid for Young Scientists from the JSPS to T.I.; the Naito Foundation (J.H.); and the Global COE (Centers of Excellence) Program from the JSPS (J.H., T.T., and T.I.).
Abbreviations used:
- Arf
ADP-ribosylation factor
- CDR
circular dorsal ruffle
- F-actin
filamentous actin
- GAP
GTPase-activating protein
- PDGF
platelet-derived growth factor
- PH
pleckstrin homology
- PI(3,4,5)P3
phosphatidylinositol 3,4,5-trisphosphate
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0017) on May 9, 2012.
REFERENCES
- Beemiller P, Hoppe AD, Swanson JA. A phosphatidylinositol-3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcgamma receptor-mediated phagocytosis. PLoS Biol. 2006;4:e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- Braun V, Deschamps C, Raposo G, Benaroch P, Benmerah A, Chavrier P, Niedergang F. AP-1 and ARF1 control endosomal dynamics at sites of FcR mediated phagocytosis. Mol Biol Cell. 2007;18:4921–4931. doi: 10.1091/mbc.E07-04-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- Campa F, Yoon HY, Ha VL, Szentpetery Z, Balla T, Randazzo PA. A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J Biol Chem. 2009;284:28069–28083. doi: 10.1074/jbc.M109.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- D'souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Daniele T, Di Tullio G, Santoro M, Turacchio G, De Matteis MA. ARAP1 regulates EGF receptor trafficking and signalling. Traffic. 2008;9:2221–2235. doi: 10.1111/j.1600-0854.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J Biol Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Arfs and membrane lipids: sensing, generating and responding to membrane curvature. Biochem J. 2008;414:e1–e2. doi: 10.1042/BJ20081438. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Gu Z, Noss EH, Hsu VW, Brenner MB. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J Cell Biol. 2011;193:61–70. doi: 10.1083/jcb.201007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha VL, Bharti S, Inoue H, Vass WC, Campa F, Nie Z, de Gramont A, Ward Y, Randazzo PA. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J Biol Chem. 2008;283:14915–14926. doi: 10.1074/jbc.M709717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Tokuda E, Tenno T, Tsujita K, Sawai H, Hiroaki H, Takenawa T, Itoh T. SH3YL1 regulates dorsal ruffle formation by a novel phosphoinositide-binding domain. J Cell Biol. 2011;193:901–916. doi: 10.1083/jcb.201012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J Cell Biol. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SA, Lee ES, Yoon HY, Randazzo PA, Lee ST. PTK6 inhibits down-regulation of EGF receptor through phosphorylation of ARAP1. J Biol Chem. 2010;285:26013–26021. doi: 10.1074/jbc.M109.088971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugmann S, et al. Identification of ARAP3, a novel PI3K effector regulating both Arf and Rho GTPases, by selective capture on phosphoinositide affinity matrices. Mol Cell. 2002;9:95–108. doi: 10.1016/s1097-2765(02)00434-3. [DOI] [PubMed] [Google Scholar]
- Krugmann S, Andrews S, Stephens L, Hawkins PT. ARAP3 is essential for formation of lamellipodia after growth factor stimulation. J Cell Sci. 2006;119:425–432. doi: 10.1242/jcs.02755. [DOI] [PubMed] [Google Scholar]
- Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nat Cell Biol. 2008;10:30–41. doi: 10.1038/ncb1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- Miura K, Jacques KM, Stauffer S, Kubosaki A, Zhu K, Hirsch DS, Resau J, Zheng Y, Randazzo PA. ARAP1: a point of convergence for Arf and Rho signaling. Mol Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Itoh T, Takenawa T. Sequential signals toward podosome formation in NIH-src cells. J Cell Biol. 2008;182:157–169. doi: 10.1083/jcb.200801042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Weller SG, McNiven MA. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 2006;66:3603–3610. doi: 10.1158/0008-5472.CAN-05-2916. [DOI] [PubMed] [Google Scholar]
- Randazzo PA, Andrade J, Miura K, Brown MT, Long YQ, Stauffer S, Roller P, Cooper JA. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc Natl Acad Sci USA. 2000;97:4011–4016. doi: 10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabe H, Onodera Y, Mazaki Y, Hashimoto S. ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr Opin Cell Biol. 2006;18:558–564. doi: 10.1016/j.ceb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Spang A, Shiba Y, Randazzo PA. Arf GAPs: gatekeepers of vesicle generation. FEBS Lett. 2010;584:2646–2651. doi: 10.1016/j.febslet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tan YC, Kreitzer GE, Nakai Y, Shan D, Zheng Y, Huang XY. G proteins G12 and G13 control the dynamic turnover of growth factor-induced dorsal ruffles. J Biol Chem. 2006;281:32660–32667. doi: 10.1074/jbc.M604588200. [DOI] [PubMed] [Google Scholar]
- Yoon HY, Lee JS, Randazzo PA. ARAP1 regulates endocytosis of EGFR. Traffic. 2008;9:2236–2252. doi: 10.1111/j.1600-0854.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HY, Miura K, Cuthbert EJ, Davis KK, Ahvazi B, Casanova JE, Randazzo PA. ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating-protein activity and binding to RhoA-GTP. J Cell Sci. 2006;119:4650–4666. doi: 10.1242/jcs.03237. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Lai T, Koh CG, LeDuc PR, Chiam KH. Investigating circular dorsal ruffles through varying substrate stiffness and mathematical modeling. Biophys J. 2011;101:2122–2130. doi: 10.1016/j.bpj.2011.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.