Abstract
The androgen receptor (AR) is a nuclear receptor that is expressed in growing follicles and involved in folliculogenesis and follicle growth. The orphan nuclear receptor, Nur77, also has an important role in steroid signaling and follicle maturation. We hypothesized that AR levels and androgen signaling through AR are regulated by Nur77 in the ovary. In the ovaries of Nur77 knockout mice (n = 5), real-time PCR results showed that the mRNA levels of AR and an androgen signaling target gene, Kitl, were decreased by 35% and 24%, respectively, relative to wild-type mice (n = 5), which suggested transcriptional regulation of AR by Nur77 in vivo. In cultured mouse granulosa cells and a steroidogenic human ovarian granulosa-like tumor cell line, KGN, mRNA and protein expression levels of AR were increased by overexpressing Nur77 but decreased by knocking down endogenous Nur77. Consistent with increased AR expression, chromatin immunoprecipitation showed that Nur77 bound to the NGFI-B response element (NBRE) in the AR promoter sequence. AR promoter activity was stimulated by Nur77 in HEK293T cells and attenuated in Nur77 knockout mouse granulosa cells (luciferase assay). Overexpression of Nur77 enhanced the androgenic induction of Kitl (200 nM; 48h), while knockout of Nur77 attenuated this induction. These results demonstrate that AR is regulated by Nur77 in the ovaries, and they suggest that the participation of Nur77 in androgen signaling may be essential for normal follicular development.
Introduction
Recently, the study of female reproductive physiology has been focused on the genesis and development of ovarian follicles. Follicular development and function are controlled by a plethora of intrinsic and endocrine factors, and are affected by interactions between multiple cell types within the ovaries [1].
The Nur77 [nerve growth factor induced-B (NGFI-B), nuclear receptor 4A1 (NR4A1)] orphan nuclear receptor is a transcription factor that activates target genes [2]. Nur77 has been shown to recognize a specific binding site called the NGFI-B response element (NBRE; AAAGGTCA), which is present in a subset of genes [3], [4], and Nur77 has been shown to have an effect on transcriptional activation. Nur77 is predominantly expressed in theca cells and granulosa cells of ovarian follicles [5]. Nur77 has a variety of functions, including promoting expression of genes involved in the endocrine, nerve, and immune systems [6]. In the endocrine system, Nur77 has been shown to participate in the regulation of the following genes related to the production of steroidogenic enzymes: Pomc [7] and Cyp17a1 [8] in rats; StAR [9] and Cyp21a1 [3] in mice; and CRH [10] and HSD3B2 [11] in humans.
The androgenic steroid hormones are mostly produced by the adrenal glands and gonads of both sexes. Although androgens are present at much lower concentrations in the circulation of females relative to males, they are essential in maintaining female physiological functions, especially intraovarian functions, such as stimulating follicular growth on early stages [12], enhancing granulosa cell apoptosis [13], and regulating the function and lifespan of the corpus luteum [14]. Androgen signal transduction requires the androgen receptor (AR), which is a member of the nuclear receptor family that has been found to be expressed in growing follicles [15]. In an AR-deficient mouse model, there is an age-dependent progression in ovarian abnormality with a premature ovary failure (POF) phenotype that includes lower follicle numbers, impaired mammary development, fewer pups per litter, and a marked increase of atretic follicles, which indicates that AR-mediated androgen signaling is indispensable for the maintenance of folliculogenesis, and that impaired androgen signaling is a potential cause of the POF syndrome [16]. Nevertheless, the direct involvement of androgen and AR in ovarian follicles is still not well-known.
Nur77 and AR are both involved in follicle maturation. Nur77 is expressed in corpora lutea and cultured ovarian granulosa cells, and it activates steroidogenic gene expression during luteinization, thus, indicating a functional relevance to stage-specific expression during follicular maturation [6]. AR is expressed in all cell types of an ovarian follicle, including oocytes, granulosa cells, and theca cells [17]. Androgen is capable of triggering oocyte maturation in Xenopus laevis [18], mouse [19], and pig [20] most likely via the classical AR mechanism. In cultured granulosa cells or follicles of various species in vitro, AR mediates the stimulatory effects of androgen on proliferation and follicular development [12], [21], [22]. However, the mechanism of AR regulation is unknown.
Therefore, this study aimed at investigating a novel role for Nur77 in AR regulation by testing the hypothesis that AR expression and signaling are affected by Nur77. The mRNA expression patterns of AR and Kitl in the ovaries were examined in Nur77 knockout and wild-type mice to determine the regulation of AR by Nur77 in vivo. The mRNA and protein levels of AR and Kitl were analyzed in cultured mouse granulosa cells and a steroidogenic human ovarian granulosa-like tumor cell line, KGN, after overexpression and knockdown of Nur77 in vitro. The interactions between AR and Nur77 were also studied using chromatin immunoprecipitation and AR promoter activity. Moreover, we showed that Nur77 regulated Kitl expression through stimulation or inhibition of AR signaling using androgen or antiandrogen flutamide, thus, demonstrating an important interaction between Nur77 and androgen signaling in the ovaries.
Materials and Methods
Animals
Nur77 +/− mice (B6.129S2-Nr4a1tm1Jmi/J) were purchased from Jackson Laboratory (Bar Harbor, Maine, USA). Three-week-old ICR mice were purchased from the Lab Animal Center of Yangzhou University (Yangzhou, China). All animals were maintained in the Animal Laboratory Center of Drum Tower Hospital (Nanjing, China) on a 12∶12-h light/dark cycle (lights off at 19∶00) with food and water available ad libitum. All animal experiments were performed according to the guidelines of the Experimental Animals Management Committee (Jiangsu Province, China).
Cell lines
The KGN cell line (a generous gift from Dr. Yiming Mu at the General Hospital of the People's Liberation Army, Beijing, China), which was established from a human GCT and expresses typical granulosa cell markers, was cultured as previously described [23]. HEK293T cells and HEK293A cells were maintained in DMEM/F12 (Gibco BRL/Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Scientific, South Logan, UT, USA), and antibiotics (100 IU/ml penicillin and 100 µg/ml streptomycin; Gibco BRL/Invitrogen) at 37°C in a humidified environment with 5% CO2.
Collection and culture of mouse primary granulosa cells (mGCs)
Mouse granulosa cells were collected from the ovaries of immature mice (21 day old) as previously described [24]. In brief, the ovaries were harvested and cleared from the surrounding fat. The ovaries were then punctured with 25 gauge needles, and the cells were collected into phenol red-free DMEM/F12 (Gibco BRL/Invitrogen) containing 0.2% BSA, 10mM HEPES, and 6.8mM EGTA. The cells were incubated for 15 min at 37°C under 95% O2:5% CO2 and centrifuged for 5 min at 250×g. The pellets were suspended in DMEM/F12 containing 10% fetal bovine serum, 1 mM pyruvic acid (Gibco BRL/Invitrogen), 2 mM glutamine and antibiotics (100 IU/ml penicillin, and 100 µg/ml streptomycin). The oocytes were filtered out with a 40-µm cell strainer. The mouse granulosa cells were cultured in the suspended medium at 37°C in a humidified environment with 5% CO2, and they were used within three passages in this study.
Construction of recombinant adenovirus
Adenoviruses harboring the full-length Nur77 gene (Ad-Flag-Nur77) and adenoviruses harboring human Nur77 siRNA oligonucleotides (5′-AAGUUGUCCGAACAGACAGCCUGA-3′; Ad-siNur77) were generated using the AdMax (Microbix Biosystems, Inc., Toronto, Canada) and pSilencer™ adeno 1.0-CMV (Ambion, Austin, TX, USA) systems as previously described [25]. Adenovirus bearing LacZ (Ad-LacZ) was obtained from BD Biosciences Clontech (Palo Alto, CA, USA). Viruses were packaged and amplified in HEK293A cells and purified using CsCl banding followed by dialysis against 10 mmol/L Tris-buffered saline with 10% glycerol. Titering was performed on HEK293A cells using the Adeno-X Rapid Titer Kit (BD Biosciences Clontech) according to the manufacturer's instructions.
RNA isolation and quantitative real-time PCR
Total RNA was extracted from tissues and cultured cells using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 1mg of purified total RNA using a PrimeScript RT reagent kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's instructions. The specific primers used for quantitative polymerase chain reaction (PCR) analysis are listed in Table 1. Each real-time PCR reaction had the following components: 1 µl of RT product, 10 µl of SYBR Green PCR Master Mix (Bio-Rad Laboratories), and 500 nM forward and reverse primers. Real-time PCR was performed on a MyiQ Single Color Real-time PCR Detection System (Bio-Rad Laboratories) for 40 cycles (95°C for 10 sec; 60°C for 30 min) after an initial 3 min incubation at 95°C. Each sample was analyzed in triplicate, and the experiment was repeated three times. Data were analyzed using the 2−△△CT method [26], and the fold change in expression of each gene was normalized to the endogenous control (18S rRNA).
Table 1. Oligonucleotide primer sequences for quantitative real-time PCR.
| Species | Genes | Primers (5′–3′) | Products size (bp) |
| Mouse | AR | GGACCATGTTTTACCCATCG | 171 |
| TCGTTTCTGCTGGCACATAG | |||
| Mouse | Kitl | TTGTTACCTTCGCACAGTGGCTGGT | 166 |
| AGCGAAGCACTCTGCTCCAACAA | |||
| Mouse | Nur77 | GCACAGCTTGGGTGTTGATG | 187 |
| CAGACGTGACAGGCAGCTG | |||
| Mouse | 18S rRNA | ATGGCCGTTCTTAGTTGGTG | 183 |
| CGGACATCTAAGGGCATCAC | |||
| Human | AR | CCTGGCTTCCGCAACTTACAC | 168 |
| GGACTTGTGCATGCGGTACTCA | |||
| Human | KITLG | TGGTGGCAAATCTTCCAAAAG | 76 |
| CAATGACTTGGCAAAACATCCA | |||
| Human | Nur77 | ACCCACTTCTCCACACCTTG | 200 |
| ACTTGGCGTTTTTCTGCACT | |||
| Human | 18S rRNA | CGGCTACCACATCCAAGGAA | 186 |
| CTGGAATTACCGCGGCT |
Western blot analysis
The proteins were prepared and separated on SDS-PAGE as previously described [27]. Cells were rinsed twice with ice-cold PBS (pH 7.4) and lysed with lysis buffer (50 mM Tris-HCl, pH 7.6; 150 mM NaCl; and 1.0% NP-40) containing protease inhibitor cocktail (Sigma, St. Louis, MO, USA). The protein concentrations were measured by the Bradford assay (Bio-Rad Laboratories). Equal amounts of total protein (40 ug) were separated on a 10% SDS-polyacrylamide gel and transferred onto polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA). Immunoblotting was performed with primary antibodies against hAR (1∶500; Bioworld Technology Inc., Minneapolis, MN, USA), Nur77 (1∶1000; Cell Signaling Technology, Danvers, MA, USA), and mAR (1∶500; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). β-actin (1∶5000; Abcam, Cambridge, MA, USA) was measured as an internal control. Immunodetection was accomplished using a goat anti-rabbit (1∶5000; Bio-Rad Laboratories) or goat anti-mouse (1∶10000; Bio-Rad Laboratories) secondary antibody, and an enhanced chemiluminescence detection kit (Millipore).
Transient transfection and luciferase reporter assays
Approximately 383 bp of the mouse AR promoter sequence (−2509/−2127) was amplified by PCR from mouse granulosa cell genomic DNA with the following primers: 5′-TATAGGTACCGGCCTCAACTGCCTTACTCACAGC-3′ and 5′-TAGCAAGCTTTGGCGTTGGTGGAGTATGCAATCA-3′. Approximately 1.2 kb of the human AR promoter sequence (−2837/−1638) was amplified by PCR from human granulosa-luteinized (hGL) cell genomic DNA with the following primers: 5′-TATAGGTACCATCATTGGTTGCCTGAGGAG-3′ and 5′-TAGCAGATCTGGCAGGATGGTAGAATGGAA-3′. The PCR products were cloned into the pGL3-basic luciferase reporter plasmid (Promega, Madison, WI, USA) and sequenced to confirm the resulting plasmid. Preconfluent (75–80%) HEK293T cells in 12-well plates were transfected with pCMV-Flag-Nur77 or pCMV-empty vector (pCMV-EV), and the AR firefly luciferase report plasmid using Nanofectin (PAA, Pasching, Austria). mGCs (70% confluent) from mice with different genotypes were transfected with the AR luciferase report plasmid using Lipofectamine 2000 (Invitrogen). All cells were cotransfected with the Renilla luciferase reporter plasmid (pRL-RSV; Promega) as a control for transfection efficiency. Luciferase activity was assayed 48 h after transfection using the Luciferase Assay System (Promega) and measured with a Centro XS3 LB 960 luminescence counter (Berthold Technologies, GmbH Co., KG, Germany). At least three transfection assays were performed to obtain statistically significant data.
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed based on the protocol provided in the kit with some modifications (ChIP assay kit; Upstate Biotechnology, Lake Placid, NY, USA) as previously described [28]. Briefly, mGCs or KGN cells (70–80% confluent) were infected with Ad-LacZ and Ad-Flag-Nur77 (10 MOI) for 48 h then washed with PBS and crosslinked with 1% formaldehyde for 15 min at room temperature. Cross-linking was stopped with the addition of glycine (0.125 M final concentration) for 10 min. The cells were washed twice with cold PBS and harvested in lysis buffer A (20 mM Tris-HCl, pH 8.0; 85 mM KCl; 1 mM EDTA; 0.5 mM EGTA; and 0.5% Nonidet P40) containing protease inhibitor cocktail (Sigma), and the cells were then pelleted by centrifugation. The cell pellets were lysed in nuclear lysis buffer B (50 mM Tris-HCl, pH 8.0; 10 mM EDTA; and 1% SDS) containing protease inhibitor cocktail. The cell samples were sonicated in ice to yield genomic DNA fragments with sizes of approximately 500–1000 bp. Then, the precleared supernatants were incubated with 30 µl of anti-FLAG M2 affinity agarose gel (Sigma) and rotated at 4°C overnight. The immune complexes were pulled down by protein A/G beads (Upstate Biotechnology) and eluted by incubation at 65°C for 30 min followed by an incubation at room temperature for 15 min in fresh elution buffer (1% SDS and 0.1 M NaHCO3). The crosslinks were reversed by incubation at 65°C for 5 h with a final concentration of 0.3M NaCl. DNA was purified by phenol: chloroform extraction and ethanol precipitation after the eluates were incubated with proteinase K. Finally, the purified DNA fragments were used as templates for PCR amplification and products were visualized on agarose gels. The specific primers were as follows: 5′-TATAGGTACCGGCCTCAACTGCCTTACTCACAGC-3′ and 5′-TAGCAAGCTTTGGCGTTGGTGGAGTATGCAATCA-3′ (spanning −2509/−2127 bp) for mouse AR promoter DNA fragments and 5′-TATAGGTACCATCATTGGTTGCCTGAGGAG-3′ and 5′-TAGCAGATCTGGCAGGATGGTAGAATGGAA-3′ (spanning −2837/−1638 bp) for human AR promoter DNA fragments.
To provide stronger evidence about Nur77 binding to NBRE in human AR promoter, independent quantitative ChIP experiments were performed. For these studies, chromatin was co-immunoprecipitated from KGN lysates with an antibody to Nur77 or irrelevant antibody (goat anti-rabbit IgG) as control. Lysates were sonicated to generate chromatin fragments of 200 bp which were amplified by quantitative real time PCR. The sets of primers included: 5′-AGTGCTCCAGGTGGAAGAGA-3′ and 5′-CTCTGGAAAGCCAGGATGAG-3′ (spanning −2553/−2401 bp), 5′-AGGAAAGAGTGGAGGGAGGA-3′ and 5′-AGGATGAGCTGACCTTTGGA-3′ (spanning −2584/−2413 bp) which both overlap putative Nur77-binding site of human AR promoter; 5′-TCCTTAAATGCAGGGTCCAC-3′ and 5′-AAATGCCCTGTGATTTCAGC-3′ (spanning −4713/−4526 bp), 5′-GAGTCGTTGGAGGACCTGAA-3′ and 5′-CCTGTCACTGTCTCCCCATT-3′ (spanning −4119/−3967 bp) as negative control region.
Avidin-biotin conjugate DNA (ABCD) precipitation assay
Double-stranded oligonucleotides were designed for ABCD assays and biotinylated at the 3′-end of the sense strand. The following primers were designed: 5′ biotin–CACCATTTTCCAAAGGTCAGCTCATCCTGG-3′ for human AR wild-type F (from −2440 to −2411 bp); and 5′ biotin–CACCATTTTCCAgAatTCAGCTCATCCTGG-3′ for human AR mutant F. KGN cells infected with Ad-LacZ and Ad-Flag-Nur77 (10MOI) for 48 h were lysed with lysis buffer (10 mM Tris–Cl, pH 7.8; 1 mM EDTA; 150 mM NaCl; and 0.1% Nonidet P40) containing protease inhibitor cocktail at 4°C. The cell extracts were incubated with 500 pmol of each double-stranded DNA immobilized on streptavidin agarose beads in binding buffer (10 mM Tris, pH 8.0; 150 mM NaCl; 0.5% TritonX-100; 0.5 mM DTT; 0.5 mM EDTA; 10% glycerol; and 20 μg/ml poly [dI–dC]) containing protease inhibitor cocktail for 4 h at 4°C. The protein complexes on the beads were separated by SDS-PAGE, transferred onto polyvinylidine fluoride membrane and probed with an anti-Flag-HRP antibody (1∶3000; Sigma). Immunodetection was performed using an enhanced chemiluminescence detection kit.
Statistical analysis
Data were expressed as the means ± SEM from at least three independent experiments. Student's t-test was performed for comparison between the mean values for two groups, and ANOVA was used to detect differences among more than two groups Values were determined to be significant when P<0.05.
Results
AR and Kitl mRNA levels decrease in Nur77−/− mice ovaries
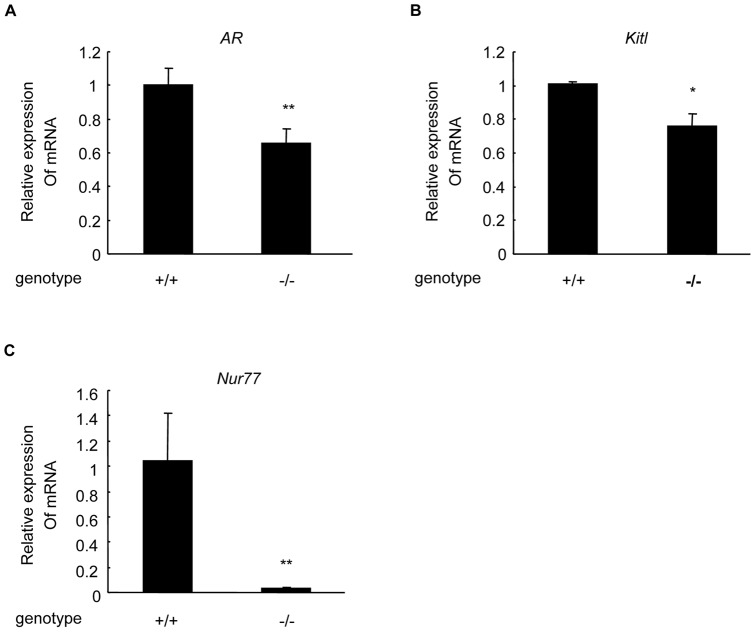
To determine if AR and Kitl in the ovaries were regulated by Nur77 in vivo, AR and Kitl mRNA expression levels were analyzed with real-time PCR in the ovaries from Nur77 +/+ and Nur77 −/− 20-week-old female mice. In comparison with Nur77 +/+ ovaries, the expression levels of AR mRNA (Figure 1A) and Kitl mRNA (Figure 1B) were decreased by 35% and 24%, respectively, in Nur77 −/− ovaries (Figure 1C).
Figure 1. AR and Kitl mRNA levels decreased in Nur77 −/− mouse ovaries.
RNA was collected from 20-week-old Nur77 +/+ and Nur77 −/− female mouse ovaries and reverse transcribed for real-time PCR. AR (A), Kitl (B) and Nur77 (C) mRNA levels of Nur77 −/− mouse ovaries (n = 5) versus Nur77 +/+ mouse ovaries (n = 5) were measured and expressed as relative quantities (*P<0.05, **P<0.01).
Nur77 increases AR and Kitl expression in mouse primary granulosa cells
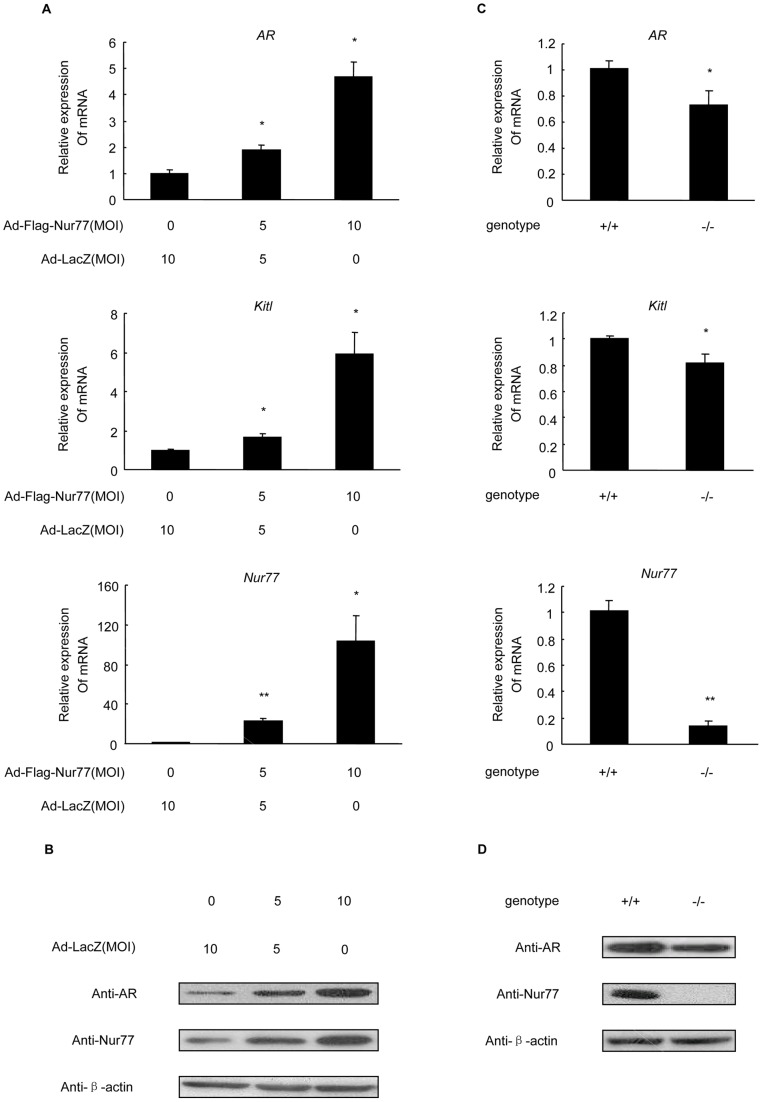
To investigate the functional role of Nur77 in the regulation of AR and Kitl expression in mGCs, we used adenoviral technology to alter the endogenous Nur77 protein expression in Nur77 +/+ mGCs at an MOI of 0, 5 and 10 for 48 h, and we collected RNA for real-time PCR analysis and protein for western blot analysis. We found that adenovirus-mediated overexpression of Nur77 in Nur77 +/+ mGCs increased both AR mRNA and protein expression levels (Figure 2A, B) in a concentration-dependent manner. Moreover, as shown in Figure 2A, Kitl mRNA expression was significantly increased by approximately 6.0-fold when Nur77 was overexpressed at an MOI of 10 in Nur77 +/+ mGCs.
Figure 2. Effects of Nur77 overexpression and knockout on mouse AR and Kitl expression. A:
Nur77 +/+ mGCs were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h. AR and Kitl mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). B: AR protein levels were measured by western blot analysis. C: Real-time PCR analysis of AR and Kitl mRNA expression in Nur77 +/+ and Nur77 −/− mGCs from 3-week-old mice. D: Western blot analysis of AR protein expression in Nur77+/+ and Nur77−/− mGCs from 3-week-old mice. The results are an average of three independent experiments performed in triplicate (*P<0.05, **P<0.01).
To further confirm the regulatory effect of Nur77 on AR and Kitl expression in vivo, we isolated mGCs from Nur77 +/+ and Nur77 −/− female mouse ovaries. As expected, both AR and Kitl mRNA expression levels were significantly decreased in Nur77 −/− mGCs as compared to wild-type mGCs (Figure 2C). Furthermore, AR protein expression decreased (Figure 2D) in purified mGCs from Nur77 −/− female mouse ovaries. Together, these findings strongly suggested that Nur77 is involved in the regulation of AR and Kitl expression in mouse ovary granulosa cells.
Nur77 binds to the mouse AR promoter and stimulates its activity
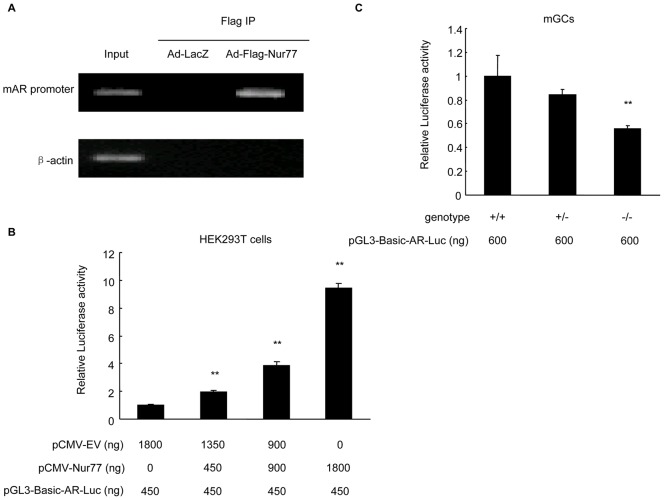
Nur77 has been reported to regulate target gene expression through direct binding to the NBRE sequence (AAAGGTCA) as monomers (4) and/or to the palindromic NurBE sequence as homodimers (7). Based on the mouse AR promoter sequence deposited in the Transcriptional Regulatory Element Database (accession No. 83828), we found a potential NBRE (TCAGGTCA) within mAR promoter core regions. To examine if Nur77 can bind to the AR promoter in vivo, we performed a ChIP-PCR assay. Confluent mGCs were infected with Ad-LacZ and Ad-Flag-Nur77 at an MOI of 10 for 48 h. Soluble chromatin from the mGCs was then immunoprecipitated with an anti-FLAG M2 affinity agarose gel, and the purified DNA fragments from the Flag-Nur77-Chromatin complexes were analyzed by PCR with the specific primer pairs designed for the NBRE region of the mAR promoter. Figure 3A showed that the AR promoter was efficiently recovered from the Flag-Nur77 immunoprecipitates but not from the control immunoprecipitates (Ad-LacZ), suggesting that Nur77 directly binds to the chromatin-associated AR promoter in mouse granulosa cells.
Figure 3. Nur77 binds to the mouse AR promoter and stimulates its activity.
A: The results of ChIP-PCR amplification are shown using primers against the mouse AR promoter region. PCR products were separated by agarose gel electrophoresis. B: HEK293T cells were transfected with pCMV-Nur77 or pCMV-empty vector plasmid and cotransfected with the mAR promoter-luciferase construct using the Nanofectin transfection reagent. After 48 h, luciferase assays were performed and normalized by constitutive Renilla luciferase. C: mGCs from Nur77 +/+, Nur77 +/− and Nur77 −/− mice were transfected with mAR promoter-luciferase construct using Lipofectamine 2000 reagent. After 48 h, luciferase assays were performed and normalized by constitutive Renilla luciferase (n = 3; *P<0.05, **P<0.01).
To test if Nurr77 directly regulates mAR promoter activity, we measured AR gene promoter activity in HEK293T cells and mGCs by transient transfection and luciferase reporter assays. As shown in Figure 3B, overexpression of Nur77 in HEK293T cells enhanced mAR promoter activity in a concentration-dependent manner. In mGCs from Nur77 −/− mice, however, the mAR promoter activity was significantly lower than that in mGCs from Nur77 +/+ mice (Figure 3C). Together, these data suggested that Nur77 binds to the mAR promoter and increases AR promoter activity.
Nur77 up-regulates AR and KITLG expression levels in human granulosa cells
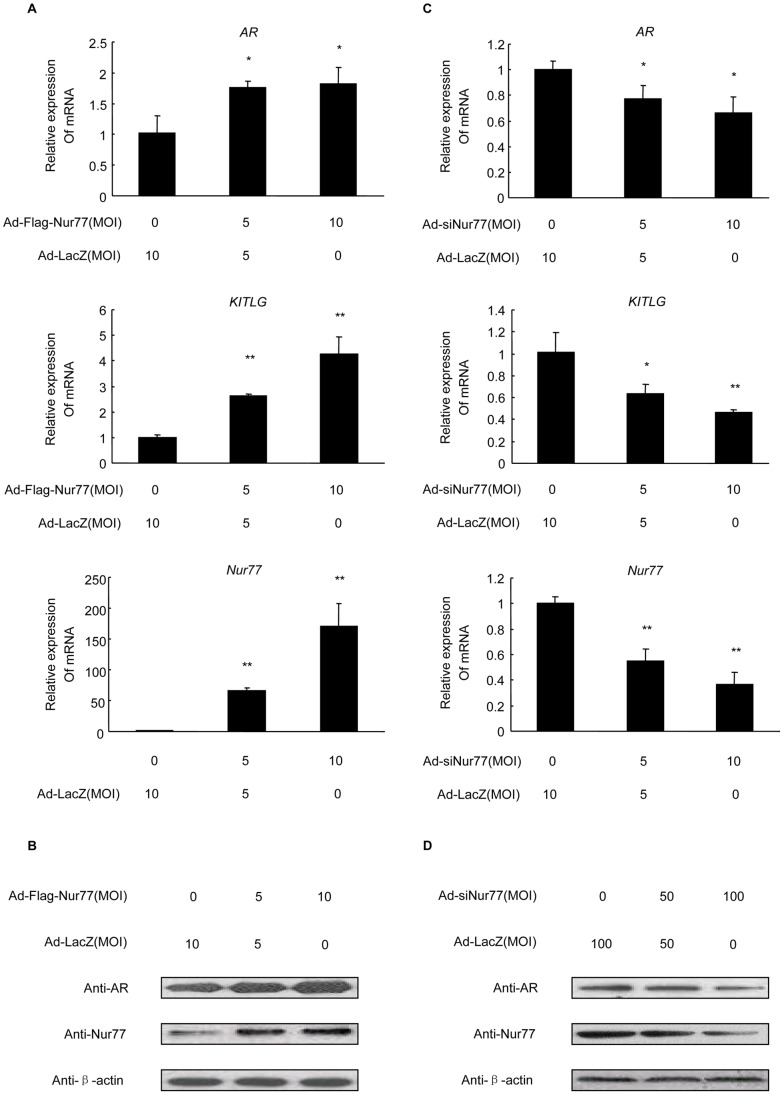
To assess if Nur77 also regulates the transcription of the human AR gene, we treated KGN cells, a human granulosa-like tumor cell line, with Ad-Flag-Nur77 at the indicated MOI for 48 h. Using real-time PCR analysis, we found that mRNA levels of hAR and hKITLG were dramatically elevated in a dose-dependent manner with increasing expression levels of Nur77 (Figure 4A). Moreover, western blot analysis indicated an increase in AR protein levels an increase in Figure 4B).
Figure 4. Effects of Nur77 overexpression and silencing on human AR and KITLG expression in KGN cells. A:
KGN cells were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h, and AR and KITLG mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). (n = 3; *P<0.05, **P<0.01). B: Western blot analysis of AR protein expression in KGN cells treated with Ad-LacZ or Ad-Flag-Nur77 at the indicted MOI for 48 h. A: Real-time PCR analysis of AR and KITLG mRNA expression levels in KGN cells infected with Ad-LacZ or Ad-siNur77 (MOI = 50 and 100) for 48 h (n = 3; *P<0.05, **P<0.01). B: Western blot analysis of AR protein expression in KGN cells treated with Ad-LacZ or Ad-siNur77 at the indicated MOI for 48h.
To further support the role of Nur77 in modulating hAR expression, we performed loss of function studies using an RNA interference technique. KGN cells were infected with Ad-siNur77 at MOIs of 0, 50 or 100, and total RNA and protein were measured by real-time PCR and western blot analysis, respectively. As expected, Ad-siNur77-infected cells showed a substantial reduction in endogenous Nur77 mRNA expression. The treatment subsequently inhibited AR and KITLG mRNA expression levels. AR mRNA was down-regulated by the inhibition of endogenous Nur77 to 54%, and KITLG expression was repressed to 42% (Figure 4C). AR protein expression was also attenuated accompanied by repression of Nur77 (Figure 4D). Thus, these results suggest that Nur77 promotes hAR expression in KGN cells.
Nur77 binds to the human AR promoter and enhances its activity
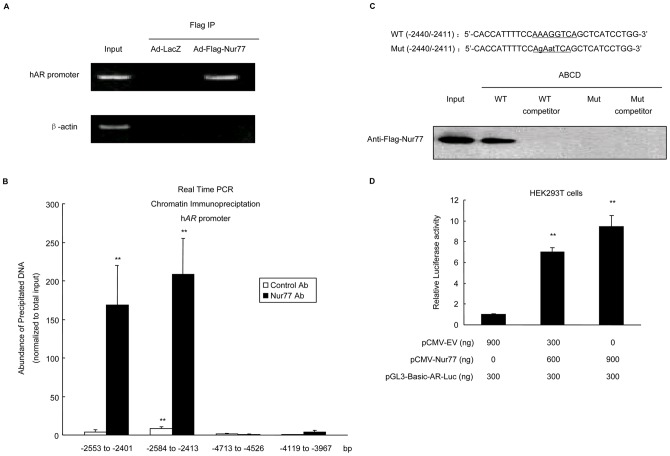
In addition, as the hAR promoter sequence has consensus AAAGGTCA binding sites located at −2429 to −2422 relative to the start site of AR transcription (Transcriptional Regulatory Element Database; accession No. 43390), we first conducted a ChIP-PCR assay as described in mGCs above, and similar results were obtained (Figure 5A). We also used quantitative chromatin immunoprecipitation to further determine if Nur77 binds to the conservative NBRE within the hAR promoter core region in vivo. For these studies, cell lysates were sonicated to generate ∼200bp chromatin fragments prior to immunoprecipitation with an antibody to Nur77 or irrelevant control antibody. Co-precipitating chromatin was amplified by real-time PCR using primers spanning the putative Nur77-binding site (−2429 to −2422bp). Primers used in these experiments were designed to generate a product of <200bp. It was found that chromatin fragments with potential NBRE, instead of negative control region, specifically co-precipitated with Nur77 from lysates of KGN cells (Figure 5B).
Figure 5. Nur77 binds to the human AR promoter and enhances its activity.
A: The results of ChIP-PCR amplification are shown using primers against the human AR promoter region. PCR products were separated by agarose gel electrophoresis. B: Quantitative ChIP was performed with an antibody to Nur77 or irrelevant control antibody. Co-precipitating chromatin fragments were analyzed by real time PCR using primer sets that span −2553/−2401 bp, −2584/−2413 bp, −4713/−4526 bp and −4119/−3967 bp in human AR promoter region. Results were normalized to total input (not precipitated) chromatin(n = 3; *P<0.05, **P<0.01). C: ABCD assays were performed using biotinylated or unbiotinylated (competitor) double-stranded AR wild-type (WT) and AR mutant (Mut) oligonucleotides in addition to KGN cell extracts after infection with Ad-LacZ or Ad-Flag-Nur77. D: HEK293T cells were transfected with pCMV-Nur77 or pCMV-empty vector and cotransfected with the hAR promoter-luciferase construct using the Nanofectin transfection reagent. Forty-eight hours after transfection, luciferase assays were performed and normalized by constitutive Renilla luciferase (n = 3; *P<0.05, **P<0.01).
We next investigated if Nur77 interacts with the human AR promoter in vitro. ABCD assays were performed using biotin conjugate, double-stranded oligonucleotides containing the sequences shown in Figure 5C. The whole cell lysates extracted from intact KGN cells infected with Ad-Flag-Nur77 or Ad-LacZ at an MOI of 10 were mixed with oligonucleotides and immobilized on streptavidin agarose beads. Nur77 strongly bound to the AR wild-type oligonucleotides but not to the mutant sequence (Figure 5C), thus, suggesting that this NBRE site interacts with Nur77 in a sequence-specific manner.
To further verify the direct transcriptional effect of Nur77 on AR in KGN cells, we generated a construct corresponding to approximately 1.2 kb of the human AR promoter sequence (−2837/−1638) in a luciferase reporter plasmid and transiently transfected HEK293T cells to measure AR gene promoter activity. Overexpression of Nur77 activated hAR promoter activity in a concentration-dependent manner (Figure 5D). These data demonstrated that the conserved NBRE (−2429 to −2422) is required for Nur77 specific binding and functional activation of human AR promoter activity.
Nur77 affects the expression of the androgen signaling target gene, Kitl, by regulating AR in granulosa cells
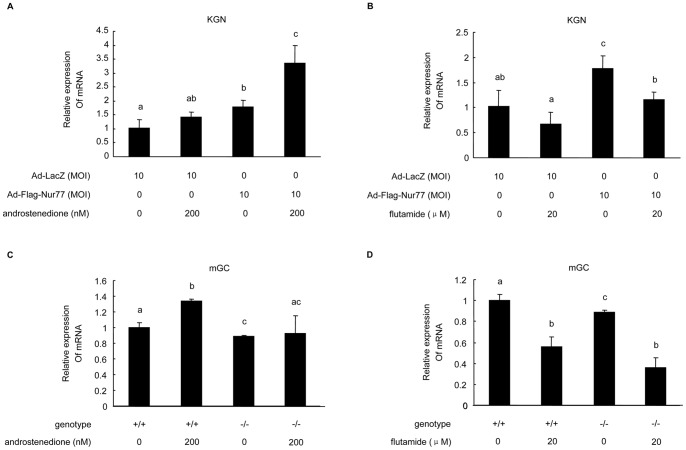
To investigate the mechanism of Kitl gene regulation by Nur77 and AR, KGN cells infected with Ad-LacZ or Ad-Flag-Nur77 at an MOI of 10 and mGCs isolated from Nur77 +/+ and Nur77 −/− female mouse ovaries were treated with androstenedione (AD) or an androgen signaling antagonist, flutamide (FL). Real-time PCR analysis showed that Kitl expression was elevated by AD in KGN cells, which is consistent with a previous report [16]. FL treatments, however, markly attenuated Kitl expression in KGN and Nur77 +/+ mGCs (Figure 6). The stimulatory effects of AD were intensified in KGN cells overexpressing Nur77 (Figure 6A). In Nur77 −/− mGCs, the positive effect of AD was almost completely inhibited (Figure 6C) while the inhibitory effect of FL was enhanced (Figure 6D). Taken together, these findings strongly suggest that Nur77 generates its physiological function in granulosa cells partly through up-regulating of AR expression.
Figure 6. Nur77 affected the expression of the androgen signaling target gene, Kitl, by regulating AR.
KGN cells were infected with Ad-LacZ or Ad-Flag-Nur77 at 10 MOI, and Nur77 +/+ or Nur77 −/− mGCs were treated with androstenedione (A, C) or the androgen signaling antagonist, flutamide (B, D). Cellular RNA was collected, and real-time PCR analysis showed Kitl expression levels with different treatments. The results are shown as a ratio over control (n = 3). Values with different superscripts (a, b, c, d) are significantly different (p<0.05) from each other.
Discussion
Previous studies have shown that Nur77 was originally identified by its rapid activation by nerve growth factor in PC12 cells [29] and by serum growth factors in fibroblasts [30]. Nur77 belongs to the NR4A family of orphan nuclear receptors along with two other members, NURR1 (NR4A2) and NOR1 (NR4A3) [2]. These family members possess similar structural features with a conserved DNA binding domain and a ligand binding domain, but these members have a variable sequence in the N-terminal AF-1 domain [3]. These receptors act as endpoint effectors in the protein kinase A (PKA) signaling pathway. The AF-1 domain of Nur77 has a key role in transcriptional activation, intramolecular interactions, intermolecular interactions, and cofactor recruitment [31], [32]. Nur77 is expressed in various systems, including ovary, testis, adrenal gland, muscle, thymus, pituitary, and central nervous system [6]. Nur77 is known to be regulated in the following ways: by luteinizing hormone in testicular Leydig cells [33]; by follicle-stimulating hormone in mouse testis tissue and human fetal Sertoli cells [34]; and by gonadotropin during follicle development in rat ovaries [8]. Nur77 regulates the expression of genes, such as Cyp21, Cyp11b2, HSD3B2 and StAR, encoding steroidogenic enzymes in the adrenal glands, testes and ovaries [35]–[37], and all of these genes are involved in androgen biosynthesis, thus, suggesting that Nur77 may be important in androgen production as well as its function through AR. Furthermore, both Nur77 and AR are particularly relevant to ovarian follicular maturation [6], [18]–[20]. In the present study, we found that Nur77 positively regulates AR expression in components of the female ovaries. Different granulosa cells, such as mGCs and KGN cells, were used in this study to allow the evaluation and confirmation of the interactions between Nur77 and AR.
The luciferase assays suggested a direct role in transcriptional regulation of AR expression by Nur77. Because a conserved NBRE element (AAAGGTCA) and a partial NBRE sequence (AGGTCA) have been observed in human and mouse AR promoters respectively, further experiments, such as ChIP and ABCD assays were conducted in the current study. These assays suggested that Nur77 was recruited to the AR promoter and that Nur77 bound to the NBRE to mediate AR promoter activation.
As the other two members of the NR4A family, Nurr1 and Nor1, have similar expression patterns and transcriptional roles as Nur77 [38]. It was proposed that there might be potential functional redundancy between NR4A family members [39], [40]. However in the thymuses of Nur77−/− mice, Nurr1 expression was proved to be faint and not increased [41]. In our study, Nurr1 level in the ovary of Nur77−/− mice seemed to have a fluctuation along with the mouse age (data not shown). In addition, expression levels of Nurr1 and Nor1 in human ovarian follicles and corpora lutea are much lower than Nur77 [11]. Thus they may not have a maximal role in regulating AR as Nur77 in the ovaries.
Kit ligand (Kitl) is a growth factor that influences target cells through binding to its tyrosine kinase receptor, Kit [42]. The granulosa cell-derived Kitl can bind to the Kit receptor expressed in oocytes [43], thus, having an important role in the following functions: primordial follicle activation [44]; oocyte growth and survival [45]; meiotic maturation of preovulatory follicles [46]; granulosa cell proliferation [47]; and regulation of ovarian steroidogenesis [48]. Antiandrogen flutamide attenuates Kitl induction by 5α-dihydrotestosterone in mouse ovaries and KGN cells. Moreover, androgen-induced transactivation of mouse and human KITLG promoters has been observed using a luciferase reporter assay in KGN, 293T, and HeLa cells [16]. These studied have established that Kitl represents a direct downstream target of androgen signaling in a regulatory cascade controlling folliculogenesis. In the current study, we found that the changes in Kitl occurred along with changes in AR after the alterations in Nur77 both in vivo and in vitro. To ensure this effect was androgen-dependent, we performed the Nur77 overexpression experiments in KGN cells and Nur77+/+mGCs maintained in complete medium (DMEM/F12 medium plus 10% FBS), phenol-free medium (phenol-free medium plus 10% FBS), or androgen-free medium (phenol-free medium with charcoal/dextran-treated FBS). The results provided evidence that Kitl was regulated by Nur77 in the presence of exogenous androgen, whereas its expression was not up-regulated in androgen-free circumstance (Figure S1, S2). Therefore we consider that Nur77, as an upstream regulator, may indirectly increase androgen-induced Kitl expression through the stimulation of AR, thus, contributing to oocyte growth and maturation.
Global AR knockout (ARKO) female mice are subfertile, and they have defective folliculogenesis. Moreover, ARKO female mice ultimately develop premature ovarian failure [16], [49]. AR signaling in granulosa cells regulates female fertility possibly by controlling preantral follicle growth and development to antral follicles and preventing follicular atresia [50]. Consistent with the function of AR stimulation mediated by Nur77, we postulate that Nur77 represents one of the primary molecules participating in female fertility and folliculogenesis. As there was only the capacity of reproduction which had been analyzed in Nur77−/− mice as a measure of reproductive function in previous studies [39], [41], additional detailed studies need to be performed in our future research.
In conclusion, AR protein expression is positively regulated by Nur77 at least at the transcriptional level through the AR promoter in mouse and human granulosa cells. AR signaling is also mediated by Nur77 as shown by changes in Kitl expression. The transcriptional down-regulation is also confirmed in the ovaries from Nur77 knockout mice in vivo. Our study provides new insights into Nur77 functions in the female reproductive system. Understanding the crosstalk between Nur77 and AR signaling helps us to understand the mechanisms of androgen signaling, follicle growth and oocyte maturation, which are the key pathological processes in diseases, such as premature ovarian failure (POF) and polycystic ovary syndrome (PCOS; hyperandrogenic anovulation).
Supporting Information
Effects of Nur77 overexpression on mouse AR and Kitl expression. Nur77 +/+ mGCs were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h. Cells were cultured in complete medium (A, DMEM/F12 medium plus 10% FBS), phenol-free medium (B, phenol-free medium plus 10% FBS), or androgen-free medium (C, phenol-free medium with charcoal/dextran-treated FBS). AR and Kitl mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). The results are an average of three independent experiments performed in triplicate (*P<0.05, **P<0.01).
(TIF)
Effects of Nur77 overexpression on human AR and KITLG expression. KGN cells were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h. Cells were cultured in complete medium (A, DMEM/F12 medium plus 10% FBS), phenol-free medium (B, phenol-free medium plus 10% FBS), or androgen-free medium (C, phenol-free medium with charcoal/dextran-treated FBS). AR and KITLG mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). The results are an average of three independent experiments performed in triplicate (*P<0.05, **P<0.01).
(TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the State Key Development Program of Basic Research of China Grant (973 Project No. 2010CB945104), the National Natural Science Foundation of China (No. 81070508, No. 30900727, No. 81070492, No. 81170570), the Special Grant for Principal Investigators from the Health Department of Jiangsu Province of PR China (No. XK201102, No. LJ201102, No. RC2011005), and the Natural Science Foundation of Jiangsu Province Grant of PR China (No. BK2009038). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296:2178–2180. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- 2.Maruyama K, Tsukada T, Ohkura N, Bandoh S, Hosono T, et al. The NGFI-B subfamily of the nuclear receptor superfamily. Int J Oncol. 1998;12:1237–1243. doi: 10.3892/ijo.12.6.1237. [DOI] [PubMed] [Google Scholar]
- 3.Wilson TE, Fahrner TJ, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Xue K, Ling J, Diao F, Cui Y, et al. The orphan nuclear receptor NR4A1 regulates transcription of key steroidogenic enzymes in ovarian theca cells. Mol Cell Endocrinol. 2010;319:39–46. doi: 10.1016/j.mce.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Ghosh S, Nishi Y, Yanase T, Nawata H, et al. The orphan nuclear receptors NURR1 and NGFI-B modulate aromatase gene expression in ovarian granulosa cells: a possible mechanism for repression of aromatase expression upon luteinizing hormone surge. Endocrinology. 2005;146:237–246. doi: 10.1210/en.2004-0889. [DOI] [PubMed] [Google Scholar]
- 7.Philips A, Lesage S, Gingras R, Maira MH, Gauthier Y, et al. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Mellon SH. Multiple orphan nuclear receptors converge to regulate rat P450c17 gene transcription: novel mechanisms for orphan nuclear receptor action. Mol Endocrinol. 1997;11:891–904. doi: 10.1210/mend.11.7.9940. [DOI] [PubMed] [Google Scholar]
- 9.Martin LJ, Tremblay JJ. Glucocorticoids antagonize cAMP-induced Star transcription in Leydig cells through the orphan nuclear receptor Nur77. J Mol Endocrinol. 2008;41:165–175. doi: 10.1677/JME-07-0145. [DOI] [PubMed] [Google Scholar]
- 10.Murphy EP, McEvoy A, Conneely OM, Bresnihan B, FitzGerald O. Involvement of the nuclear orphan receptor NURR1 in the regulation of corticotropin-releasing hormone expression and actions in human inflammatory arthritis. Arthritis Rheum. 2001;44:782–793. doi: 10.1002/1529-0131(200104)44:4<782::AID-ANR134>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Havelock JC, Smith AL, Seely JB, Dooley CA, Rodgers RJ, et al. The NGFI-B family of transcription factors regulates expression of 3β-hydroxysteroid dehydrogenase type 2 in the human ovary, Mol Hum Reprod. 2005;11:79–85. doi: 10.1093/molehr/gah139. [DOI] [PubMed] [Google Scholar]
- 12.Vendola KA, Zhou J, Adesanya OO, Weil SJ, Bondy CA. Androgens stimulate early stages of follicular growth in the primate ovary. J Clin Invest. 1998;101:2622–2629. doi: 10.1172/JCI2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billig H, Furuta I, Hsueh AJ. Estrogens inhibit and androgens enhance ovarian granulosa cell apoptosis. Endocrinology. 1993;133:2204–2212. doi: 10.1210/endo.133.5.8404672. [DOI] [PubMed] [Google Scholar]
- 14.Sheri HP, Neal BW, Robert MB, Richard LS. Localization of androgen receptor in the follicle and corpus luteum of the primate ovary during the menstrual cycle. Biol Reprod. 1991;44:561–568. doi: 10.1095/biolreprod44.3.561. [DOI] [PubMed] [Google Scholar]
- 15.Tetsuka M, Whitelaw PF, Bremner WJ, Millar MR, Smyth CD, et al. Developmental regulation of androgen receptor in rat ovary. J Endocrinol. 1995;145:535–543. doi: 10.1677/joe.0.1450535. [DOI] [PubMed] [Google Scholar]
- 16.Shiina H, Matsumoto T, Sato T, Igarashi K, Miyamoto J, et al. Premature ovarian failure in androgen receptor-deficient mice. Proc Natl Acad Sci U S A. 2006;103:224–229. doi: 10.1073/pnas.0506736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan SD, Moenter SM. Prenatal androgens alter GABA ergic drive to gonadotropin-releasing hormone neurons: implications for a common fertility disorder. Proc Natl Acad Sci U S A. 2004;101:7129–7134. doi: 10.1073/pnas.0308058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutz LB, Cole LM, Gupta M K, Kwist KW, Auchus RJ, et al. Evidence that androgens are the primary steroids produced by Xenopus laevis ovaries and may signal through the classical androgen receptor to promote oocyte maturation. Proc Natl Acad Sci U S A. 2001;98:13728–13733. doi: 10.1073/pnas.241471598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill A, Jamnongjit M, Hammes SR. Androgens promote maturation and signaling in mouse oocytes independent of transcription: a release of inhibition model for mammalian oocyte meiosis. Mol Endocrinol. 2004;18:97–104. doi: 10.1210/me.2003-0326. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Schatten H, Sun Q. Androgen receptor's destiny in mammalian oocytes: a new hypothesis. Mol Hum Reprod. 2009;15:149–154. doi: 10.1093/molehr/gap006. [DOI] [PubMed] [Google Scholar]
- 21.Sen A, Hammes SR. Granulosa cell-specific androgen receptors are critical regulators of ovarian development and function. Mol Endocrinol. 2010;24:1393–1403. doi: 10.1210/me.2010-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickey TE, Marrocco DL, Gilchrist RB, Norman RJ, Armstrong DT. Interactions between androgen and growth factors in granulosa cell subtypes of porcine antral follicles. Biol Reprod. 2004;71:45–52. doi: 10.1095/biolreprod.103.026484. [DOI] [PubMed] [Google Scholar]
- 23.Nish Y, Yanase T, Mu Y, Oba K, Ichino I, et al. Establishment and characterization of a steroidogenic human granulosa-Like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology. 2001;142:437–445. doi: 10.1210/endo.142.1.7862. [DOI] [PubMed] [Google Scholar]
- 24.Kipp JL, Mayo KE. Use of reporter genes to study the activity of promoters in ovarian granulosa cells. Methods Mol Biol. 2009;590:177–193. doi: 10.1007/978-1-60327-378-7_11. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Hu Y, Zhao J, Zhen X, Yan G, et al. The orphan nuclear receptor Nur77 regulates decidual prolactin expression in human endometrial stromal cells. Biochem Biophys Res Commun. 2011;404:628–633. doi: 10.1016/j.bbrc.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Yan G, You B, Chen SP, Liao JK, Sun J. Tumor necrosis factor-alpha downregulates endothelial nitric oxide synthase mRNA stability via translation elongation factor 1-alpha 1. Circ Res. 2008;103:591–597. doi: 10.1161/CIRCRESAHA.108.173963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, Chen L, Yan G, Wang R, Diao Z, et al. HOXA10 suppresses p/CAF promoter activity via three consecutive TTAT units in human endometrial stromal cells. Biochem Biophys Res Commun. 2009;379:16–21. doi: 10.1016/j.bbrc.2008.11.144. [DOI] [PubMed] [Google Scholar]
- 29.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 30.Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci U S A. 1988;85:8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maira M, Martens C, Batsche E, Gauthie Y, Drouin J. Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol Cell Biol. 2003;23:763–776. doi: 10.1128/MCB.23.3.763-776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wansa KD, Harris JM, Yan G, Ordentlich P, Muscat GE. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. J Biol Chem. 2003;278:24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- 33.Song KH, Park JI, Lee MO, Soh J, Lee K, et al. LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology. 2001;142:5116–5123. doi: 10.1210/endo.142.12.8525. [DOI] [PubMed] [Google Scholar]
- 34.Ding L, Yan G, Ge Q, Yu F, Zhao X, et al. FSH acts on the proliferation of type A spermatogonia via Nur77 that increases GDNF expression in the Sertoli cells. FEBS Lett. 2011;585:2437–2444. doi: 10.1016/j.febslet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Kelly SN, McKenna TJ, Young LS. Modulation of steroidogenic enzymes by orphan nuclear transcriptional regulation may control diverse production of cortisol and androgens in the human adrenal. J Endocrinol. 2004;181:355–365. doi: 10.1677/joe.0.1810355. [DOI] [PubMed] [Google Scholar]
- 36.Bassett MH, Suzuki T, Sasano H, De Vries CJ, Jimenez PT, et al. The orphan nuclear receptor NGFIB regulates transcription of 3beta-hydroxysteroid dehydrogenase. Implications for the control of adrenalfunctional zonation. J Biol Chem. 2004;279:37622–37630. doi: 10.1074/jbc.M405431200. [DOI] [PubMed] [Google Scholar]
- 37.Martin LJ, Boucher N, Brousseau C, Tremblay JJ. The orphan nuclear receptor Nur77 regulates hormone-induced StAR transcription in Leydig cells through cooperation with Ca2+/calmodulin-dependent protein kinase I. Mol Endocrinol. 2008;22:2021–2037. doi: 10.1210/me.2007-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, et al. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 39.Crawford PA, Sadovsky Y, Woodson K, Lee SL, Milbrandt J. Adrenocortical function and regulation of the steroid 21-hydroxylase gene in NGFI-B-deficient mice. Mol Cell Biol. 1995;15:4331–4336. doi: 10.1128/mcb.15.8.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16:1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, et al. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77). Science. 1995;269:532–535. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 42.Karla JH, Eileen AM, Michael KH. KIT/KIT Ligand in Mammalian Oogenesis and Folliculogenesis: Roles in Rabbit and Murine Ovarian Follicle Activation and Oocyte Growth. Biol Reprod. 2006;75:421–433. doi: 10.1095/biolreprod.106.051516. [DOI] [PubMed] [Google Scholar]
- 43.Thomas FH, Vanderhyden BC. Oocyte-granulosa cell interactions during mouse follicular development: regulation of kit ligand expression and its role in oocyte growth. Reprod Biol Endocrinol. 2006;4:19. doi: 10.1186/1477-7827-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrott JA, Skinner MK. Kit-ligand/stem cell factor induces primordial follicle development and initiates folliculogenesis. Endocrinology. 1999;140:4262–4271. doi: 10.1210/endo.140.9.6994. [DOI] [PubMed] [Google Scholar]
- 45.Packer AI, Hsu YC, Besmer P, Bachvarova RF. The ligand of the KIT receptor promotes oocyte growth. Dev Biol. 1994;161:194–205. doi: 10.1006/dbio.1994.1020. [DOI] [PubMed] [Google Scholar]
- 46.Ye Y, Kawamura K, Sasaki M, Kawamura N, Groenen P. Kit ligand promotes first polar body extrusion of mouse preovulatory oocytes. Reprod Biol Endocrinol. 2009;7:26. doi: 10.1186/1477-7827-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otsuka F, Shimasaki S. A negative feedback system between oocyte bone morphogenetic protein 15 and granulosa cell kit ligand: its role in regulating granulosa cell mitosis. Proc Natl Acad Sci U S A. 2002;99:8060–8065. doi: 10.1073/pnas.122066899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutt K, McLaughlin EA, Holland MK. KL and KIT have diverse role during mammalian oogenesis. Mol Human Reprod. 2006;12:61–69. doi: 10.1093/molehr/gal010. [DOI] [PubMed] [Google Scholar]
- 49.Walters KA, Allan CM, Jimenez M, Lim PR, Davey RA, et al. Female mice haploinsufficient for an inactivated androgen receptor (AR) exhibit age-dependent defects that resemble the AR null phenotype of dysfunctional late follicle development, ovulation, and fertility. Endocrinology. 2007;148:3674–3684. doi: 10.1210/en.2007-0248. [DOI] [PubMed] [Google Scholar]
- 50.Aritro S, Stephen RH. Granulosa Cell-Specific Androgen Receptors Are Critical Regulators of Ovarian Development and Function. Mol Endocrinol. 2010;24:1393–1403. doi: 10.1210/me.2010-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effects of Nur77 overexpression on mouse AR and Kitl expression. Nur77 +/+ mGCs were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h. Cells were cultured in complete medium (A, DMEM/F12 medium plus 10% FBS), phenol-free medium (B, phenol-free medium plus 10% FBS), or androgen-free medium (C, phenol-free medium with charcoal/dextran-treated FBS). AR and Kitl mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). The results are an average of three independent experiments performed in triplicate (*P<0.05, **P<0.01).
(TIF)
Effects of Nur77 overexpression on human AR and KITLG expression. KGN cells were infected with Ad-LacZ or Ad-Flag-Nur77 at the indicated MOI for 48 h. Cells were cultured in complete medium (A, DMEM/F12 medium plus 10% FBS), phenol-free medium (B, phenol-free medium plus 10% FBS), or androgen-free medium (C, phenol-free medium with charcoal/dextran-treated FBS). AR and KITLG mRNA levels were measured by real-time PCR and shown as a ratio over control (Ad-LacZ). The results are an average of three independent experiments performed in triplicate (*P<0.05, **P<0.01).
(TIF)