Abstract
The ability of bats and toothed whales to echolocate is a remarkable case of convergent evolution. Previous genetic studies have documented parallel evolution of nucleotide sequences in Prestin and KCNQ4, both of which are associated with voltage motility during the cochlear amplification of signals. Echolocation involves complex mechanisms. The most important factors include cochlear amplification, nerve transmission, and signal re-coding. Herein, we screen three genes that play different roles in this auditory system. Cadherin 23 (Cdh23) and its ligand, protocadherin 15 (Pcdh15), are essential for bundling motility in the sensory hair. Otoferlin (Otof) responds to nerve signal transmission in the auditory inner hair cell. Signals of parallel evolution occur in all three genes in the three groups of echolocators—two groups of bats (Yangochiroptera and Rhinolophoidea) plus the dolphin. Significant signals of positive selection also occur in Cdh23 in the Rhinolophoidea and dolphin, and Pcdh15 in Yangochiroptera. In addition, adult echolocating bats have higher levels of Otof expression in the auditory cortex than do their embryos and non-echolocation bats. Cdh23 and Pcdh15 encode the upper and lower parts of tip-links, and both genes show signals of convergent evolution and positive selection in echolocators, implying that they may co-evolve to optimize cochlear amplification. Convergent evolution and expression patterns of Otof suggest the potential role of nerve and brain in echolocation. Our synthesis of gene sequence and gene expression analyses reveals that positive selection, parallel evolution, and perhaps co-evolution and gene expression affect multiple hearing genes that play different roles in audition, including voltage and bundle motility in cochlear amplification, nerve transmission, and brain function.
Author Summary
The convergent origins of laryngeal echolocation in two groups of bats (Yangochiroptera and Rhinolophoidea) and toothed whales have long been a focus of interest for biologists. We screened three candidate genes—Cdh23, Pcdh15, and Otof—involved in different steps in the echolocation system. Signals of parallel evolution occurred in all three genes in the three groups of echolocators. Cdh23 and Pcdh15 constitute part of the mechanical link within the hair bundle of the ear. Both genes showed signals of both convergent evolution and positive selection, which implied they may have co-evolved to optimize cochlear amplification. Further, three lines of evidence suggest that Otof plays an important role in the transmission of signals in the brain during echolocation. First, the gene is more highly expressed in the auditory cortex of the brain in echolocating adult bats than in other cerebral cortexes. Second, this expression is higher in adult echolocating female bats than in their embryos, which do not use echolocation. Third, echolocators also have a higher level of expression in their cerebral cortexes than do non-echolocating bats. Taken with other evidence, the independent origins of echolocation involve the same genes that have evolved in precisely identical ways.
Introduction
The ability of echolocation using ultrahigh frequency sounds occurs in two groups of bats (Yangochiroptera and Rhinolophoidea) and in toothed whales including dolphins [1]–[3]. These mammals use this complex bio-sonar system to assist with orientation and feeding [4], [5]. Echolocation by bats and dolphins provides an iconic example of either parallel or convergent evolution via natural selection.
Previous molecular studies on echolocation have mainly focused on the Organ of Corti. In this organ, the motor protein prestin plays a key role in voltage motility [6]–[8]. It appears to have undergone sequence convergence between bats and dolphins [1], [2], as well as within laryngeal echolocating bats [9]. Further, the voltage-gated potassium channel gene KCNQ4 underwent parallel evolution in echolocating bats [3], [10]. Mammalian audition requires not only voltage motility, but also hair bundle motility, which is executed by outer hair cells in the cochlea [11]. Proteins encoded by the genes Cdh23 and Pcdh15 are essential to hair bundle motility [12]–[14], and their malfunctions in humans cause deafness in newborns and progressive retinitis pigmentosa (Usher syndrome type I) [15]. Homodimers of Cdh23 and Pcdh15 directly link to each other via their amino termini; they form the upper and lower part of tip-links, respectively (Figure S1), which lie between the stereocilia within the hair bundle [14], [16], [17]. The auditory system involves the perception and enhancement of sound signals, as well as transformation of the mechanical signals to ion fluxes in inner hair cells. Management of the electric signals to the brain involves a series of nerve channel openings [18]. Genetic mutations in the gene encoding otoferlin (Otof) cause a clinical, autosomal recessive nonsyndromic form of prelingual and sensorineural deafness [19]–[21]. This protein that may act as the major Ca2+ sensor that triggers membrane fusion at the ribbon synapse of the auditory inner hair cell [22]. Although the above functions are involved in the conversion of sound signals into electrical impulses in the inner ear, the expression of Otof also occurs in neurons and nerve fibers in the brain [23].
The molecular mechanism of voltage motility in echolocation is widely studied. Echolocation is a complex system that includes signal reception by hair cells in the Organ of Corti, nerve transmission, and signal processing in the brain [24]. Therefore, herein we investigate the gene sequence evolution of Cdh23, Pcdh15, and Otof. These proteins function in different steps during echolocation. Because the brain modulates sensory information from peripheral sensory organs [25], and because Otof is involved in transferring sound signal by electrical impulses, we also examine the expression patterns of Otof in the cerebral cortexes of different species. We synthesize evidence from sequences and expressions to study the convergent evolution of echolocation in bats and dolphins.
Results
Tree analyses
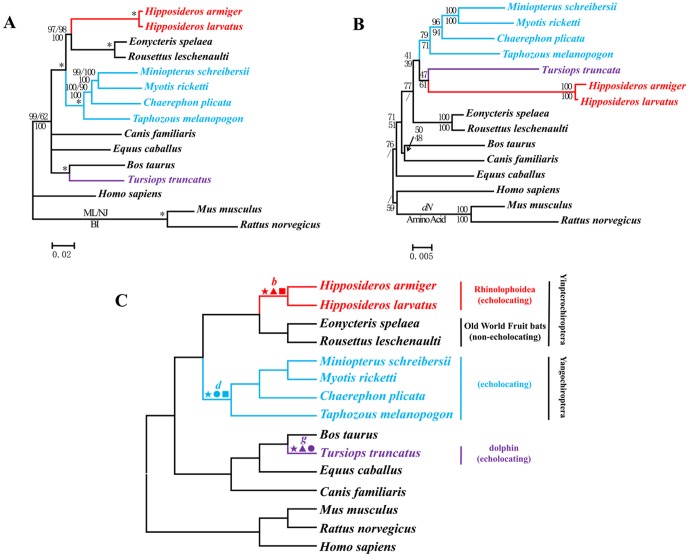
We built ML, BI, and NJ trees based on both nucleotide and amino acid sequences. The length of aligned nucleotides for Cdh23 was 9657 base pairs (bp). The gene trees for Cdh23 based on nucleotide sequences (Figure 1A) were basically the same as the well-accepted species tree (Figure 1C) [26]–[28] in all methods of tree-building. Echolocating Hipposideros clustered with Old World (OW) fruit bats, which represented the Yinpterochiroptera. The genera Taphozous, Chaerephon, Miniopterus, and Myotis clustered together, forming the Yangochiroptera, the sister group of Yinpterochiroptera. However, the topologies based on amino acid sequences (Figure 1B) differed substantially from those based on nucleotide sequences. All echolocators incorrectly grouped together and then became the sister-group of the OW fruit bats, which do not possess the ability of laryngeal echolocation. The topology of the tree based on synonymous sites of Cdh23 was nearly the same as that based on nucleotide sequences, as well as the species-tree. The NJ tree using nonsynonymous changes was congruent with the amino acid tree (Figure 1B).
Figure 1. Parallel evolution of Cdh23 in bats and dolphins.
(A) Gene tree of Cdh23 based on nucleotide sequences that is consistent with the species tree. Numbers above the branches are the ML and NJ bootstrap values, respectively. Numbers below the branches are Bayesian posterior probabilities. * indicates all values equal 100. (B) Gene tree of Cdh23 based on nonsynonymous mutations and the amino acid sequences. Numbers along the branches are NJ bootstrap values. (C) Species tree based on previous studies [26]–[28]. Symbols above the branches correspond to amino acid replacements. ★ indicates a parallel amino acid replacement presenting on branches b, d, and g: R204Q. ▴ indicates parallel amino acid replacements presenting on branches b and g: R204Q, D517N, P518A, S639N, N737S, S747T, A1080S, K1141T, S1314T, A1382S, I1673V, N1697D, L1960F, L1974I, A2146V, G2229S, V2427I, T2439R, R2639K, Q2725L, and N3180S. • indicates a parallel amino acid replacement presenting on branches d and g: R204Q. ▪ indicates parallel amino acid replacements presenting on branches b and d: R204Q, R535K, T904I, and V1691I.
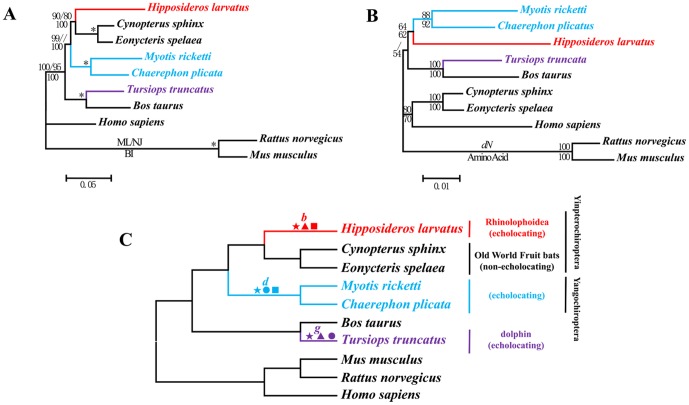
For Pcdh15, the aligned length was 5835 bp. The tree based on its nucleotide sequences (Figure 2A) depicted the well-accepted species tree (Figure 2C) and the nodes received high bootstrap values. In contrast, the amino acid trees (Figure 2B) clustered all echolocating bats together, and this arrangement differed from the nucleotide trees. As with Cdh23, the topology of the tree based on synonymous sites was virtually identical to that based on nucleotide sequences, while the tree based on the nonsynonymous changes was congruent with the amino acid tree (Figure 2B).
Figure 2. Parallel evolution of Pcdh15 in bats and dolphins.
(A) Gene tree of Pcdh15 based on nucleotide sequences. Numbers above the branches are the ML and NJ bootstrap values, respectively. Numbers below the branches are Bayesian posterior probabilities. * indicates all values equal 100. (B) Gene tree of Pcdh15 based on nonsynonymous changes and amino acid sequences. Numbers along the branches are NJ bootstrap values. (C) Species tree based on the previous studies [26]–[28]. Symbols above the branches correspond to amino acid replacements. ★ indicates parallel amino acid replacements presenting on branches b, d, and g: M946I and D1278E. ▴ indicates parallel amino acid replacements presenting on branches b and g: N218D, Q310E, E393V, T427S, A433V, I438V, T490I, V546F, I643V, N666K, K820R, I853V, K856T, M946I, V952A, R999L, T1139R, F1160L, A1173S, K1275R, D1278E, and I1404V. • indicates parallel amino acid replacements presenting on branches d and g: A726D, M946I, and D1278E. ▪ indicates parallel amino acid replacements presenting on branches b and d: L423V, Q468P, H765Y, F876L, M946I, F984S, V1019I, and D1278E.
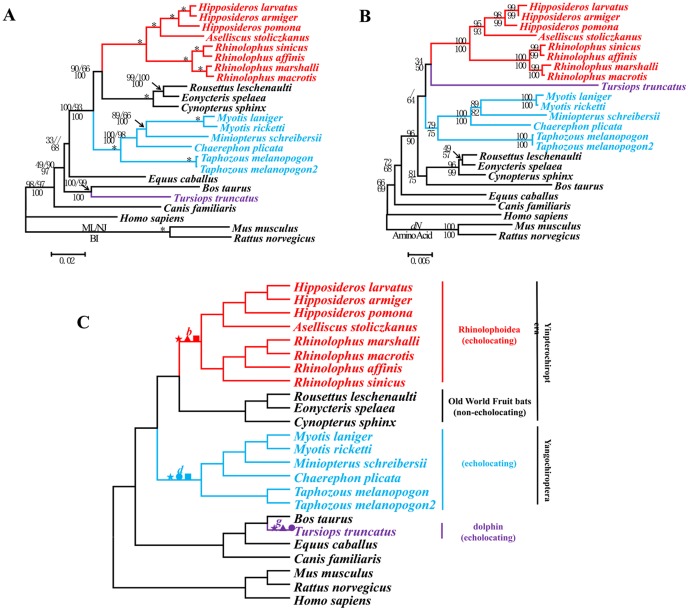
The sequenced coding region of Otof varied from 5363 to 5645 bp. As common in the Yinpterochiroptera, a 20 bp deletion occurred from bp site 1248 to 1267. The trees for Otof showed patterns similar to those of the previous two genes; the amino acid trees and NJ topology for nonsynonymous sites incorrectly clustered all echolocators (Figure 3B), and this association conflicted with the nucleotide trees (Figure 3A). Again, the nucleotide trees were consistent with the species tree (Figure 3C).
Figure 3. Parallel evolution of Otof in bats and dolphins.
(A) Gene tree of Otof based on nucleotide sequences. Numbers above the branches are the ML and NJ bootstrap values, respectively. Numbers below the branches are Bayesian posterior probabilities. * indicates all values equal 100. (B) Gene tree of Otof based on the nonsynonymous changes and corresponding amino acid sequences. Numbers on the branches are NJ bootstrap values. (C) Species tree based on the previous studies [26]–[28]. Symbols above the branches correspond to amino acid replacements. ★ indicates a parallel amino acid replacement presenting on branches b, d, and g: D396E. ▴ indicates parallel amino acid replacements presenting on branches b and g: P191H, G213A, D396E, and V440M. • indicates parallel amino acid replacements presenting on branches d and g: D396E and R1238H. ▪ indicates a parallel amino acid replacement presenting on branches b and d: D396E.
Analyses of convergent/parallel evolution
Branches b, d, and g in the species tree (Figure 1C, Figure 2C, Figure 3C) lead to mammals that have the ability to echolocate. Branch b represented the common ancestor of the Rhinolophoidea, d of the Yangochiroptera, and g of the dolphin. We reconstructed ancestral nodes and mapped amino acid changes along these branches. For Cdh23, branches b, d, and g shared one amino acid change (R204Q). Branches b and g shared the following 21 parallel mutations: R204Q, D517N, P518A, S639N, N737S, S747T, A1080S, K1141T, S1314T, A1382S, I1673V, N1697D, L1960F, L1974I, A2146V, G2229S, V2427I, T2439R, R2639K, Q2725L, and N3180S. Parallel evolution of these two branches statistically differed from random expectations (P<0.001). Branches b and d had four parallel mutations (R204Q, R535K, T904I, and V1691I). Again, parallel evolution was statistically significant (P<0.001). All changed sites were mapped in Figure 1C. The positions of these sites in the domain structure of Cdh23 were mapped in Figure S2. We constructed a BI tree from the aligned amino acids of Cdh23 while excluding all parallel-evolved sites. The BI tree agreed with the species tree (Figure S3).
Two parallel mutations in Pcdh15 (I946M and E1278D) were shared by branches b, d, and g. Parallel evolution was statistically significant (P<0.001) between branches b and g for the following 22 parallel mutations: N218D, Q310E, E393V, T427S, A433V, I438V, T490I, V546F, I643V, N666K, K820R, I853V, K856T, M946I, V952A, R999L, T1139R, F1160L, A1173S, K1275R, D1278E, and I1404V. Parallel evolution was also statistically significant (P<0.001) between branches d and g for three parallel mutations: A726D, M946I, and D1278E. Finally, eight parallel mutations occurred between branches b and d, including L423V, Q468P, H765Y, F876L, M946I, F984S, V1019I, and D1278E (Figure 2C). Parallel evolution between these two branches was statistically significant (P<0.001). The positions of these sites in the domain structure of Pcdh15 were mapped in Figure S4. The BI tree based on amino acids excluding all parallel-evolved sites differed somewhat with the species tree, but it did not group echolocators together (Figure S5).
Otof had one amino acid change shared among echolocators (D396E). The four parallel-evolved sites along branches b and g were P191H, G213A, D396E, and V440M (Figure 3C), and parallel evolution was statistically significant (P<0.001) between these two branches. Parallel evolution of branch d and g was also significant (P<0.001). The conserved sites were shown in Figure S6. The BI tree based on amino acids while excluding all parallel-evolved sites agreed with the species tree (Figure S7).
Selective pressure analysis
Selective pressure was evaluated using the PAML package and test results were presented in Table 1 and Table S1. The one-ratio model obtained an average ω (Ka/Ks ratio) of 0.0546 (lnL = −35796.7768) for Cdh23. For specific branches, we alternatively set the echolocating bats (Rhinolophoidea and Yangochiroptera; branches b and d in Figure 1C, respectively) or dolphin (branch g in Figure 1C) as foreground branch. In both conditions, although the ratios of ω for the foreground branches were greater than background branches, they were less than 1 (ωbranch (b+d) = 0.1107 while ω0 = 0.0500; ωbranch g = 0.1145 while ω0 = 0.0516). When considering all echolocators together, ωecholocators was 0.1126 for the foreground branch compared with 0.0465 for background branch (2Δl = 660.6044, df = 1, P<0.001). For the branch-site models, test 2 (Model A vs. null model) was used to control the false positive signals. Branch b was detected to have undergone significant positive selection (P<0.01), and eight sites with BEB values >0.90 (213 A 0.984, 692 F 0.984, 1165 N 0.989, 1171 S 0.958, 1256 D 0.989, 1356 I 0.990, 1687 T 0.925, and 2492 L 0.970). When we set the dolphin as the foreground branch (branch g), significantly positive selection was also detected (2Δl = 10.7224, df = 1, P<0.01), and six sites (580 S 0.974, 840 H 0.973, 1011 H 0.973, 1014 T 0.978, 2133 T 0.952, and 2224 T 0.974) with BEB values >0.90. When all echolocating species were combined as the foreground branch, again significant positive selection signals were obtained (2Δl = 8.6179, df = 1, P<0.01). The positions of these positively selected sites were mapped in Figure S2.
Table 1. Summary of selective pressure analysis for the hearing genes Cdh23, Pcdh15, and Otof.
| Gene | branch | Parameter Estimated | 2ΔlnLP value | Positively Selected Sites (BEB Analysis) | ||||
| site class | 0 | 1 | 2a | 2b | ||||
| Cdh23 | branch b | proportion | 0.92097 | 0.05178 | 0.02580 | 0.00145 | 10.4158 P<0.01 | 213 A, 692 F, 1165 N, 1171 S, 1256 D, 1356 I, 2492 L |
| background ω | 0.03156 | 1.00000 | 0.03156 | 1.00000 | ||||
| foreground ω | 0.03156 | 1.00000 | 3.85446 | 3.85446 | ||||
| branch d | proportion | 0.94313 | 0.05687 | 0.00000 | 0.00000 | 0 | ||
| background ω | 0.03347 | 1.00000 | 0.03347 | 1.00000 | ||||
| foreground ω | 0.03347 | 1.00000 | 1.00000 | 1.00000 | ||||
| branch g | proportion | 0.92922 | 0.05575 | 0.01418 | 0.00085 | 10.7224 P<0.01 | 580 S, 840 H, 1011 H, 1014 T, 2133 T, 2224 T | |
| background ω | 0.03168 | 1.00000 | 0.03168 | 1.00000 | ||||
| foreground ω | 0.03168 | 1.00000 | 5.94936 | 5.94936 | ||||
| Pcdh15 | Branch b | proportion | 0.79672 | 0.11161 | 0.08040 | 0.01126 | 2.6591P = 0.1 | 454 H, 816 T |
| background ω | 0.02983 | 1.00000 | 0.02983 | 1.00000 | ||||
| foreground ω | 0.02983 | 1.00000 | 2.00038 | 2.00038 | ||||
| branch d | proportion | 0.86720 | 0.12926 | 0.00308 | 0.00046 | 6.4259 P<0.05 | 573 T 0.921 | |
| background ω | 0.03721 | 1.00000 | 0.03721 | 1.00000 | ||||
| foreground ω | 0.03721 | 1.00000 | 24.77802 | 24.77802 | ||||
| Branch g | proportion | 0.81368 | 0.11630 | 0.06127 | 0.00876 | 2.9014P = 0.09 | 1584D | |
| background ω | 0.03403 | 1.00000 | 0.03403 | 1.00000 | ||||
| foreground ω | 0.03403 | 1.00000 | 2.72477 | 2.72477 | ||||
| Otof | Branch b | proportion | 0.95114 | 0.03918 | 0.00930 | 0.00038 | 0.7680P<0.5 | |
| background ω | 0.02313 | 1.00000 | 0.02313 | 1.00000 | ||||
| foreground ω | 0.02313 | 1.00000 | 3.10544 | 3.10544 | ||||
| branch d | proportion | 0.95968 | 0.04032 | 0.00000 | 0.00000 | 0P = 1 | ||
| background ω | 0.02352 | 1.00000 | 0.02352 | 1.00000 | ||||
| foreground ω | 0.02352 | 1.00000 | 1.00000 | 1.00000 | ||||
| Branch g | proportion | 0.92038 | 0.03758 | 0.04039 | 0.00165 | 0P = 1 | 944 L | |
| background ω | 0.02221 | 1.00000 | 0.02221 | 1.00000 | ||||
| foreground ω | 0.02221 | 1.00000 | 1.00000 | 1.00000 | ||||
For Pcdh15, we implemented the same series of analysis as for Cdh23. All branch models were significantly better than the null model that fixed ω of the foreground branch to 1, although the value of ω never exceeded 1 (ωbranch (b+d) = 0.3211, ω0 = 0.0998, 2Δl = 83.0381, df = 1, P<0.001; ωbranch g = 0.4469, ω0 = 0.1109, 2Δl = 16.6620, df = 1, P<0.001; and ωecholocators = 0.3561, ω0 = 0.0851, 2Δl = 96.3532, df = 1, P<0.001). For the branch-site models, a significant signal of positive selection was detected on branch d and one site had a BEB value >0.90 (P<0.05). The position of this positively selected site was mapped in Figure S4.
For Otof, the ω ratios in branch models were greater than ratios of the background branches, but less than 1 (ωbranch (b+d) = 0.0572, ω0 = 0.0364; ωbranch g = 0.0886, ω0 = 0.0352; and ωecholocators = 0.0732 with ω0 = 0.0340). Branch-site models did not detect any signals of positive selection in the echolocators (Table 1).
Gene expression pattern analysis
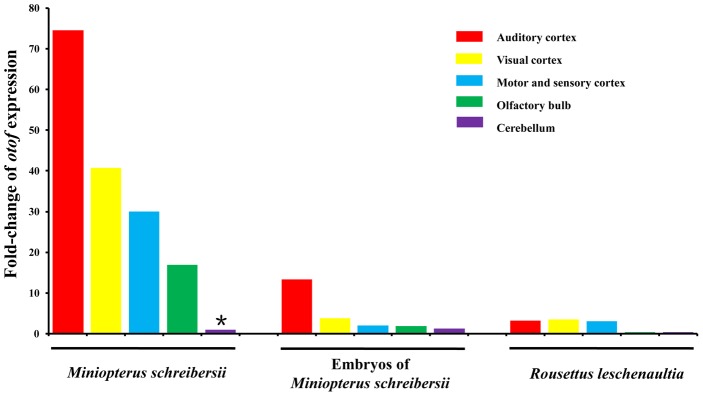
Real-Time PCR was used to assess expression patterns of Otof in the brain. We set the mean value of gene expression in the cerebellum of the adult Common Bent-wing Bat (Miniopterus schreibersii) as the baseline unit (marked * in Figure 4), and then compared expression patterns of Otof in different cortexes of the brain (Figure 4 and Table S2). The level of expression in the auditory cortex was more than 70-fold that of the baseline value. The levels of expression from the visual cortex, and motor and sensory cortex were more than 40-fold and 30-fold greater than the baseline value, respectively, whereas the expression in the olfactory bulb was nearly 17-fold greater. We compared this expression pattern with that of embryos. Expression levels of Otof in the auditory cortexes of three embryonic Common Bent-wing Bats were nearly 13-fold the baseline value and the visual cortex was over 3-fold. In contrast, expression levels in motor and sensory cortex, olfactory bulb, and cerebellum were similar to the baseline values. In adult Old World Fruit Bats (Rousettus leschenaultii), which do not echolocate, expression levels in the auditory cortex, visual cortex, and motor and sensory cortex were all around 3-fold greater than the baseline value, and expression levels in the olfactory bulb and cerebellum were less than the baseline value.
Figure 4. Expression patterns of the gene Otof in different cerebral cortexes of bats.
The * indicates baseline value.
Discussion
Morphological development is very complex and involves a suite of genes. Some mammals have independently developed similar features, such as the ability to echolocate objects. Echolocation by bats and dolphins provides an extreme example of parallel or convergent evolution. Although the morphologies of sending and receiving sonic signals differ greatly, the hearing of ultrasonic sounds and the mechanisms of decoding signals are shared [29], [30]. Thus, the genes coding for the auditory system become ideal candidates for adaptive evolution at the molecular level during echolocation.
The genes Cdh23 and Pcdh15 are essential to hair bundle motility [12]–[14], [31], [32]. Otof encodes a protein that may act as the major Ca2+ sensor to trigger membrane fusion at the auditory inner hair cell ribbon synapse [22]. The three genes are involved in different steps of the hearing system. Their nucleotide gene trees are largely congruent with the species tree and the nodes enjoy high support. In contrast, the amino acid trees conflict with the species tree; all unite the echolocators. Further, trees based on nonsynonymous sites have topologies similar to those based on corresponding amino acids, and yet the trees based on synonymous mutations do not show this pattern. Clearly, the difference in branching order is the result of amino acid changes (nonsynonymous mutations).
Three independent lineages of mammals can echolocate and it is important to identify whether this involves convergent or parallel evolution. If echolocation involves convergent evolution, then similar traits or functions independently emerge in two or more lineages from different ancestral states. Parallel evolution differs in that similar ancestral traits descend into similar extant states in different lineages [33].
Reconstructions of the ancestral sequences of the internal nodes can detect which amino acid changes cause the incongruence between the amino acid and species trees. A suite of sites show parallel changes in these echolocators, and these changes have a statistically significant signal of parallel evolution. Twenty-one amino acid sites converge on the same residue in branch g and branch b and with a highly significant probability (P<0.001). One convergent site appears in branches g and d (P<0.001), and four parallel sites occur in branches b and d (P<0.001) in Cdh23. In Pcdh15, 22 and three convergent sites occur between branch g and branches b and d, respectively (P<0.001 in both cases) and eight parallel sites have been identified between branches b and d (P<0.001). Four parallel sites in Otof occur on branches g and b (P<0.001). Convergent evolution is not accommodated by current phylogenetic methods and it can strongly mislead phylogenetic inference [34]. Upon excluding the parallel-evolved amino acid sites, the reconstructed amino acid trees did not incorrectly unite all echolocators (Figures S3, S5, S7). Thus, the discovery of multiple parallel-evolving amino acid sites explains the unnatural uniting of echolocators in the amino acid trees.
In addition to the parallel evolution of Prestin [1], [2], [9] and KCNQ4 [3], our analyses document a high level of complexity in a large-scale, multigene adaptation. Functional assays have proven that parallel and convergent amino acid changes are responsible for parallel and convergent functional changes [35], [36]. Thus, parallel evolution of gene sequences may have driven phenotypic and functional convergence in echolocating bats and dolphins. Further functional assays are needed to affirm this association.
Our analyses of selection pressure detect signals of positive selection in Cdh23 on the branches leading to the Rhinolophoidea and dolphin. The same occurs in Pcdh15 along the branch leading to Yinpterochiroptera (see Table 1 and Table S1). Positive selection appears to have acted on these genes to fit the requirements for echolocation. Whereas outer hair cells amplify sound by somatic [37], [38] or ciliary [39], [40] mechanisms, inner hair cells are passive detectors of the amplified vibratory signal. The signals are converted into electrical impulses by activating fibers of the cochlear (auditory) nerve [41], which then sends the signals to higher auditory processing centers in the brain. Prestin mediates the voltage somatic motility unique to mammals [42]–[44]. Part of the cochlear amplification, Cdh23 and Pcdh15 participate in hair bundle motility [39], [45], [46]. Otof participates by releasing neurotransmitter to nerves [22]. Convergent evolution and positive selection on these genes reflect the pathway from receipt of signal to signal amplification, and then to neural transduction. All parts of the pathway are involved in the auditory system, and thus may play important roles in echolocation.
Homodimers of Cdh23 and Pcdh15 directly link to each other via their amino termini, and they constitute the upper and lower part of tip-links, respectively, that lie between the stereocilia within the hair bundle [14], [16], [17]. Sequence alignments suggest that Cdh23 and Pcdh15 have 27 and 11 extracellular cadherin repeats, respectively [47]. Functional evidence encompassing a classical genetic approach shows that mutations at these two cadherin proteins can interact to cause hearing loss in digenic heterozygotes of both mice and humans [48]. Cdh23 and Pcdh15 appear to have undergone both convergent functional evolution and positive selection in echolocators. This finding suggests a strong interaction between the proteins and co-evolution is likely to have optimized their function in cochlear amplification for the development of echolocation.
Dolphins and bats employ different tools to echolocate. Whereas the larynx generates sound in echolocating bats [29], the monkey lip/dorsal bursa complex does the same in dolphins [30]. In bats, the sound involves a constant frequency (CF) and frequency modulation (FM), but dolphins use FM and amplitude modulation (AM) [49]. Convergence occurs via both auditory systems being adapted to receiving and processing ultrahigh frequency sounds. Our study discovers evidence of parallel sequence evolution in three genes involved in hearing and this may indicate a genetic basis for echolocation.
Sequence evolution only tells a part of the story. Expression pattern is usually a strong indicator of protein-demand and function [50], [51]. The central auditory system plays the crucial role of receiving impulses from auditory nerves and sending messages back to the cochlea [52]. Because Otof plays a role in neural signal transmission, we have evaluated its patterns of expression in different cerebral cortexes. Otof is most highly expressed in the auditory cortex of echolocating adult female Common Bent-Wing Bats. Adult females, which frequently use ultrasonic sounds to explore their environments, have higher levels of expression than their embryos, which do not use echolocation. Further, a comparison of expression of Otof between the brains of an adult echolocating bat (Common Bent-Wing Bat, Miniopterus schreibersii) and a non-echolocating bat (Old World Fruit Bat, Rousettus leschenaultii) reveals a higher level of expression in the former. Indeed, the gene exhibits great differences in levels of expression in both different cerebral cortexes and in species with or without the ability to echolocate.
Echolocation signals begin at the hair cells in the Organ of Corti, continue along the auditory nerve, and terminate in the auditory cortex of the brain [24]. Combined with sequence data and expression data, we conclude that multiple instances of parallel sequence evolution are involved in genes in different parts of auditory system between the three groups of echolocators. This occurs not only in well-studied voltage motility, but also bundle motility, and not only in cochlear amplification, but also in neural transduction. Further, co-evolution optimizes the function of homodimers of Cdh23 and Pcdh15 in cochlear amplification. The expression pattern of Otof in different cerebral cortexes implies that the evolution of gene expression might be required for echolocation. In conclusion, we synthesize gene sequence and gene expression analyses and conclude that positive selection, convergent evolution, and perhaps co-evolution and gene expression evolution play roles in audition (voltage motility and bundle motility in cochlear amplification, nerve transmission, and brain) during the independent origins of echolocation in bats and dolphins.
Materials and Methods
Ethics statement
All research involving animals used in this study followed the guidelines and bylaws on animal experimentation. Bats were anesthetized using an intraperitoneal injection of sodium pentobarbital (C11H17N2NaO3) at a dosage of 100 mg per kg body weight. Following anesthesia, bats were euthanized and then their brains were sampled. Protocols were approved by the Ethics and Experimental Animal Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences.
Source of data and primary treatments
Sixteen species of bats were used in our analysis (Table S3). Total RNA was isolated from the brain using a RNAiso Plus Kit (Takara, China), and RT-PCR was performed on 2 ug of RNA using the PrimeScript RT-PCR Kit (Takara, China) to obtain cDNA. Subsequently, genes were amplified from cDNA using gene-specific primers (Table S4). PCR products were purified on a 1% agarose gel and a Watson Gel Purification Kit (Watson BioTechnology, Shanghai), and finally transformed into the pMD18-T vector (Takara, China). Each strand was sequenced in both directions with an ABI 3730 sequencer. RNA samples of brain were not available for the dolphin (Tursiops truncatus), so genomic sequences were amplified from total genomic DNA, which was extracted from muscle tissue using a standard 3-step phenol/chloroform extraction method [53].
Raw nucleotide sequences were edited using Lasergene SeqMan software (DNASTAR Inc., Madison, WI, USA). Newly determined sequences were deposited in GenBank (Accession numbers JF808081–JF808094, JQ284400–JQ284430). The sequences of background species came from the Ensembl database (Release 66) and those of high quality were used (Table S3). All the sequences were aligned using ClustalX 1.81 [54] and then visually checked for accuracy (the aligned sequences are available by request).
Phylogenetic and molecular evolutionary analyses
The best-fit models were selected by jModeltest v0.1.1 [55], [56] for nucleotide sequences and ProtTest 3.0 beta [57] for amino acid sequences. Maximum likelihood (ML) trees were reconstructed by PAUP* [58] with 1,000 replications, and Bayesian inference (BI) trees were reconstructed by MrBayes 3.1.2 with 1,000,000 replications [59], [60].
Neighbor-joining (NJ) phenograms were based on Kimura 2-parameter corrected distances for nucleotide sequences and uncorrected P-distances for amino acid sequences, each with 1,000 bootstrap replications. We implemented the Li-Wu-Luo method [61] to reconstruct NJ trees based on both synonymous and nonsynonymous sites.
The sequences of the internal nodes were reconstructed using distance-based Bayesian methods, which included the branch lengths estimated by the least squares method and the ancestral amino acids inferred by the Bayesian approach. These data were used to obtain an unbiased estimate of the true probability [62]. Convergent and parallel amino acid substitutions along each lineage were detected. The statistical significance of these amino acid changes was tested with the method developed by Zhang and Kumar [63].
The CODEML program in PAML 4 [64] was used to detect selective pressure. The species tree [26]–[28] was used as guide tree for analysis. Four models of evolution were used: one-ratio model, free-ratio model, branch models, and branch-site models. For the last two models, three groups of echolocators were set as the foreground branch to detect whether or not they had undergone positive selection.
Expressional data analysis
We sampled auditory cortex, visual cortex, motor and sensory cortex, olfactory bulb, and cerebellum from euthanized adult and embryonic Miniopterus schreibersii, and adult Rousettus leschenaultii, which represented the adult echolocating bats, embryonic echolocating bats, and adult non-echolocating bats, respectively. Tissue was selected according the human brain atlas [65]. These tissues were stored in liquid nitrogen. Total RNA was isolated using a RNAiso Plus kit (Takara, China). Next, DNA-free RNA samples were condensed using a RNAqueous-4PCR Kit (Applied Biosystems, US). We synthesized cDNA using a PrimeScript RT-PCR Kit (Takara, China), and then used it in Real-Time PCR. TaqMan Gene Expression Assays were custom designed by Applied Biosystems based on our sequencing data of Otof and Actb in bats. The assay details were listed in Table S5. Sequences of Otof and Actb were amplified and detected using an ABI PRISM 7000 Sequence Detection System with a PCR profile as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. The products were purified and sequenced in both directions with an ABI 3730 sequencer to insure the assays' specificity. Real-Time PCR was performed on 96-well reaction plates in a 20 µl reaction volume containing 100 ng of cDNA per reaction with TaqMan Universal Master Mix II (Applied Biosystems, US). For each group, three individuals were used and each sample was performed three times. Expression data for the target gene Otof were normalized relative to the housekeeping gene Actb. Raw data were obtained and analyzed using the 7000 SDS 1.1 software (Applied Biosystems, US). The comparative CT method (ΔΔCt) was chosen to further calculate the relative expressions between different groups.
Supporting Information
Domain structure of Cdh23 (SS, signal sequence; EC, ectodomain; HTM, transmembrane domain), and the positions of the positively selected sites and parallel-evolving sites.
(TIF)
The BI tree for Cdh23 based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Domain structure of Pcdh15 (SS, signal sequence; EC, ectodomain; HTM, transmembrane domain), and the positions of the positively selected sites that excluded all parallel-evolving sites.
(TIF)
The BI tree for Pcdh15 based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Parallel-evolved sites of Otof.
(TIF)
The BI tree for Otof based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Details of the selective pressure analyses on the three hearing genes.
(DOCX)
Expression levels of Otof in the Common Bent-wing Bat and Old World Fruit Bat.
(DOCX)
Species and their accession numbers for the genes Cdh23, Pcdh15, Otof, and Actb used in this research.
(DOCX)
The primers used for amplifying and sequencing the three hearing genes.
(DOCX)
Details of the TaqMan gene expression assays.
(DOCX)
Acknowledgments
We thank Jing-Kuan Wei from the Laboratory of Sensory Motor Integration in the Kunming Institute of Zoology for his help in sampling different cerebral cortexes from bats. We thank the Ethics and Experimental Animal Committee of the Kunming Institute of Zoology for approving our experiments. We also thank the three anonymous reviewers, Christopher Blair, and Ross MacCulloch for their help in improving the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the National Natural Science Foundation of China (30621092 and 31172080) and the Bureau of Science and Technology of Yunnan Province. RWM was supported by a Visiting Professorship for Senior International Scientists from the Chinese Academy of Sciences, and manuscript preparation was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant A3148. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Li Y, Liu Z, Shi P, Zhang J. The hearing gene Prestin unites echolocating bats and whales. Curr Biol. 2010;20:R55–R56. doi: 10.1016/j.cub.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Cotton JA, Shen B, Han X, Rossiter SJ, et al. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 2010;20:R53–R54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Li S, Wang W, Xu D, Murphy RW, et al. Parallel evolution of KCNQ4 in echolocating bats. PLoS ONE. 2011;6:e26618. doi: 10.1371/journal.pone.0026618. doi: 10.1371/journal.pone.0026618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Speakman JR. The evolution of echolocation for predation. Symp zool Soc Lond. 1993;65:39–63. [Google Scholar]
- 5.Arch VS, Narins PM. ‘Silent’ signals: selective forces acting on ultrasonic communication systems in terrestrial vertebrates. Anim Behav. 2008;76:1423–1428. doi: 10.1016/j.anbehav.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fettiplace R. Active hair bundle movements in auditory hair cells. J Physiol. 2006;576:29–36. doi: 10.1113/jphysiol.2006.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashmore J, Avan P, Brownell WE, Dallos P, Dierkes K, et al. The remarkable cochlear amplifier. Hear Res. 2010;266:1–17. doi: 10.1016/j.heares.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J, Shen W, He DZZ, Long KB, Madison LD, et al. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Wang J, Rossiter SJ, Jones G, Cotton JA, et al. The hearing gene Prestin reunites echolocating bats. Proc Natl Acad Sci U S A. 2008;105:13959–13964. doi: 10.1073/pnas.0802097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Han N, Franchini LF, Xu H, Pisciottano F, et al. The voltage-gated potassium channel subfamily KQT member 4 (KCNQ4) displays parallel evolution in echolocating bats. Mol Biol Evol. 2012;29:1441–1450. doi: 10.1093/molbev/msr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallos P. Cochlear amplification, outer hair cells and prestin. Curr Opin Neurobiol. 2008;18:370–376. doi: 10.1016/j.conb.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Palma F, Pellegrino R, Noben-Trauth K. Genomic structure, alternative splice forms and normal and mutant alleles of cadherin 23 (Cdh23). Gene. 2001;281:31–41. doi: 10.1016/s0378-1119(01)00761-2. [DOI] [PubMed] [Google Scholar]
- 13.Bolz H, Von Brederlow B, Ramírez A, Bryda EC, Kutsche K, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 14.Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, et al. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed ZM, Riazuddin S, Wilcox ER. The molecular genetics of Usher syndrome. Clin Genet. 2003;63:431–444. doi: 10.1034/j.1399-0004.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 16.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 17.Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, et al. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- 18.Zak M, Pfister M, Blin N. The otoferlin interactome in neurosensory hair cells: significance for synaptic vesicle release and trans-Golgi network (Review). Int J Mol Med. 2011;28:311–314. doi: 10.3892/ijmm.2011.716. [DOI] [PubMed] [Google Scholar]
- 19.Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, et al. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet. 1999;21:363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- 20.Houseman MJ, Jackson AP, Al-Gazali LI, Badin RA, Roberts E, et al. A novel mutation in a family with non-syndromic sensorineural hearing loss that disrupts the newly characterised OTOF long isoforms. J Med Genet. 2001;38:E25. doi: 10.1136/jmg.38.8.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga R, Kelley PM, Keats BJ, Starr A, Leal SM, et al. Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J Med Genet. 2003;40:45–50. doi: 10.1136/jmg.40.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roux I, Safieddine S, Nouvian R, Grati M, Simmler M, et al. Otoferlin, defective in a human deafness form, is essential for exocytosis at the auditory ribbon synapse. Cell. 2006;127:277–289. doi: 10.1016/j.cell.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 23.Schug N, Braig C, Zimmermann U, Engel J, Winter H, et al. Differential expression of otoferlin in brain, vestibular system, immature and mature cochlea of the rat. Eur J Neurosci. 2006;24:3372–3380. doi: 10.1111/j.1460-9568.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 24.Fay RR, Popper AN. Evolution of hearing in vertebrates: the inner ears and processing. Hear Res. 2000;149:1–10. doi: 10.1016/s0378-5955(00)00168-4. [DOI] [PubMed] [Google Scholar]
- 25.Seidman MD, Ahmad N, Bai U. Molecular mechanisms of age-related hearing loss. Ageing Res Rev. 2002;1:331–343. doi: 10.1016/s1568-1637(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 26.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 27.Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 28.Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, et al. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 29.Adams RA, Pedersen SC. Ontogeny, Functional Ecology, and Evolution of Bats. New York: Cambridge University Press; 2000. 138 [Google Scholar]
- 30.Perrin WF, Würsig B, Thewissen JGM. Encyclopedia of Marine Mammals. San Diego, CA: Academic Press; 2008. 360 [Google Scholar]
- 31.Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, et al. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci. 2006;26:7022–7034. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, et al. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 33.Zakon HH. Convergent evolution on the molecular level. Brain Behav Evol. 2002;59:250–261. doi: 10.1159/000063562. [DOI] [PubMed] [Google Scholar]
- 34.Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet. 2006;38:819–823. doi: 10.1038/ng1812. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama S, Radlwimmer FB. The ‘five-sites’ rule and the evolution of red and green color vision in mammals. Mol Biol Evol. 1998;15:560–567. doi: 10.1093/oxfordjournals.molbev.a025956. [DOI] [PubMed] [Google Scholar]
- 37.Dallos P, Wu X, Cheatham MA, Gao J, Zheng J, et al. Prestin-based outer hair cell motility is necessary for mammalian cochlear amplification. Neuron. 2008;58:333–339. doi: 10.1016/j.neuron.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellado Lagarde MM, Drexl M, Lukashkina VA, Lukashkin AN, Russell IJ. Outer hair cell somatic, not hair bundle, motility is the basis of the cochlear amplifier. Nat Neurosci. 2008;11:746–748. doi: 10.1038/nn.2129. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy HJ, Crawford AC, Fettiplace R. Force generation by mammalian hair bundles supports a role in cochlear amplification. Nature. 2005;433:880–883. doi: 10.1038/nature03367. [DOI] [PubMed] [Google Scholar]
- 40.Chan DK, Hudspeth AJ. Ca2+ current-driven nonlinear amplification by the mammalian cochlea in vitro. Nat Neurosci. 2005;8:149–155. doi: 10.1038/nn1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rauschecker JP, Shannon RV. Sending sound to the brain. Science. 2002;295:1025–1029. doi: 10.1126/science.1067796. [DOI] [PubMed] [Google Scholar]
- 42.Liberman MC, Gao J, He DZ, Wu X, Jia S, et al. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–304. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- 43.Albert JT, Winter H, Schaechinger TJ, Weber T, Wang X, et al. Voltage-sensitive Prestin orthologue expressed in zebrafish hair cells. J Physiol. 2007;580:451–461. doi: 10.1113/jphysiol.2007.127993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheatham MA, Huynh KH, Gao J, Zuo J, Dallos P. Cochlear function in Prestin knockout mice. J Physiol. 2004;560:821–830. doi: 10.1113/jphysiol.2004.069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Assad JA, Corey DP. An active motor model for adaptation by vertebrate hair cells. J Neurosci. 1992;12:3291–3309. doi: 10.1523/JNEUROSCI.12-09-03291.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howard J, Hudspeth AJ. Compliance of the hair bundle associated with gating of mechanoelectrical transduction channels in the bullfrog's saccular hair cell. Neuron. 1988;1:189–199. doi: 10.1016/0896-6273(88)90139-0. [DOI] [PubMed] [Google Scholar]
- 47.Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structural determinants of cadherin-23 function in hearing and deafness. Neuron. 2010;66:85–100. doi: 10.1016/j.neuron.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, et al. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14:103–111. doi: 10.1093/hmg/ddi010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrin WF, Würsig B, Thewissen JGM. Encyclopedia of Marine Mammals. San Diego, CA: Academic Press; 2008. pp. 1126–1127. [Google Scholar]
- 50.Shanahan CM, Cary NR, Metcalfe JC, Weissberg PL. High expression of genes for calcification-regulating proteins in human atherosclerotic plaques. J Clin Invest. 1994;93:2393–2402. doi: 10.1172/JCI117246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uguccioni M, Mackay CR, Ochensberger B, Loetscher P, Rhis S, et al. High expression of the chemokine receptor CCR3 in human blood basophils. Role in activation by eotaxin, MCP-4, and other chemokines. J Clin Invest. 1997;100:1137–1143. doi: 10.1172/JCI119624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chermak GD, Musiek FE. Central Auditory Processing Disorders: New Perspectives. San Diego: Singular Publishing Group Press; 1997. [Google Scholar]
- 53.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 54.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 56.Posada D. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 2009;537:93–112. doi: 10.1007/978-1-59745-251-9_5. [DOI] [PubMed] [Google Scholar]
- 57.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swofford DL. PAUP*: phylogenetic analysis using parsimony (* and other methods). Version 4.0. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- 59.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 60.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 61.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44:S139–146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- 63.Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- 64.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 65.Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. New York: Thieme Press; 1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Domain structure of Cdh23 (SS, signal sequence; EC, ectodomain; HTM, transmembrane domain), and the positions of the positively selected sites and parallel-evolving sites.
(TIF)
The BI tree for Cdh23 based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Domain structure of Pcdh15 (SS, signal sequence; EC, ectodomain; HTM, transmembrane domain), and the positions of the positively selected sites that excluded all parallel-evolving sites.
(TIF)
The BI tree for Pcdh15 based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Parallel-evolved sites of Otof.
(TIF)
The BI tree for Otof based on the amino acid sequences excluding all parallel-evolved sites.
(TIF)
Details of the selective pressure analyses on the three hearing genes.
(DOCX)
Expression levels of Otof in the Common Bent-wing Bat and Old World Fruit Bat.
(DOCX)
Species and their accession numbers for the genes Cdh23, Pcdh15, Otof, and Actb used in this research.
(DOCX)
The primers used for amplifying and sequencing the three hearing genes.
(DOCX)
Details of the TaqMan gene expression assays.
(DOCX)