GTPase-activating proteins (GAPs) control Rab activity along the endomembrane system. In addition to its role in exocytosis, the GAP Msb3 also acts along the endocytic pathway on Rab GTPases Vps21 and Ypt7. The combined data suggest that GAPs can have multiple substrates and can function in a context- or membrane-specific manner.
Abstract
Fusion of organelles in the endomembrane system depends on Rab GTPases that interact with tethering factors before lipid bilayer mixing. In yeast, the Rab5 GTPase Vps21 controls fusion and membrane dynamics between early and late endosomes. Here we identify Msb3/Gyp3 as a specific Vps21 GTPase-activating protein (GAP). Loss of Msb3 results in an accumulation of Vps21 and one of its effectors Vps8, a subunit of the CORVET complex, at the vacuole membrane in vivo. In agreement, Msb3 forms a specific transition complex with Vps21, has the highest activity of all recombinant GAPs for Vps21 in vitro, and is found at vacuoles despite its predominant localization to bud tips and bud necks at the plasma membrane. Surprisingly, Msb3 also inhibits vacuole fusion, which can be rescued by the Ypt7 GDP–GTP exchange factor (GEF), the Mon1–Ccz1 complex. Consistently, msb3∆ vacuoles fuse more efficiently than wild-type vacuoles in vitro, suggesting that GAP can also act on Ypt7. Our data indicate that GAPs such as Msb3 can act on multiple substrates in vivo at both ends of a trafficking pathway. This ensures specificity of the subsequent GEF-mediated activation of the Rab that initiates the next transport event.
INTRODUCTION
Transport of proteins and lipids in the secretory and endocytic transport requires vesicular carriers that form at one membrane and fuse with the acceptor organelle. Fusion relies on a conserved machinery that mediates the initial contact between the two membranes before bilayer mixing mediated by membrane-anchored soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) proteins. For each fusion event, a specific Rab GTPase has been identified. Rabs are switch-like proteins, which exist in an inactive, GDP-bound and an active, GTP-bound form (Barr and Lambright, 2010). Because Rabs are incomplete enzymes, the transition between the two forms depends on specific activating proteins. GDP–GTP exchange factors (GEFs) bind to the GDP-Rab and promote exchange of the GDP for the much more abundant cellular GTP. Only in their GTP form are two specific regions within the Rab, termed switches I and II, stabilized such that interaction partners or effectors can bind the Rab-GTP. On the contrary, GAPs provide a missing arginine to the active site of the Rab, which triggers the otherwise slow GTP hydrolysis rate and inactivates the Rab. In their GDP form, Rabs can then be extracted from membranes by the GDP dissociation inhibitor, which binds the GTPase domain and simultaneously shields the hydrophobic C-terminal dual prenyl anchor (Goody et al., 2005).
The active Rab-GTP binds to several effectors. For fusion reactions, the tethering proteins are the most important interactors. Tethering proteins can come in two forms: as long, coiled-coil proteins like the early endosome protein EEA1 (Vac1 in yeast), or as multisubunit tethering complexes (MTCs; Bröcker et al., 2010; Yu and Hughson, 2010). Either tethering factor is believed to initiate contact between membranes by binding the Rab GTP on one or both membranes that will subsequently fuse, which is supported by our structural data on HOPS and Ypt7 (Bröcker et al., 2012). MTCs are believed to combine Rab binding and membrane tethering with the chaperoning of SNAREs.
Several GEFs and GAPs have been identified. Whereas GEFs have escaped identification due to their variable structure, GAPs have a defined TBC (Tre2/Bub2/Cdc16) domain, which contains the aforementioned “arginine” and “glutamine” finger, and were identified on the sequence level early on. Despite their early identification in yeast, it has been challenging to match each GAP to its corresponding Rab, because the isolated TBC domain often revealed overlapping activities for multiple Rabs (Albert and Gallwitz, 1999; Albert et al., 1999). However, functional and in vivo studies identified Gyp7 as the GAP for Ypt7 and additional Rab–GAP pairs (Du and Novick, 2001; Gao et al., 2003; Sciorra et al., 2005; Brett et al., 2008). A series of studies identified GAPs for mammalian Rabs regulating a range of trafficking pathways, including RabGAP5 in endocytosis (Haas et al., 2005; Fuchs et al., 2007; Yoshimura et al., 2007).
We are interested in the control of the endocytic pathway and previously identified the CORVET complex as a novel effector of the Rab5 GTPase Vps21 (Peplowska et al., 2007). Vps21 is activated by the Rabex5 homologue Vps9 during endocytosis or on early endosomes (Hama et al., 1999). We and others postulated that endosomal maturation includes an exchange of Vps21 by the Rab7-like Ypt7, which is required for fusion of late endosomes with the vacuole (Rink et al., 2005; Spang, 2009; Abenza et al., 2010; Huotari and Helenius, 2011; Lachmann et al., 2011). Such a cascade should include the inactivation of Vps21 by a GAP and its release from membranes once Ypt7 has been converted to the GTP form. Indeed, evidence for such a cascade exists for the exocytic pathway (Rivera-Molina and Novick, 2009). However, even though the mammalian and the Caenorhabditis elegans Rab5 GAP is known, the equivalent in yeast is lacking. Here we identify Msb3 as a specific Vps21 GAP that binds Vps21 and is required for Vps21 recycling from the endosome. Msb3 is localized primarily to the plasma membrane and cytosol and was previously assigned to the exocytic Rab Sec4 together with its homologue Msb4 (Albert et al., 1999; Albert and Gallwitz, 2000; Gao et al., 2003). Because Msb3 can also inactivate Ypt7, our combined data argue for an additional role of Msb3 in the endocytic pathway.
RESULTS
Identification of the Vps21 GAP
Yeast contains 11 Rabs and eight GAPs, indicating a mismatch and potential overlapping functions of the respective GAPs. By sequence comparison, three yeast GAPs—Mdr1/Gyp2, Msb3/Gyp3, and Msb4/Gyp4—fall in the same clade as mammalian RabGAP-5 and C. elegans TBC-2 and could be potential homologues that act on Vps21 (Gao et al., 2008). Previous biochemical studies often used truncated GAPs or proteins extracted from yeast to determine specificity. However, the isolated TBC domain of Gyp1 is, for instance, an efficient GAP of many Rabs, including Ypt7 (Eitzen et al., 2000), which indicates the challenge in defining Rab–GAP pairs.
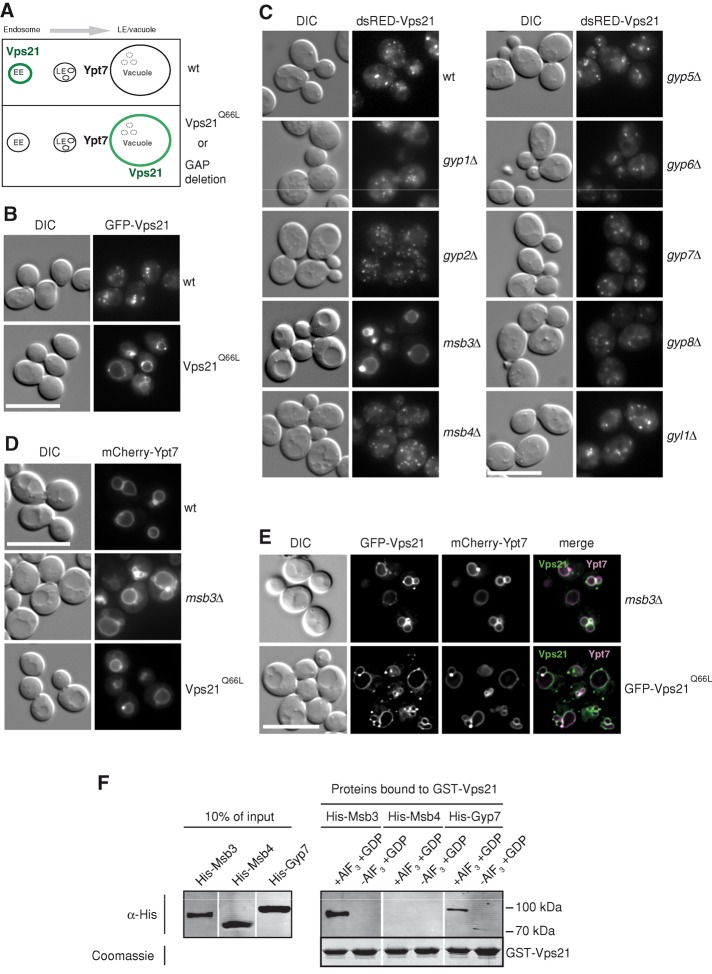
We thus devised an assay to identify the Vps21-specific Rab. We reasoned that a permanently activated Rab should not be extracted by Gdi1 and thus migrate to the next organelle, that is, the vacuole for Vps21 (Figure 1A). As predicted, the GTP-locked Vps21 Q66L indeed accumulated on the vacuole rim, whereas green fluorescent protein (GFP)– or red fluorescent protein–tagged wild-type Vps21 was present in multiple endosomal dots (Markgraf et al., 2009; Figure 1B). Similarly, we speculated that Vps21 should be found on the vacuole if the specific GAP were missing. We therefore analyzed all GAP deletions. Only in the msb3∆ strain was Vps21 found on the vacuole (Figure 1C), thus mimicking the GTP-locked Vps21 version analyzed before (Figure 1B). The shift of Vps21 to the vacuole, either in msb3∆ or in the Vps21 Q66L cells, did not affect the vacuole localization of Ypt7 (Figure 1D). Loss of Msb3 thus results in a compartment with two active Rabs, which points to a missing inactivation of the upstream Vps21 (Figure 1E).
FIGURE 1:
Identification of Msb3 as a Vps21-specific GAP. (A) A screen to identify a Vps21-specific GAP in yeast. In wild type, Vps21 is found in endosomes, whereas it is expected to shift to the vacuole if maintained in the GTP form. See text for details. (B) Localization of GTP-locked Vps21 to the vacuole. GFP-tagged Vps21 and its Q66L variant were introduced into a vps21∆ strain by integration of a linearized plasmid in which Vps21 is under the control of the NOP1 promotor. Yeast cells were monitored by fluorescence microscopy. DIC, differential interference contrast. (C) Localization of Vps21 in all GAP deletion strains. The indicated strains were transformed with a single-copy (CEN) plasmid encoding an additional copy of N-terminal dsRED-tagged Vps21 and analyzed as in A. (D) Ypt7 localization. mCherry-tagged Ypt7 was expressed as an additional copy in wild type, msb3∆, and Vps21Q66L strains and analyzed as in A. (E) Colocalization of GFP-Vps21 and mCherry-Ypt7 in the indicated strains by fluorescence microscopy. The grayscale pictures were two-dimensionally deconvolved with AutoQuant X software to reduce background fluorescence and were combined to an overlay with the GFP signal in the green channel and the mCherry signal in the red and blue channel. Merged signals can be observed in white. (F) In vitro interaction of Vps21 with GAPs. Purified His-Msb3, His-Msb4, or His-Gyp7 was incubated in the presence or absence of AlF3 with GST-Vps21, which was preloaded with GDP and coupled to GSH agarose. Bound proteins were eluted by boiling, resolved on SDS–PAGE, and detected with antibodies to the histidine tag (see Materials and Methods for details). A Coomassie-stained loading control of GST-Vps21 is shown.
Msb3 and its homologue Msb4 have been assigned to the exocytic Rab Sec4 at the plasma membrane (Gao et al., 2003). We therefore used an alternative assay to determine the specific interaction of Msb3 with Vps21 (Figure 1F). During activation of GTP hydrolysis the Rab binds to the GAP via the arginine-finger motif (Pan et al., 2006). For the Ras-GTPase, this interaction was previously stabilized by the addition of GDP and AlF3, which mimics a transition state in the hydrolysis reaction and is sufficiently strong to crystallize a Ras–GAP and Rab–GAP complex (Scheffzek et al., 1997; Pan et al., 2006). We then incubated immobilized glutathione S-transferase (GST)–Vps21 in the GDP form with different recombinantly purified GAPs either in the absence or presence of AlF3 and analyzed the corresponding eluates. Of importance, Msb3 was strongly retained with Vps21-GDP only in the presence of AlF3, whereas neither its homologue Msb4 nor Gyp7 bound significantly to Vps21 (Figure 1F).
Vps21-GTP on vacuoles retains its effector Vps8
Our data did not clarify whether Vps21 on the vacuole was indeed active. Along the endocytic pathway, Vps21-GTP interacts with the early endosomal Vac1 protein and the CORVET subunits Vps8 and Vps3 (Tall et al., 1999; Markgraf et al., 2009; Plemel et al., 2011). We previously observed that elevated Vps21 levels affected the localization of Vps8 but not Vps3 (Markgraf et al., 2009). We therefore asked whether either of the two interactors would shift to the vacuole with Vps21 (which we assume to be in its GTP-loaded state) due to prolonged binding. We thus expressed Vps8 as an additional GFP-tagged copy in msb3∆ or Vps21 Q66L cells and observed that Vps8 clearly followed Vps21 to the vacuole (Figure 2A). We previously showed that the GEF of Ypt7—the Mon1–Ccz1 complex—is mislocalized in cells lacking Vps21 and Vps8 (Nordmann et al., 2010). This observation suggested cross-talk between Vps21 and the Mon1–Ccz1 complex as the activator of the downstream Ypt7 Rab in the process of endosome maturation. When we analyzed functional GFP-tagged Mon1, we observed more protein on vacuoles of msb3∆ and Vps21 Q66L–expressing cells (Figure 2B). This indicates that active Vps21 also affects Mon1–Ccz1 localization, either by directly binding Mon1–Ccz1 or by affecting the endosomal surface composition, to relocate Vps21 to the vacuole, in agreement with our model (Nordmann et al., 2010).
FIGURE 2:
Effect of msb3 deletion and mutants on the localization of Vps21 and effectors. (A–E) Wild-type, msb3∆, and Vps21 Q66L strains were transformed with a single-copy CEN plasmid expressing Vps8-GFP under the control of a NOP1 promotor (A), dsRED-Pep12 under the control of a PHO5 promotor (D), and Vps3-GFP under the control of a NOP1 promotor (E). Mon1 (B) and Vac1 (C) were genomically tagged with GFP or tomato, respectively. All strains were analyzed by fluorescence microscopy (see Materials and Methods). (F) Localization of GFP-tagged Msb3 and dsRED-tagged Vps21 was analyzed by fluorescence microscopy. (G) Localization of Vps21 in Msb3 active-site mutants. Variations of MSB3 were generated by QuikChange mutagenesis and stably integrated in the yeast genome of msb3∆ strains via a pRS shuttle vector. The proteins were expressed as GFP fusions under the control of the NOP1 promotor. dsRED-Vps21 was expressed from a single-copy CEN plasmid under the control of the PHO5 promotor. Cells were analyzed by fluorescence microscopy as before. DIC, differential interference contrast.
In contrast, the localization of the endosomal Vps21 effector Vac1 was unaffected, probably due to the restricted binding of Vac1 to endosomal phosphoinositol-3-phosphate via its FYVE domain (Figure 2C). Similarly, endosomes as marked by Pep12 did not change in msb3∆ cells. These controls point to a functional recycling of endosomal factors and no increased unspecific fusion of the endosome with the vacuole (Figure 2D). Of interest, the loss of Msb3 had no effect on Vps3 (Figure 2E), even though it is part of the CORVET complex (Peplowska et al., 2007), also interacts with Vps21-GTP in vitro (Plemel et al., 2011), and requires Vps21 for localization (unpublished data). This suggests that the functional cooperation of the two Rab-specific subunits Vps3 and Vps8 with the Rab5 Vps21 differs in vivo.
Msb3 requires its conserved arginine to affect Vps21 localization
To analyze Msb3 in vivo, we first examined the functionality of the tagged protein using the Vps21 localization (Figure 1, A and B) as a readout. As shown in Figure 2F, Msb3 tagged at either the N- or the C-terminus with GFP localized to the plasma membrane, the bud neck, and cytosol, in agreement with previous studies (Gao et al., 2003; Tcheperegine et al., 2005). Moreover, Vps21 localized as in wild-type cells if Msb3 was tagged, indicating that tagged Msb3 is functional.
Msb3 binds to many proteins of the polarisome, as well as to regulators of cytoskeleton organization (Tcheperegine et al., 2005). We therefore asked whether the msb3 deletion could have a secondary effect on Vps21 localization, and we therefore generated active-site point mutants. A replacement of the arginine finger is considered to be sufficient to disrupt GAP activity (Haas et al., 2005). For Msb3, a mutation of R282 to lysine or phenylalanine yielded an Msb3 variant that was correctly localized but behaved like msb3∆ for Vps21 localization (Figure 2G). We thus conclude that the GAP activity of Msb3 is required for correct Vps21 localization by inactivation and recycling of the Rab GTPase during maturation.
In vitro activity of yeast GAPs on individual Rabs
We next tested whether we could find the same specificity of Msb3 for Vps21 in vitro. For this purpose, we isolated all recombinant full-length Rabs and GAPs (except for Gyp5) to determine their relative specificity in an in vitro GAP assay that monitors phosphate release after GTP hydrolysis. Here we focus on the three relevant Rabs for our study: Vps21, Sec4, and Ypt7 (Figure 3). All GAP–Rab data are summarized in Table 1. To compare the specificity of all GAPs for a particular Rab, we set the entire hydrolysis rate within the assay as 100% and plot all GAP–Rab hydrolysis rates in relation to this value. Within our data set, we largely confirmed the previously reported GAP values (Figure 3C; Albert et al., 2000; see also Brett et al., 2008, and references therein). Of importance, the most active GAP for Vps21/Ypt51 and its homologue Ypt52 was again Msb3 (Figure 3A), whereas the homologous Ypt53 protein was unaffected by the same GAP. Moreover, our MBP-Gyp8 construct showed very high activity on many Rabs and had the highest relative activity on Ypt7 (Figure 3B) but acted poorly on Vps21, in support of a specific Msb3–Vps21 interaction for Vps21 inactivation. Our data nicely fit with the role of Msb3 in Vps21 localization and its interaction in vitro (Figure 1) and are also in good agreement with a previous study (Albert and Gallwitz, 1999). As also noticed before, GAPs can act on multiple substrates, and Msb3 also shows strong activity for Sec4 but none for Ypt7 (Figure 3, B and C). Gyp2 acts equally well as Msb3 on Sec4 (Figure 3C), whereas Msb4 shows no activity in our assays. Surprisingly, the identified GAP of Ypt7, Gyp7, also does not show any activity for Ypt7 in our in vitro assay. It has been reported that the full-length Gyp7 is in general of poor activity, and therefore truncated versions were preferred for in vitro assays (Albert and Gallwitz, 1999; Eitzen et al., 2000). It should also be noted that Gyp8 has a transmembrane domain and is believed to be an endoplasmic reticulum (ER) protein, which would restrict it to Rabs that can bind to the ER. We thus consider it likely that some GAPs require the right membrane context for Rab inactivation.
FIGURE 3:
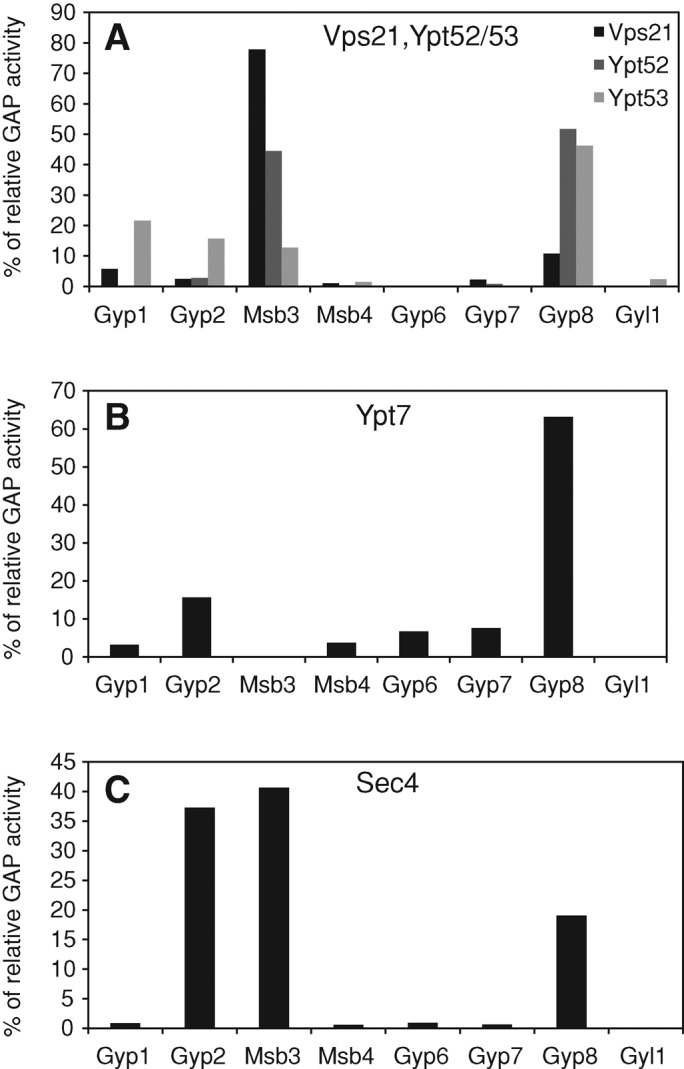
Activity of Msb3 and its interaction with Vps21. (A–C) Relative GTP-hydrolysis activity of the analyzed Rab–GAP pairs. All GAPs and Rabs were purified as full-length proteins. Rabs were precharged with [γ-32P]GTP, and release of radioactive 32P was analyzed over time as described in Materials and Methods. The entire GTP-hydrolysis of the respective Rab in all reactions with GAPs was set to 100%, and all Rab–GAP values are shown as a fraction of this hydrolysis rate. Each value was determined in duplicate and in three repetitions. All other GAP activities for each Rab are shown in Table 1.
TABLE 1:
Summary of in vitro GAP assays.
| Rab/GAP | Gyp1 | Gyp2 | Msb3 | Msb4 | Gyp6 | Gyp7 | Gyp8 | Gyl1 |
|---|---|---|---|---|---|---|---|---|
| Ypt1 | 6.1 | 0.6 | 6.2 | 6.5 | 4.8 | 0 | 48 | 0 |
| Ypt6 | 0 | 9.5 | 7.4 | 0 | 100/129 | 0 | 25 | 67 |
| Ypt7 | 3.5 | 3.1 | 0 | 100/118 | 20 | 7 | 25 | 0 |
| Ypt10 | 100/349 | 17 | 35 | 68 | 0 | 100/432 | 0 | 0 |
| Ypt11 | 0 | 6.3 | 16 | 8.8 | 0 | 0.5 | 0 | 52 |
| Ypt31 | 0 | 1.1 | 4.3 | 3.3 | 0 | 0 | 5.4 | 0 |
| Ypt32 | 0 | 2.4 | 9.0 | 7.7 | 0 | 0 | 12 | 0 |
| Vps21/Ypt51 | 13 | 1.0 | 30 | 1.9 | 0 | 4.2 | 8.8 | 0 |
| Ypt52 | 0 | 2.3 | 34 | 0 | 0 | 3.0 | 85 | 0 |
| Ypt53 | 4.1 | 0.5 | 0 | 0 | 0 | 0 | 3.0 | 32 |
| Sec4 | 13 | 100/1952 | 100/2200 | 32 | 38 | 7.6 | 100/999 | 0 |
| Tem1 | 1.2 | 0 | 0 | 0 | n.d. | 0 | 0 | 100/5 |
A GAP screen of all Rab GTPases. The respective GAP was added to all purified Rabs that were precharged with [γ-32P] GTP, and release of radioactive 32P was analyzed over time as described in Materials and Methods. For every GAP the maximal stimulation of GTP hydrolysis reached in this assay was set here to 100%, and all other activities were referred to this. The total amount of hydrolyzed GTP (pmol/h) at maximal activity is noted on the right side of the slash. We focused on Vps21, and these results are depicted in bold letters. Gyl1 has a TBC domain lacking the critical arginine and served as a negative control for this assay. Note that the values in Figure 3 are shown as relative GTP hydrolysis rates.
Msb3 interacts with Vps21 in vivo
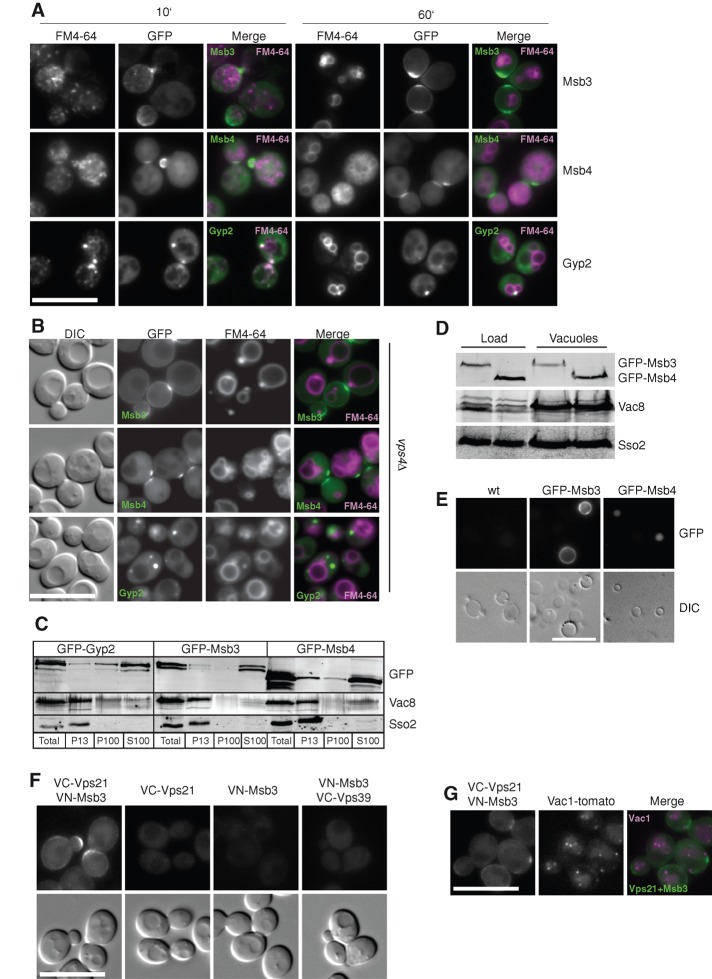
We were surprised that a deletion or mutation of Msb3 caused such a strong defect on localization of Vps21 and its effector Vps8. In wild-type cells, the majority of Msb3 is localized to the plasma membrane, and here in particular to the bud tip and neck (Figure 4A), even though we also detect a large cytosolic portion in vivo (Figure 4, A–C). It is possible that Msb3 was transiently present on FM4-64–positive endosomal structures. We thus followed cells labeled with this lipophilic dye, which passes through the endocytic pathway to the vacuole, but did not detect any significant colabeling. Similarly, the accumulation of MVBs in the vps4 mutant, which leads to an accumulation of several endocytic proteins, such as Vps21 or endosomal sorting complex required for transport (ESCRT) subunits, did not affect Msb3 localization (Figure 4B). Similar results were obtained for Msb4. Of interest, a control GAP protein, Gyp2, was found in FM4-64–positive structures (Figure 4A) and partially accumulated in endosomes of the vps4 mutant (Figure 4B), which suggests a role in endosomal transport, most likely in recyling from endosomes to the Golgi, as described previously (Lafourcade et al., 2003).
FIGURE 4:
Colocalization of Msb3 and Vps21 in vivo. (A) Colocalization of GAPs with endosomal membranes. Cells expressing the indicated GFP-tagged GAPs were labeled with FM4-64 and washed, and the dye was monitored after 10 and 60 min by fluorescence microscopy. (B) Localization of GAPs in the vps4 deletion. FM4-64 labeling and microscopy of the indicated strains was done as before. (C) Subcellular fractionation of cells expressing GFP-tagged Gyp2, Msb3, and Msb4. Cells of the indicated strains were lysed as described in Materials and Methods, and proteins were separated into a low-speed (P13) and a high-speed (P100) pellet. The pellets and the supernatants of the high-speed pellets (S100) were subjected to TCA precipitation and analyzed by Western blotting against the GFP tag. The vacuolar membrane protein Vac8 and the plasma membrane marker Sso2 served as control. We assume that the additional bands of Msb4 and Msb3 represent degradation products that appear due to the preparation, although we cannot exclude possible posttranslational modifications. (D, E) Analysis of Msb3 and Msb4 in isolated vacuole fractions. Vacuoles were purified from BY strains carrying GFP-tagged Msb3 and Msb4 (see Materials and Methods), and 40 μg of the isolated vacuoles was analyzed by SDS–PAGE and Western blotting against the GFP tag (D). In addition, vacuoles were directly analyzed by fluorescence microscopy (E). (F) Split-YFP analysis of Vps21 and Msb3. Vps21 and Msb3 were N-terminally tagged with the C-terminal (VC) or N-terminal (VN) Venus fragment, respectively (see Materials and Methods). Individually expressed proteins, as well as both in the same strain, were monitored by fluorescence microscopy. N-Terminally tagged VC-Vps39, together with VN-Msb3, was included as a control for unspecific binding. (G) Colocalization of the Vps21-Msb3 signal with genomically tomato-tagged Vac1 at its C-terminus by fluorescence microscopy.
Our data did not, however, exclude an additional role of Msb3 in the endocytic pathway due to a transient interaction with Vps21. We therefore selected two additional approaches. First, we examined cells with GFP-tagged GAPs by subcellular fractionation (Figure 4C). Most of the GAPs were found in the cytosol, consistent with our in vivo analysis, although a small fraction of each GAP was also recovered in the vacuole-containing P13 fraction (Figure 4C). In addition, Gyp2 was enriched in the P100 fraction, indicative of an endosomal pool (Figure 4B). For the P13 fraction, we cannot exclude the plasma membrane as one origin of the detected Msb3, since we found the marker protein Sso2 in this fraction as well. However, Msb3 is present in a large cytosolic pool, which could have access to the endocytic pathway. To examine this in more detail, we isolated vacuoles and vacuole-associated organelles by gradient flotation and analyzed protein fractions for the presence of selected GAPs. As shown in Figure 4D, Msb3 and Msb4 were copurified with vacuoles, suggesting that a portion of Msb3 is associated with vacuoles or vacuole-associated endosomes. For Gyp2, we observed strong degradation in the vacuole-enriched fraction (unpublished data). However, due to the impurity of the preparation with portions of the plasma membrane, we also detected Sso2 in the floated vacuole fraction. Therefore, we also analyzed the same vacuoles by fluorescence microscopy and found Msb3 at the vacuole membrane (Figure 4E). This clearly indicates that the endosomal and vacuole population of Msb3 is likely masked by the cytosolic background and the predominant plasma membrane pool when we localize Msb3 in vivo (Figure 2, F and G) or biochemically (Figure 4, C and D). In contrast, GFP-Msb4 was found in the vacuolar lumen, possibly due to its degradation in the vacuole.
Second, we used the split-YFP system (Sung and Huh, 2007) to ask whether Vps21 encounters Msb3 in vivo. When either protein was fused to the YFP-half, no fluorescent signal was observed. However, coexpression of both resulted in a clear signal close to the plasma membrane (Figure 4F), suggesting that Vps21 and Msb3 can interact in vivo. We were surprised to find this interaction at the plasma membrane in patches where Msb3 normally localizes instead of seeing it relocalized to the endosomes. To exclude unspecific binding, we used a Vps39 construct fused to one-half of YFP, which interacts efficiently with the Ypt7 that carries the opposite half (unpublished data). When this Vps39 construct was expressed together with tagged Msb3, we observed no interaction (Figure 4F), indicating that the Vps21–Msb3 interaction is specific. We then asked whether the split-YFP would stabilize the interaction of Vps21 and Msb3 and therefore cluster the endosomal compartment at the plasma membrane. When we colocalized the Msb3–Vps21 interaction site with Vac1, we observed a clear separation, indicating that no unspecific recruitment of Vps21 together with endosomes occurred (Figure 4G). We conclude that Msb3 and Vps21 do interact transiently in vivo and that an active fraction of Msb3, but not Msb4, is found on endosomes and vacuoles.
Msb3 deletion does not affect protein trafficking to the vacuole
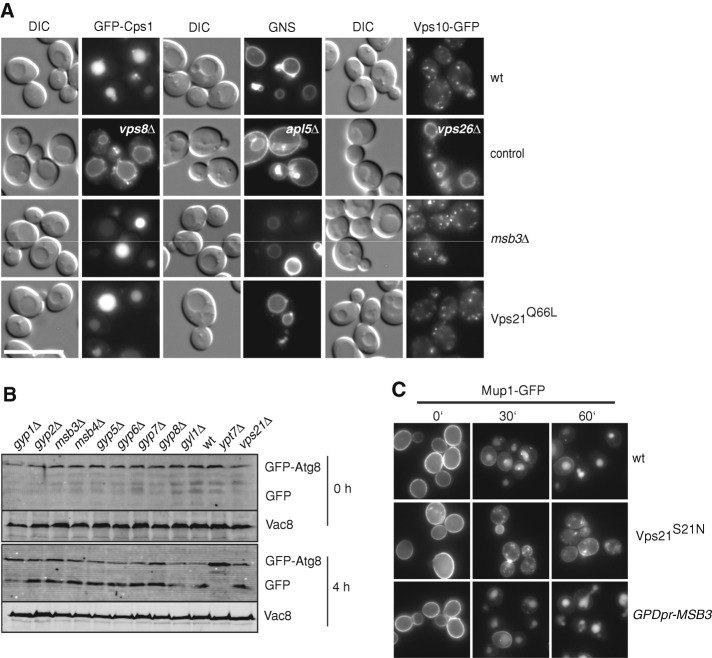
The Msb3 deletion has a clear effect on the relocalization of Vps21 to the vacuole, although it had not been detected in any previous screen as a protein involved in endocytic trafficking. We decided to readdress this issue by analyzing central trafficking reactions. Initially, we followed sorting of GFP-tagged Cps1, a substrate of the ESCRT pathway at the endosome, which is sorted to the vacuole lumen in wild-type cells. In ESCRT deletions (Odorizzi et al., 1998), but also in vps21∆ cells (unpublished data), GFP-Cps1 is found on the vacuole rim. However, neither in msb3∆ cells nor in cells expressing only activated Vps21 Q66L did we detect any defect in sorting (Figure 5A). Similarly, the AP-3 pathway as marked by the GFP-Nyv1-Snc1 fusion protein (Reggiori et al., 2000) or the retrograde transport from the Golgi, which we followed by tagging the CPY-receptor Vps10 with GFP, was unaffected in both mutants (Figure 5A). In addition, we observed no defect in autophagy, which we followed by GFP-Atg8 processing (Figure 5B). The latter assay relies on the clipping of Atg8 to free GFP, which is observed in wild-type (wt) but not in the absence of the vacuolar Ypt7 Rab GTPase (Figure 5B). Of interest, GAP proteins have been implicated in mammalian autophagy (Behrends et al., 2010), although we have not yet found evidence of this.
FIGURE 5:
Protein sorting in msb3∆ or overproduction strains. (A) Analysis of biosynthetic cargo, AP-3, and retrograde cargo to the vacuole. Wild-type, msb3∆, and Vps21 Q66L strains were transformed with a Cen-plasmid encoding an additional copy of N-terminal GFP-tagged Cps1, the GNS construct (GFP-Nyv1-Snc1; Reggiori et al., 2000), or the C-terminal GFP-tagged Vps10. As a positive control, we analyzed marker constructs in deletion strains. Vps8 is a class D subunit of the CORVET tethering complex, and its deletion causes Cps1 missorting to the vacuole membrane. Apl5 is a subunit of the AP-3 protein coat, causing missorting of the GNS construct to the plasma membrane upon deletion (Reggiori et al., 2000), and Vps10-GFP is missorted if retrograde transport is interrupted as in the vps26 mutant, a subunit of the retromer. (B) Effect of GAP deletions on macroautophagy. GAP deletions were transformed with a CEN plasmid harboring GFP-Atg8, and the cells were grown to mid exponential phase in a synthetic full medium lacking uracil. The cells were shifted to synthetic starvation medium and harvested at indicated time points, and protein extract was analyzed on a Western blot using anti-GFP antibodies. (C) Sorting of endocytic cargo to the vacuole lumen. The methionine transporter Mup1 was GFP tagged in wt cells, cells expressing inactive Vps21 S22N, or those carrying MSB3 under the control of the strong GPD promoter (Janke et al., 2004). Cells were grown in the absence of methionine (0 min). Methionine was added to a final concentration of 20 μg/ml, and Mup1-GFP was monitored immediately and 30 or 60 min thereafter.
Finally, we analyzed trafficking of the methionine transporter Mup1 from the plasma membrane. In the absence of methionine, the transporter is stable at the plasma membrane and is sorted to the vacuole in the presence of the amino acid (Figure 5C). This transport is defective if Vps21 is inactive, as shown for the Vps21 S21N mutant (Figure 5C). If Msb3 were overexpressed, we would expect a similar effect. However, sorting appeared unaffected if Msb3 expression was driven from the GPD promoter, suggesting that Vps21 was not completely inactivated under these conditions. We thus conclude that the mislocalization of Vps21 to the vacuole in msb3∆ cells does not affect protein trafficking to the vacuole and that Msb3 overexpression does not affect the overall Vps21 function.
Msb3 acts on both Ypt7 and Vps21
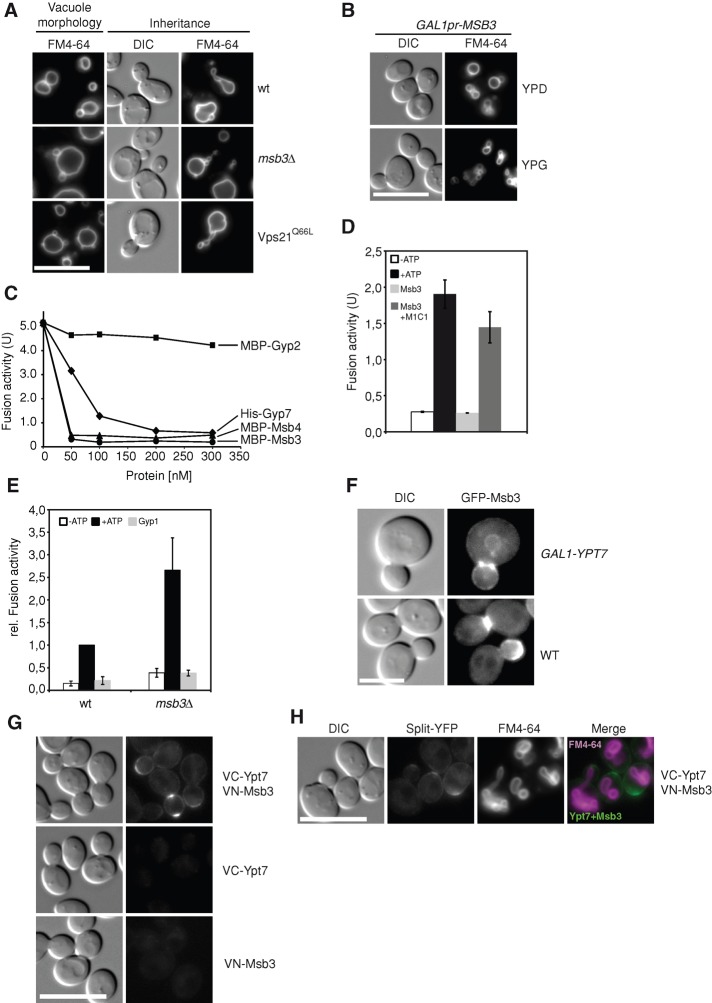
During our in vivo analyses, we noticed that vacuoles in msb3∆ and Vps21 Q66L mutants were enlarged and often showed very small structures adjacent to the main vacuole structure, reminiscent of the described class F phenotype (Raymond et al., 1992). Whereas wild-type vacuoles are normally stretched toward the daughter cell during inheritance, both mutants displayed fragmented inheritance structures, although the process itself is functional (Figure 6A). The alteration in morphology thus suggested a shift in the fusion and fission equilibrium and a potential role of Msb3 in vacuole biogenesis.
FIGURE 6:
Influence of Msb3 on vacuole morphology and fusion. (A) Typical vacuole morphology and morphological structures during inheritance. Vacuoles of wild-type, msb3∆, and Vps21 Q66L strains from a BY background (vacuole morphology) and a BJ background (inheritance) were labeled with FM4-64 as before and analyzed by fluorescence microscopy. BJ vacuoles have consistently large vacuoles and thus facilitate the scoring of possible inheritance defects. (B) Vacuole morphology upon Msb3 overexpression. Cells without (YPD) or with overexpressed (YPG) Msb3 were stained with FM4-64 and monitored by fluorescence microscopy. (C) Titration of purified GAPs into the vacuole fusion reaction. Vacuoles from the two tester strains were isolated as described in Materials and Methods and incubated in the presence of ATP and the indicated amounts of recombinant Gyp2 or Msb3. Fusion was determined after 90 min at 26°C (see Materials and Methods). (D) Recovery of fusion activity by Mon1–Ccz1. Fusion of wild-type vacuoles was done as in C. Where indicated, 0.5 μM Msb3 was added, and Mon1–Ccz1 (300 nM; Nordmann et al., 2010) was included in the presence of the same amounts of Msb3. (E) Fusion activity of msb3∆ vacuoles. Fusion of wild-type tester vacuoles was performed in parallel to tester vacuoles without Msb3. Gyp1-46 (0.5 μM) was added where indicated. (F) Localization of Msb3 in the absence and presence of overproduced Ypt7. Ypt7 was overproduced from the GAL1-promoter in cells expressing GFP-tagged Msb3. Cells were monitored by fluorescence microscopy. (G) Split-YFP analysis of Ypt7 and Msb3. Ypt7 and Msb3 were N-terminally tagged with the C-terminal (VC) or N-terminal (VN) Venus fragment, respectively (see Materials and Methods). Individually expressed proteins, as well as both in the same strain, were monitored by fluorescence microscopy. (H) Colocalization of the Ypt7-Msb3 signal with vacuoles. Vacuoles of a strain expressing both VC-Ypt7 andVN-Msb3 were labeled with FM4-64 as before and analyzed by fluorescence microscopy.
In agreement with this observation in the msb3∆ mutant, it was previously observed that overexpression of Msb3 or Msb4 interferes with vacuole morphology (Brett et al., 2008), which we could reproduce when Msb3 was placed under the control of the strong GAL1 promoter (Figure 6B). We then wondered how we could explain that the overexpression of Msb3 caused a vacuole morphology defect, and we considered the possibility that Msb3 might also affect Ypt7. We therefore titrated Msb3 into the vacuole fusion reaction, which uses isolated vacuoles from two tester strains. Lumenal mixing of the two vacuole types results in activation of immature alkaline phosphatase present in one type by the protease of the other type (Cabrera and Ungermann, 2008). Of interest, Msb3 addition strongly inhibited vacuole fusion, whereas similar amounts of highly active Gyp2 (Figure 3B) were far less active in this assay (Figure 6C). We asked whether it was indeed Ypt7 that was inactivated by Msb3. As shown in Figure 6D, the Ypt7-specific GEF, the Mon1–Ccz1 complex, rescued fusion activity of vacuoles that were pretreated with Msb3, showing that Msb3 acts on Ypt7. This was surprising, as Msb3 had almost no activity on Ypt7 in vitro (Figure 3; Brett et al., 2008). We therefore consider it likely that GAPs such as Msb3 act in a context-specific manner on organellar membranes and might require activation to act on Ypt7. If Msb3 were a GAP for both Ypt7 and Vps21, we would expect that vacuoles fuse better in its absence. This was indeed observed (Figure 6E). Vacuoles obtained from msb3∆ cells were about double as fusogenic than the corresponding wild-type vacuoles, and both vacuole types were sensitive to the general fusion inhibitor Gyp1-46. Our data thus suggest that Msb3 can act on Vps21 and Ypt7 along the endocytic pathway to dampen Rab activity and promote membrane fusion specificity and dynamics.
We finally asked whether we also could observe an interaction of Msb3 and Ypt7 in vivo. We reasoned that an excess of Ypt7 should affect the relative localization of Msb3. Indeed, we could shift a portion of GFP-tagged Msb3 to the vacuole by overexpressed Ypt7 (Figure 6F). To analyze the interaction of both proteins further, we used the split-YFP assay. As for Vps21, we observed an interaction of the two proteins at the plasma membrane (Figure 6G). In addition, both proteins colocalized at some vacuoles during inheritance (Figure 6H), although inheritance was not affected in the msb3 mutant, as discussed. In sum, we take the combined findings as an indication of an encounter of Msb3 GAP and two Rabs, Vps21 and Ypt7, within the endocytic pathway, which is consistent with our in vitro interactions (Figure 1) and in vivo localization of Vps21 in msb3∆ (Figure 2).
DISCUSSION
On the basis of an extensive mapping of all yeast full-length GAPs versus all Rabs and a comparison of Vps21 localization in all GAP deletion strains, we were able to identify Msb3, but not its homologue Msb4, as a Vps21-specific GAP. We demonstrated the specificity at several levels. First, Msb3 interacts with Vps21-GDP in the presence of AlF3, which stabilizes the transition state of GTP hydrolysis. Second, Msb3 binds Vps21 in vivo. Third, in the absence of Msb3, activated Vps21 remains on endosomes, is translocated to the vacuole, and can interact there with its effector Vps8. This is a specific effect that was only observed for the Msb3 GAP deletion. Finally, Msb3 can also inactivate Ypt7 on isolated vacuoles (Figure 6) and presumably in vivo (Figure 6B; Brett et al., 2008)), suggesting that the Rab GAPs have a broader specificity in the cell than previously anticipated. In agreement with our study, Merz and colleagues identified Msb3 independently as the Vps21 GAP, using a completely different approach of endosomal signaling activity as a read-out (Nickerson et al., 2012).
Msb3 takes part in the organization of polarized growth on several levels by regulating exocytosis via its activity on Sec4, interacting with and possibly coordinating Rho GTPases, components of the polarisome, and the actin cytoskeleton (Bi et al., 2000; Gao et al., 2003; Tcheperegine et al., 2005). Msb3 was thus not our primary candidate for a Vps21 GAP. Like its homologue Msb4, Msb3 is primarily found at the plasma membrane, and here in particular at the growing bud tip and bud neck (Gao et al., 2003; this study). The predominant localization of Msb3 at the plasma membrane suggested that it also acts predominantly there, which is most likely true for the Sec4 inactivation (Gao et al., 2003). The situation is likely different for Vps21 and Ypt7. Msb3 is partially cytosolic, suggesting that it could be transiently recruited to selected organelles such as endosomes to inactivate Vps21. Such activities are known as “moonlighting” and reflect additional functions of a specific enzyme, and they likely apply to Msb3. Consistent with this notion, we identified a portion of Msb3 on isolated vacuoles, although the primary Vps21–Msb3 interaction as measured by the split-YFP assay occurred proximal to the plasma membrane. We cannot be certain that the interaction of Vps21 or Ypt7 with Msb3 as detected by split-YFP corresponds to the enzymatic Rab–GAP intermediate. Such a complex should form only very transiently, consistent with the very weak interaction of Rabs and GAPs with affinities in the micromolar range (Albert and Gallwitz, 1999; Albert et al., 1999). As such, the split-YFP interaction of Msb3 and Vps21 or Ypt7 at the plasma membrane may correspond to a prolonged interaction of the Rab and GAP mediated by the two halves of the fluorophore. With regard to this, the Rab GTPase may then follow Msb3 to the plasma membrane before being recycled very slowly. It is thus possible that this finding does not represent the physiological place of interaction. How Msb3 then diverts its activities also to the late endosome and vacuole is not clear.
Our data on the GAP localization are not without precedent. No data on the direct physical interaction of a Rab GTPase with its GAP in living cells have been available, and, along the same lines, some Rab–GAP pairs seem to localize at different compartments. For instance, the mammalian Rab43 localizes to the Golgi and endoplasmic reticulum to regulate retrograde transport from the endocytic pathway to the Golgi and has an additional function in the effective uptake of Shigella toxin at the plasma membrane (Fuchs et al., 2007; Haas et al., 2007). Of interest, the Rab43-specific GAP RN-tre localizes mainly to the cell surface, possibly implicating a similar mechanism. RN-tre has also been proposed to be a GAP for Rab5 (Lanzetti et al., 2004), which then would raise exactly the same problem of the Rab5–GAP interaction site.
We noticed that only massive overproduction of Msb3 resulted in impaired vacuole biogenesis, and overproduction from a constitutive promoter did not interfere with the endocytosis of the methionine transporter Mup1. At this stage, we can exclude a redundant function of the Vps21 paralogues Ypt52 and Ypt53, since we analyzed endocytosis after Msb3 overproduction also in a double-deletion mutant (unpublished data). It is possible that an additional factor that regulates Msb3 localization is thus limiting to locally inactivate Vps21.
Our data suggest that Msb3 is not the only GAP that acts in the endocytic pathway. Gyp2 is also found in the same clade of Rab5-GAP–like proteins with Msb3 and Msb4 (Gao et al., 2008), affects vacuole morphology if overproduced (Brett et al., 2008), and accumulates at late endosomes in vps4∆ mutants. However, the inhibition of Gyp2 of in vitro vacuole fusion was by far less efficient than the activity of Msb3. It is thus possible that Gyp2 cooperates with Msb3 along the endocytic pathway, even though we were not able to identify a specific transport defect in gyp2∆ cells. Moreover, Gyp2 was described previously as a GAP for Ypt31 and Ypt6 in vivo, again pointing to a redundant function of GAPs in vivo (Lafourcade et al., 2003; Sciorra et al., 2005).
It is assumed that a Rab cascade in which the downstream Rab (such as Ypt7) replaces the previous one (Vps21) on the same organelle requires the recruitment of the respective Vps21 GAP at a specific time point. Such a scenario has been postulated for the forward reaction as well, in that the GEF of the downstream Rab would be recruited by the upstream Rab (del Conte-Zerial et al., 2008; Hutagalung and Novick, 2011). Indeed, Vps21-GTP seems to affect Mon1–Ccz1 localization (Figure 2). Several studies on yeast exocytosis (Wang and Ferro-Novick, 2002; Rivera-Molina and Novick, 2009; Mizuno-Yamasaki et al., 2010) and the endocytic pathway (Rink et al., 2005; Abenza et al., 2010; Nordmann et al., 2010; Poteryaev et al., 2010) also support such models, although details on the exact mechanism are lacking. A Rab cascade model would be an attractive explanation for consecutive fusion reactions along the endomembrane system. For Msb3, we also asked whether we could detect an interaction between Ypt7 and Msb3, and we detected a signal proximal to the plasma membrane (Figure 6). The relevance of this interaction is not clear. It is possible that Msb3 act primarily, but not exclusively, on Ypt7 during vacuole inheritance.
How do we explain that Msb3 has such a broad substrate specificity also in vivo? We favor a model in which Rab GAPs mark both ends of a trafficking pathway to sharpen the specificity of the active Rab (Figure 7). As such, Rabs may act in a membrane-specific manner in addition to their substrate specificity. This would explain the influence of multiple GAPs in the endocytic pathway observed by us and others and why so few data on a specific in vivo GAP activity are available in general. Whereas the GAP system in yeast may reveal more redundancies, higher specificity is expected in mammalian cells. However, also in mammalian cells the >60 Rabs face ∼30 TBC-domain proteins, demonstrating an obvious mismatch. In the case of Msb3, the predominant plasma membrane localization would inactivate the Rab GTPase Sec4, which was initially present on exocytotic vesicles. Furthermore, Msb3 could also act on Vps21-GTP, which might be sorted to the plasma membrane via recycling vesicles from the endocytic compartment. Because a portion of Msb3 also localizes to the late endosomal/vacuolar compartment (Figures 4E and 6F), it would promote turnover of Vps21 and perhaps of Ypt7. In this manner, it helps to regulate exocytosis and define the boundaries of Vps21 activity along the endosomal pathway. In this context, Msb3 seems to be the primary GAP of Vps21, as Vps21 accumulates at the vacuole in its absence, although we cannot exclude additional contributions by Gyp2, for instance. As such, Msb3 would be specific enough to maintain a constant Rab turnover and dampen the Rab-GTP pool. In turn, this strengthens the importance of the Vps21 and Ypt7 GEF and consequently sharpens organelle boundaries (Figure 7). Having overlapping GAP activities may be advantageous as long as the GEF activity is defined and specific enough. Msb3 may thus be a paradigm of GAP function along one defined pathway.
FIGURE 7:
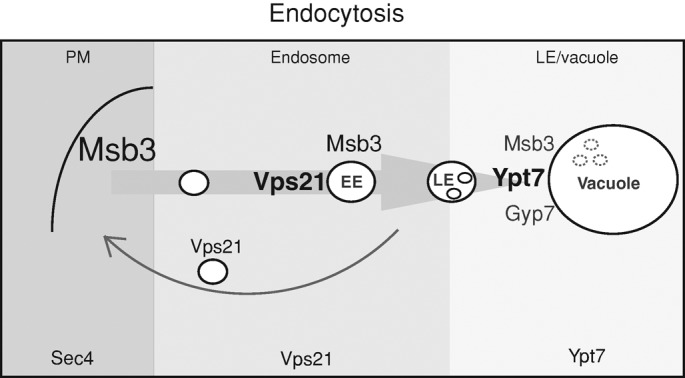
Working model. The role of Msb3 in the endocytic pathway. Relative amounts of Msb3, Vps21, and Ypt7 along the endocytic pathway are indicated by the font size. The different shades of gray indicate the predominant zones of each Rab.
MATERIALS AND METHODS
Yeast strains and molecular biology
Saccharomyces cerevisiae strains used in this study are summarized in Supplemental Table S1. Deletion of genes, promotor exchange, and tagging were done by homologous recombination with PCR-amplified fragments (Janke et al., 2004). Vps21 was genomically tagged at the N-terminus as described (Markgraf et al., 2009) or expressed as a dsRED-tagged version under the control of the PHO5 promotor from a pRS411 plasmid. Mutant versions of VPS21 (Markgraf et al., 2009) were expressed from a genomically reintegrated pRS406 or pRS403 vector under the control of the NOP1 promotor with or without an N-terminal GFP fusion. TPI1pr-mCherry-Ypt7, NOP1pr-VPS8-GFP, NOP1pr-VPS3-GFP, and PHO5pr-dsRED-Pep12 were expressed from a pRS-based CEN vector. Plasmids with pGNS416 expressing GFP-NYV1-SNC1 and pCu-GFP-ATG8 were kind gifts from Fulvio Reggiori (University Medical Center Utrecht, Utrecht, Netherlands). For split-YFP, VPS21 or YPT7 were set under the control of the CET1 promotor and N-terminally tagged with the VC half of YFP (Sung et al., 2007). Then, MSB3 or VPS39 was placed under the control of the CET1 promoter and tagged with the VN half at its N-terminus. Point mutations of the arginine 282 of Msb3 were generated by using the QuikChange Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). All mutations were confirmed by sequencing. Mutants of Msb3 or the wild-type version were subcloned into a pRS406 vector and genomically reintegrated in a deletion background. Expression was performed under the control of the NOP1 promotor.
Rab GTPases and GAPs were amplified from S. cerevisiae genomic DNA using the Pfu polymerase (Stratagene, Santa Clara, CA) and cloned into a pCRII-TOPO vector (Invitrogen, Carlsbad, CA). GYP8 was amplified as an N-terminal fragment of the first 426 amino acids lacking the transmembrane domain. The Rab GTPases were subcloned into the T7 polymerase hexahistidine–GST expression vector pFAT2 or a modified pET24d. The GAPs were subcloned into a pQE32 vector (Qiagen, Valencia, CA), except for GYP2, GYP6 and GYP8 (1-426), which were introduced into the pMalC2 vector (New England BioLabs, Ipswich, MA).
Microscopy
Cells were grown to mid logarithmic phase in yeast extract/peptone medium (YP) containing 2% glucose (YPD) or galactose (YPG). To maintain plasmids, cells were grown in synthetic complete medium lacking selected amino acids or nucleotides (SDC). They were harvested by centrifugation, washed once with synthetic complete medium supplemented with all amino acids, and visualized at room temperature. For FM4-64 (Invitrogen) labeling of the vacuoles, cells were treated as described before (LaGrassa and Ungermann, 2005). To follow Mup1-GFP sorting, we grew cells in SDC-Met to mid logarithmic phase. Methionine, 20 μg/ml, was added, and images were taken at indicated time points. Images were acquired using a Leica DM5500 B microscope (Leica, Mannheim, Germany) with a SPOT Pursuit-XS camera (Diagnostic Instruments, Sterling Heights, MI) using filters for GFP, dsRED, mCherry, FM4-64, and YFP. Where indicated, the data were subjected to a two-dimensional deconvolution with AutoQuant X Software (Media Cybernetics, Bethesda, MD) to improve resolution.
Total protein extraction from yeast
The cells were grown in YPD, and 1 OD600 unit was lysed in 0.25 M NaOH, 140 mM β‑mercaptoethanol, and 3 mM phenylmethylsulfonyl fluoride (PMSF) on ice. The samples were subjected to trichloroacetic acid (TCA) precipitation (13% final concentration of TCA), centrifuged, and washed with 1 ml of ice-cold acetone. The pellet was resuspended in SDS sample buffer, and equal amounts of protein extracts were analyzed by SDS–PAGE and Western blotting.
Protein expression and purification from bacteria
Plasmids were transformed into Escherichia coli BL21 (DE3) or JM109 strains, and protein expression was induced with 0.5 mM of isopropyl-β-d-thiogalactoside overnight at 18°C. Cells were harvested via centrifugation and resuspended in lysis buffer containing 20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5 mM β‑mercaptoethanol, 1 mM PMSF, and 0.1-fold protease inhibitor cocktail (LaGrassa and Ungermann, 2005). Lysis was performed by sonification or with the help of a Microfluidizer (Microfluidics, Newton, MA). Lysates were then centrifuged, and the supernatants were incubated for 1 h with nickel–nitriloacetic acid agarose (Qiagen, Hilden, Germany) or amylose resin (New England BioLabs, Frankfurt, Germany) at 4°C. Beads were washed with 10 column volumes of lysis buffer with or without 20 mM imidazole. The proteins were eluted with buffer containing 0.2 M imidazole or 10 mM maltose. Proteins were then dialyzed overnight or desalted via a PD-10 column (GE Healthcare, Piscataway, NJ) against storage buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM dithiothreitol, and 10% glycerol. Aliquots were frozen in liquid nitrogen and stored at −80°C.
GST-Rab pull-down
A 1-μmol amount of recombinant GST-Vps21 or GST-Ypt7 was incubated in 500 μl of 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)/NaOH, pH 7.4, 20 mM EDTA, 100 mM NaCl, 5 mM β‑mercaptoethanol, and 1 mM GDP. After an incubation for 20 min at 30°C, MgCl2 was added to a final concentration of 27 mM. The samples were then loaded onto 50 μl of prewashed GSH Sepharose (GE Healthcare) and incubated for 1 h at 4°C in a buffer containing 20 mM HEPES/NaOH, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 5 mM β‑mercaptoethanol, 5 mg/ml bovine serum albumin, and 0.1% NP-40. After incubation, the loaded beads were pelleted by centrifugation and washed twice with the same buffer. The beads were then incubated with 40–150 μg of Msb3, Msb4, or Gyp7 in a total volume of 375 μl in the presence of 1 mM GDP, 0.5 mM AlCl3, and 50 mM NaF. As a control, the incubation was performed without AlCl3 and NaF. After 1 h at 4°C, the beads were harvested by centrifugation and washed three times with or without AlCl3 and NaF. Proteins were then eluted by boiling in SDS-sample buffer and analyzed by SDS–PAGE and Western blot.
GTP hydrolysis assay
The assay was performed similar to the previous description (Haas et al., 2005). The Rab GTPases were preloaded by mixing 10 μl of assay buffer, 74 μl of H2O, 5 μl of 10 mM EDTA, pH 8.0, 5 μl of 1 mM GTP, 1 μl of [ γ-32P]GTP (10 mCi/ml; 5000 Ci/mmol; GE Healthcare), and 100 nmol of Rab protein on ice. The loading reaction was then performed for 15 min at 30°C and stored on ice. The GAP reactions were started by adding 10 pmol to 7 nmol of the GAP. Reactions were then incubated for 60 min at 30°C. A 2.5-μl amount of the reaction was directly placed into a scintillation counter to measure the specific activity in cpm/pmol GTP. A duplicate sample of 5 μl was added to 795 μl of activated charcoal (5% in 50 mM NaH2PO4; Sigma-Aldrich, St. Louis, MO) and stored on ice for 1 h. The charcoal was pelleted by centrifugation, and 400 μl of the supernatant was counted. The amount of hydrolyzed GTP was calculated from the specific activity of the reaction mixture.
Isolation of yeast vacuoles and in vitro fusion
Vacuoles were purified from the tester strains BJ3505 and DKY6281 with or without Msb3 or from BY4728 as described previously (Cabrera and Ungermann, 2008). Purified vacuoles were used in the vacuole fusion assay and analyzed by Western blotting or by microscopy. Fusion reactions containing 3 μg of each vacuole type were done in fusion reaction buffer (10 mM 1,4-piperazinediethanesulfonic acid/KOH, pH 6.8, 5 mM MgCl2, 125 mM KCl, 0.2 M sorbitol), containing 10 μM CoA, 10 μg of His-Sec18, and an ATP-regenerating system (0.5 mM ATP, 40 mM creatine phosphate, 0.1 mg/ml creatine kinase). Reactions were incubated for 90 min at 26°C and then developed (LaGrassa and Ungermann, 2005). Purified Mon1–Ccz1 (300 nM) (Nordmann et al., 2010) and a recombinant fragment of Gyp1 (Gyp1-46; 150 nM) were added to the reaction where indicated.
Membrane fractionation
Fractionation was performed as described previously (LaGrassa and Ungermann, 2005). After lysis of spheroblasts, the samples were centrifuged for 15 min at 10,000 × g at 4°C. The supernatant was then centrifuged for 1 h at 100,000 × g, resulting in a P100 pellet and an S100 supernatant fraction. The pellet fractions and the S100 fractions were subjected to TCA precipitation, washed with acetone, and resuspended in SDS sample buffer. Proteins were subsequently analyzed by SDS–PAGE and Western blotting.
Supplementary Material
Acknowledgments
This work was supported by Sonderforschungsbereich (SFB) 944 (Project P11) and by the Hans-Mühlenhoff Foundation (to C.U.). F.A.B. was supported by a Wellcome Senior Investigator Award (097769/Z/11/Z).
Abbreviations used:
- CORVET
class C core vacuole/endosome tethering
- GAP
GTPase-activating protein
- GDI
GDP diplacement factor
- GDP
guanosine diphosphate
- GEF
guanosine exchange factor
- GFP
green fluorescent protein
- GTP
guanosine triphosphate
- GTPase
guanosine triphosphatase
- HOPS
homotypic fusion and vacuole protein sorting
- MTCs
multisubunit tethering complexes
- TBC
Tre2/Bub2/Cdc16
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1030) on May 16, 2012.
REFERENCES
- Abenza JF, Galindo A, Pantazopoulou A, Gil C, de Los Ríos V, Peñalva MA. Aspergillus RabB Rab5 integrates acquisition of degradative identity with the long distance movement of early endosomes. Mol Biol Cell. 2010;21:2756–2769. doi: 10.1091/mbc.E10-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert S, Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J Biol Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Albert S, Gallwitz D. Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol Chem. 2000;381:453–456. doi: 10.1515/BC.2000.059. [DOI] [PubMed] [Google Scholar]
- Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol. 2010;22:461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Chiavetta JB, Chen H, Chen GC, Chan CS, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol Biol Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett CL, Plemel RL, Lobinger BT, Vignali M, Fields S, Merz AJ. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase. J Cell Biol. 2008;182:1141–1151. doi: 10.1083/jcb.200801001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker C, Engelbrecht-Vandré S, Ungermann C. Multisubunit tethering complexes and their role in membrane fusion. Curr Biol. 2010;20:R943–52. doi: 10.1016/j.cub.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Bröcker C, Kuhlee A, Gatsogiannis C, Kleine Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci USA. 2012;109:1991–1996. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Ungermann C. Purification and in vitro analysis of yeast vacuoles. Methods Enzymol. 2008;451:177–196. doi: 10.1016/S0076-6879(08)03213-8. [DOI] [PubMed] [Google Scholar]
- del Conte-Zerial P, Brusch L, Rink JC, Collinet C, Kalaidzidis Y, Zerial M, Deutsch A. Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Mol Syst Biol. 2008;4:206–209. doi: 10.1038/msb.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Novick P. Yeast Rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol Biol Cell. 2001;12:1215–1226. doi: 10.1091/mbc.12.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen G, Will E, Gallwitz D, Haas A, Wickner W. Sequential action of two GTPases to promote vacuole docking and fusion. EMBO J. 2000;19:6713–6720. doi: 10.1093/emboj/19.24.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Haas AK, Spooner RA, Yoshimura S-I, Lord JM, Barr FA. Specific Rab GTPase-activating proteins define the Shiga toxin and epidermal growth factor uptake pathways. J Cell Biol. 2007;177:1133–1143. doi: 10.1083/jcb.200612068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Jin C, Xue Y, Yao X. Computational analyses of TBC protein family in eukaryotes. Protein Pept Lett. 2008;15:505–509. doi: 10.2174/092986608784567483. [DOI] [PubMed] [Google Scholar]
- Gao X-D, Albert S, Tcheperegine SE, Burd CG, Gallwitz D, Bi E. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J Cell Biol. 2003;162:635–646. doi: 10.1083/jcb.200302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody R, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci. 2005;62:1657–1670. doi: 10.1007/s00018-005-4486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Haas AK, Yoshimura S-I, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- Hama H, Tall G, Horazdovsky B. Vps9p is a guanine nucleotide exchange factor involved in vesicle- mediated vacuolar protein transport. J Biol Chem. 1999;274:15284–15291. doi: 10.1074/jbc.274.21.15284. [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Lachmann J, Ungermann C, Engelbrecht-Vandré S. Rab GTPases and tethering in the yeast endocytic pathway. Small GTPases. 2011;2:182–186. doi: 10.4161/sgtp.2.3.16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafourcade C, Galan J-M, Peter M. Opposite roles of the F-box protein Rcy1p and the GTPase-activating protein Gyp2p during recycling of internalized proteins in yeast. Genetics. 2003;164:469–477. doi: 10.1093/genetics/164.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrassa TJ, Ungermann C. The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J Cell Biol. 2005;168:401–414. doi: 10.1083/jcb.200407141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Ahnert F, Arlt H, Mari M, Peplowska K, Epp N, Griffith J, Reggiori F, Ungermann C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol Biol Cell. 2009;20:5276–5289. doi: 10.1091/mbc.E09-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P. Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev Cell. 2010;18:828–840. doi: 10.1016/j.devcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, Russel MRG, Lo S-Y, Chapin HC, Milnes J, Merz AJ. Msb3/Gyp3 GAP selectively opposes Rab5 signaling at endosomal organelles. Traffic. 2012 (in press) [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz CW, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr S. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright D. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Plemel RL, Lobingier BT, Brett CL, Angers CG, Nickerson DP, Paulsel A, Sprague D, Merz AJ. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Raymond C, Howald-Stevenson I, Vater C, Stevens T. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Black M, Pelham H. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell. 2000;11:3737–49. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc Nat Acad Sci USA. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD. Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/rab-GTPase signaling pathway. Mol Biol Cell. 2005;16:776–793. doi: 10.1091/mbc.E04-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A. On the fate of early endosomes. Biol Chem. 2009;390:753–759. doi: 10.1515/BC.2009.056. [DOI] [PubMed] [Google Scholar]
- Sung M-K, Huh W-K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24:767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- Tall G, Hama H, DeWald D, Horazdovsky B. The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol Biol Cell. 1999;10:1873–1889. doi: 10.1091/mbc.10.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcheperegine SE, Gao X-D, Bi E. Regulation of cell polarity by interactions of Msb3 and Msb4 with Cdc42 and polarisome components. Mol Cell Biol. 2005;25:8567–8580. doi: 10.1128/MCB.25.19.8567-8580.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ferro-Novick S. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol Biol Cell. 2002;13:3336–3343. doi: 10.1091/mbc.01-12-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S-I, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I-M, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.