Abstract
AIM: To investigate the effect of chloride intracellular channel 1 (CLIC1) on the cell proliferation, apoptosis, migration and invasion of gastric cancer cells.
METHODS: CLIC1 expression was evaluated in human gastric cancer cell lines SGC-7901 and MGC-803 by real time polymerase chain reaction (RT-PCR). Four segments of small interference RNA (siRNA) targeting CLIC1 mRNA and a no-sense control segment were designed by bioinformatics technology. CLIC1 siRNA was selected using Lipofectamine 2000 and transfected transiently into human gastric cancer SGC-7901 and MGC-803 cells. The transfected efficiency was observed under fluorescence microscope. After transfection, mRNA expression of CLIC1 was detected with RT-PCR and Western blotting was used to detect the protein expression. Proliferation was examined by methyl thiazolyl tetrazolium and apoptosis was detected with flow cytometry. Polycarbonate membrane transwell chamber and Matrigel were used for the detection of the changes of invasion and migration of the two cell lines.
RESULTS: In gastric cancer cell lines SGC-7901 and MGC-803, CLIC1 was obviously expressed and CLIC1 siRNA could effectively suppress the expression of CLIC1 protein and mRNA. Proliferation of cells transfected with CLIC1 siRNA3 was enhanced notably, and the highest proliferation rate was 23.3% (P = 0.002) in SGC-7901 and 35.55% (P = 0.001) in MGC-803 cells at 48 h. The G2/M phase proportion increased, while G0/G1 and S phase proportions decreased. The apoptotic rate of the CLIC1 siRNA3 group obviously decreased in both SGC-7901 cells (62.24%, P = 0.000) and MGC-803 cells (52.67%, P = 0.004). Down-regulation of CLIC1 led to the inhibition of invasion and migration by 54.31% (P = 0.000) and 33.62% (P = 0.001) in SGC-7901 and 40.74% (P = 0.000) and 29.26% (P = 0.002) in MGC-803. However, there was no significant difference between the mock group cells and the negative control group cells.
CONCLUSION: High CLIC1 expression can efficiently inhibit proliferation and enhance apoptosis, migration and invasion of gastric cancer cells in vitro. CLIC1 might be a promising target for the treatment of gastric cancer.
Keywords: Chloride intracellular channel 1, Gastric carcinoma, Small interference RNA, Apoptosis, Invasion, Migration
INTRODUCTION
Gastric cancer represents the second most common cause of cancer-related deaths in the world, and the incidence is higher in Asia than in other geographical areas[1,2]. Although progress is made in early detection and adjuvant therapy, the precise mechanism underlying gastric cancer remains unclear.
Recently, the roles of ion transporters have been studied in cancer cells[3,4] and various types of ion transporters have been found in cancers of digestive organs. Chloride intracellular channel (CLIC) proteins are components or regulators of novel intracellular anion channels in mammalian cells. To date, seven distinct members of CLICs have been identified: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64 and parchorin[5]. These genes have a high degree of homology at their carboxyl termini. In terms of their molecular function in vitro, it is clear that all CLIC proteins and the invertebrate CLIC-like proteins exist as both soluble globular proteins and integral membrane proteins that possess ion channel activity. The transition between these two forms is influenced by pH and redox conditions under most instances[6]. CLIC1 is the first cloned human member of the CLIC family, and is a 241 amino acid ion channel protein. Like other members of the family, CLIC1 is highly conserved across a wide range of species and its relatives can be identified in the genome of all vertebrates so far sequenced. CLIC1 was initially found to localize in the cell nucleus and intracellular vesicles[7]. Ulmasov et al[8] found that CLIC1 was expressed in the apical domains of several simple columnar epithelia, including glandular stomach, small intestine, colon, bile ducts, pancreatic ducts, airway, the tail of the epididymis and renal proximal tubule. Chen et al[9] found that a trace amount of CLIC1 was expressed in normal gastric tissues, but it was overexpressed in gastric cancer. Elevated CLIC1 expression was strongly correlated with lymph node metastasis, lymphatic invasion, perineural invasion and pathological staging, which suggested that it was a potential prognostic marker[9]. However, its role in gastric cancer deserves further studies.
In the present study, we used the RNA interference (RNAi) technology, with a Lipofectamine 2000, in order to deliver small interference RNA (siRNA) molecules that target CLIC1 gene of the gastric cancer cells. RNAi is a highly specific, homology dependent suppression of gene expression by small double-stranded RNA (dsRNA)[10]. Long dsRNAs are cleaved by the endoribonuclease Dicer into short dsRNA duplexes or siRNA. siRNAs are loaded onto RNA-induced silencing complex (RISC)[11]. RISC contains argonaute 2 (Ago-2) which cleaves and releases one strand from the dsRNA, resulting in an activated form of RISC with a single-strand RNA (guide siRNA) that directs the specificity of the target mRNA recognition through complementary base pairing[12]. Ago-2 then cleaves the target mRNA between bases 10 and 11 related to the 5’ end of the siRNA antisense strand, thereby causing mRNA degradation and gene silencing[12,13], which occurs at a post-transcriptional level[14]. Here, we employed CLIC1 siRNA to knockdown the expression of CLIC1, and investigated its effects on cell proliferation, cycle, apoptosis, migration, invasion and the mechanisms involved.
MATERIALS AND METHODS
Cell cultures
Human gastric cancer cell lines SGC-7901 (adenocarcinomas) and MGC-803 (adenocarcinomas) were obtained from Shanghai cell bank (http://www.cellbank.org.cn), Chinese Academy of Sciences (Shanghai, China). The cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, United States) , supplemented with 10% (v/v) fetal bovine serum (FBS) (HyClone, Logan City), penicillin G (100 units/mL) (Sigma, United States) and streptomycin (100 μg/mL) (Sigma, United States), which was termed complete medium. Cells were grown in monolayer culture at 37 °C in humidified air with 5% CO2.
Real-time polymerase chain reaction assays
Total RNA was extracted from human gastric cancer cell lines SGC-7901 and MGC-803 using TRIzol reagent (Invitrogen, CA). Total RNA (1-2 μg) was reversely transcribed using the Fermentas Kit (MBI, United State). Primers were based on sequences reported at Genebank (NM_001288.4). CLIC1 sense sequence was 5’-AATCAAACCCAGCACTCAATG-3’ and anti-sense sequence was 5’-CAGCACTGGTTTCATCCACTT-3’. The expected product size of CLIC1 cDNA was 114 bp. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sense sequence was 5’-CAACGACCCCTTCATTGACC-3’ and anti-sense sequence was 5’-CGCCAGTAGACTCCACGACAT-3’. The expected product size of GAPDH cDNA was 203 bp. Polymerase chain reaction (PCR) amplification was performed in 50 μL reaction volumes containing 0.2 mmol/L each dNTP, 0.1 RM of each oligonucleotide primer, and 1.25 U Tag polymerase in PCR buffer. cDNA was amplified on a PCR thermal controller with an initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 20 s, 59 °C for 30 sec, and 68 °C for 30 s and a final extension step of 72 °C for 10 min. The amount of starting cDNA was adjusted using GAPDH intensity.
siRNA synthesis and cell transfection assays
This study included four human siRNAs (GenePharma, Shanghai, China) designed against CLIC1 (GenBank NM_001288.4). One negative random siRNA (GenePharma, Shanghai, China) exhibiting no significant sequence similarity to human, mouse or rat gene sequence, served as a negative control. The sequences for CLIC1 siRNAs and control siRNA were: siRNA1: CLIC1-Homo-131 (sense: 5’-GAGCUUGUGUUGUGCUGAATT-3’ antisense: 5’-UUCAGCACAACACAAGCUCTT-3’); siRNA2: CLIC1-Homo-630 (sense: 5’-CAGCACUCAAUGACAAUCUTT-3’ antisense: 5’-AGAUUGUCAUUGAGUGCUGTT-3’); siRNA3: CLIC1-Homo-805 (sense: 5’-GCCAAAGUUACACAUAGUATT-3’ antisense: 5’-UACUAUGUGUAACUUUGGCTT-3’); siRNA4: CLIC1-Homo-1195 (sense: 5’-GGACAACAUAUUUCAGUAATT-3’ antisense: 5’-UUACUGAAAUAUGUUGUCCTT-3’); and the negative random siRNA 5 (sense: 5’-UUCUCCGAACGUGUCACGUTT-3’ anti-sense: 5-ACGUGACACGUUCGGAGAATT-3’). The cells were split into seven groups, including the mock group supplemented with only the transfection reagent, the negative control group added with a non-targeting control siRNA and the transfection reagent, and the other five groups supplemented with different CLIC1 siRNA and the transfection reagents. For cell transfection, cells were plated on 96-well (5 × 103 cells for SGC-7901 and 3 × 103 cells for MGC-803) and 6-well plates (5 × 105 cells for SGC-7901 and 3 × 105 cells for MGC-803) in DMEM with 10% FBS and were allowed to attach for 24 h, and then treated with 5 pmol and 100 pmol siRNA per well. Equimolar amounts of siRNAs were incubated with Lipofectamine 2000 Transfection Reagent from Invitrogen (Madison, WI, United States) according to the manufacturer’s instructions. Transfected cells were grown at 37 °C for 6 h, followed by incubation with complete medium. Cells were maintained for 24 h, 48 h and 72 h before experiments, unless otherwise described.
Quantitative real-time PCR assays
Total RNA was extracted from the human gastric cancer cell lines SGC-7901 and MGC-803 using TRIzol reagent (Invitrogen, United States). After reverse transcription of the total RNA, the first-strand cDNA was then used as template for detection of CLIC1 expression using quantitative real-time PCR with the SYBR Green Master Mix (Roche, Germany). β-actin was used as control. The primers of CLIC1 sense sequence was 5’-AATCAAACCCAGCACTCAATG-3’ and CLIC1 anti-sense sequence 5’-CAGCACTGGTTTCATCCACTT-3’(product size of 114 base pairs). β-actin sense sequence was 5’-ACACTGTGCCCATCTACG-3’ and anti-sense sequence 5’-TGTCACGCACGATTTCC-3’ (product size of 153 base pairs). The cycling conditions included a holding step at 95 °C for 10 min, and 42 cycles of 95 °C for 20 s, 59 °C for 30 s and 68 °C for 30 s. A dissociation protocol was added to verify that the primer pair produced only a single product at 95 °C for 15 s, 60 °C for 1min, and 95 °C for 31s. Melting curves also showed a single sharp peak indicating one PCR product. Quantitative real-time PCR analysis was performed using an ABI 7500 Sequence Detector (ABI, Warrington, United Kingdom) according to the manufacturer’s protocol, and the results were analyzed by the 2-ΔΔct. These experiments were performed in triplicate and repeated in three independent experiments.
Western blotting assays
Total protein extracts were separated by 12% sodium dodecylsulfate polyacrylamide gel electrophoresis (20 mg per lane), and transferred onto a polyvinylidene fluoride membrane. After blocking with 5% bovine serum albumin, the membrane was then incubated with antibodies specific for CLIC1 (Sigma; 1:500), and β-actin (Abmart; 1:5000) at 4 °C overnight. The secondary antibody (KPL; 1:5000) was infrared at room temperature for 1 h. The density of the bands was quantified by densitometric analysis using the two-color infrared laser imaging analyzer system of Odyssey (United States). The inhibitory rate of CLIC1 protein expression was calculated as follows: inhibitory rate = [1-(siRNA CLIC1 density/siRNA β-actin density)/(untransfected CLIC1 density/untransfected β-actin density)] × 100%.
Cell growth and viability assay
The effect of CLIC1 specific siRNA on the viability of cells was determined by the methyl thiazolyl tetrazolium (MTT) assay. Briefly, cells were plated in 96-well microtitre plates. Cell viability was determined after transfection for 24 h, 48 h and 72 h. Then 20 μL MTT (10 mg/mL in PBS stock, diluted to a working concentration of 1 mg/mL with media) was added to each well and incubated for 4 h. After careful removal of the medium, 200 μL dimethyl sulfoxide was added to each well and shaken carefully for 10 min. The absorbance was recorded on the iMark Microplate Reader (Bio-Rad, United Kingdom) at a 570 nm wavelength. The effect of CLIC1 siRNA on cell growth inhibition was assessed as percentage cell viability where vehicle-treated cells were taken as 100% viable.
Cell cycle analysis and annexin V staining assays
For flow cytometric cell cycle analysis, the cells treated with siRNA were collected, washed with PBS, fixed in cold 70% ethanol, and stored at 4 °C until staining. After fixation, the cells were washed with PBS and incubated with 100 μL RNaseA (Sigma, United States) for 30 min at 37 °C, before staining with 400 μL propidium iodide (Sigma, United States). Apoptotic cells in early and late stages were detected using an annexin V-FITC Apoptosis Detection Kit from BioVision (Mountain View, CA, United States). In brief, culture media and cells were collected and centrifuged. After washing, cells were resuspended in 500 μL annexin V binding buffer, followed by the addition of 5 μL annexin V-FITC and 5 μL propidium iodide. The samples were incubated in the dark for 5 min at room temperature and analyzed using flow cytometry (FCM).
Cell migration assay
The ability of cells to migrate through filters was measured using Polycarbonate Membrane Transwell Inserts (Corning, United State). At 24 h after transfection, cells were trypsinised. Cell culture inserts with an 8 μm pore size polycarbonate membrane were used according to the protocol of the manufacturer. The bottom chamber included medium (0.5 mL) containing 5% FBS, whereas mock, negative control or CLIC1 siRNA transfected cells (1.0 × 106 per mL suspended in 0.1 mL of medium containing 0.5% FBS) were seeded into the upper chamber and incubated 24 h at 37 °C in a humidified atmosphere containing 5% CO2. The remaining cells on the upper surface were mechanically removed. Membranes were then washed, fixed, and stained by Methyl Violet (Medion Diagnostics, Germany). The migration ability of the cells was determined by counting the cells that had migrated to the lower side of the filter with a microscope. Experiments were performed in triplicate, and 3 fields were counted in each experiment.
Matrigel invasion assay
Matrigel invasion assay was performed using polycarbonate membrane transwell (Corning, United States) coated with the matrigel (BD Biosciences, San Jose, CA, United States). The density of cells that were seeded into the upper chamber was 3.0 × 105/mL for SGC-7901 and 1.5 × 105/mL for MGC-803. Other treatments were the same with the cell migration assay.
Statistical analysis
Data were analyzed using SPSS16.0 software. All data were expressed as mean ± SD, and analyzed by one-way analysis of variance and post-hoc testing comparing means by Student-Newman-Keuls’ test. P < 0.05 was considered statistically significant.
RESULTS
Expression of CLIC1 in cultured gastric cancer cells
We first evaluated the endogenous expression of CLIC1 in the human gastric cancer cell lines SGC-7901 and MGC-803 by RT-PCR, and found that there was CLIC1 mRNA in the human gastric cancer cell lines SGC-7901 and MGC-803 (Figure 1).
Figure 1.
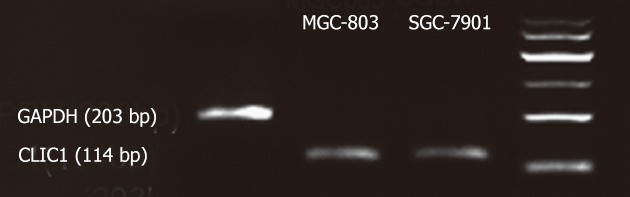
Expression of chloride intracellular channel 1 in human gastric cancer cell lines SGC-7901 and MGC-803. Specific reverse transcription polymerase chain reaction analysis for chloride intracellular channel 1 (CLIC1).Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as loading control.
Downregulation of CLIC1 expression effectively suppresses CLIC1 protein and mRNA expression in gastric cancer cells
To examine the possible roles of CLIC1 in gastric cancer cells, we knocked down the expression of CLIC1 using siRNA. Western blotting and quantitative real-time PCR were performed to examine the effect of siRNA transfection on CLIC1 protein and mRNA expression levels in SGC-7901 and MGC-803 cells. When SGC-7901 and MGC-803 cells were transfected with CLIC1 siRNA, the CLIC1 mRNA and protein levels were decreased in different degrees in siRNA1, siRNA2, siRNA3 and siRNA4 transfected cells as compared with siRNA5, mock and negative control cells at different time points (P < 0.05). There were no significant differences among siRNA5, mock and negative control cells (P > 0.05) (Figure 2). The results showed that these siRNAs designed for CLIC1, especially the CLIC1 siRNA3, successfully exerted a silencing effect for CLIC1 expression in vitro. Therefore, the CLIC1 siRNA3 was chosen for the subsequent in vitro experiments.
Figure 2.
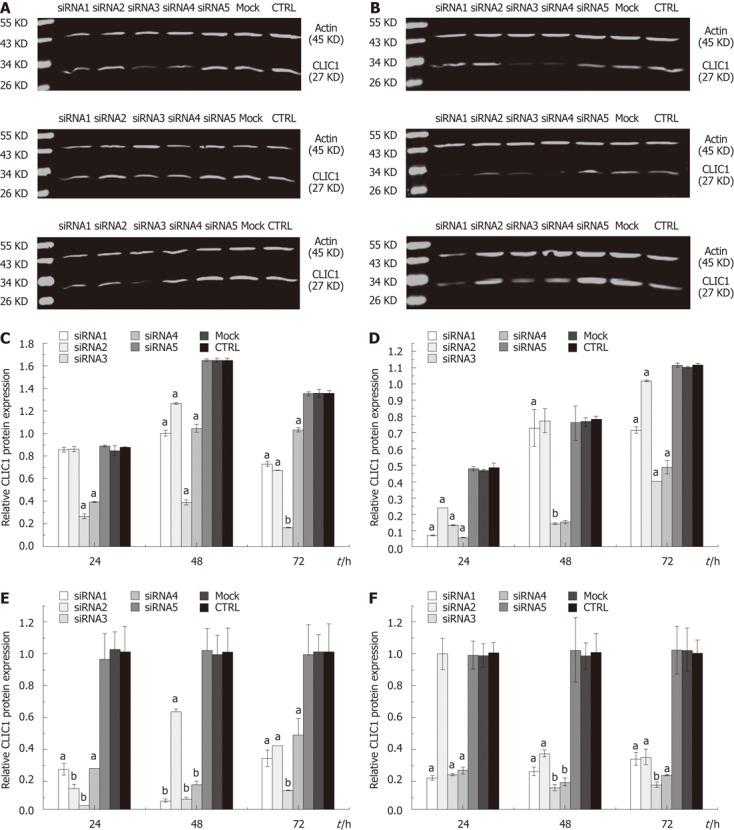
Effect of chloride intracellular channel 1 knockdown in SGC-7901 and MGC-803 cells. Chloride intracellular channel 1 (CLIC1) protein expression was detected by Western blotting using a rat polyclonal CLIC1 antibody (1:500). β-actin was used as loading control. CLIC1 mRNA expression was detected by quantitative real-time polymerase chain reaction. β-actin was used as loading control. The figure represents one of the three experiments performed independently with similar results. A: CLIC1 protein expression in SGC-7901 by Western blotting; B: CLIC1 protein expression in MGC-803 by Western blotting; C: Relative CLIC1 protein expression level in SGC-7901; D: Relative CLIC1 protein expression level in MGC-803; E: Relative CLIC1 mRNA expression level in SGC-7901; F: Relative CLIC1 mRNA expression level in MGC-803. The experiment was done in triplicate and the value obtained from the control (CTRL) group set as 100%. aP < 0.05, bP < 0.01 vs CTRL and mock.
Downregulation of CLIC1 expression enhances growth of gastric cancer cells in vitro
We investigated whether CLIC1 expression could decrease the survival of gastric cancer cells. Downregulation of CLIC1 expression enhanced notably proliferation of gastric cancer cells in a time-dependent manner, and the highest growth rates were 25.60% (P = 0.002) in SGC 7901 for CLIC1 siRNA3 at 24h and 35.55% (P = 0.001) in MGC-803 cells for CLIC1 siRNA3 at 48 h. The cells transfected with CLIC1 siRNA3 survived at increased rates compared with the mock group cells and the negative control group cells. Our findings demonstrated that downregulation of CLIC1 expression enhances growth of SGC-7901 and MGC-803 cells in vitro (Figure 3).
Figure 3.
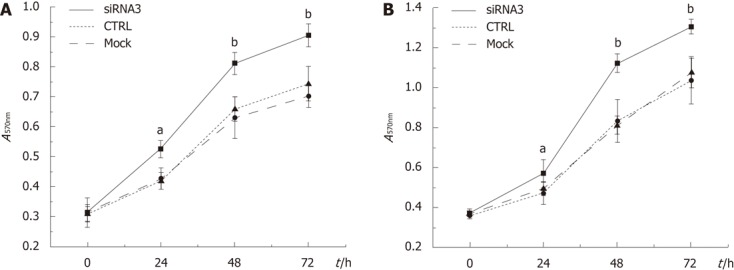
Effects of chloride intracellular channel 1 knockdown on cell growth in gastric cancer cell lines. Basal growth was determined in complete medium by the methyl thiazolyl tetrazolium assay after 0, 24, 48 and 72 h. A: Data obtained from SGC-7901 cells; B: Data obtained from MGC-803 cells. Results were shown as mean absorbance of the respective SGC7901 and MGC-803 cell lines. The experiment was done in triplicate. aP < 0.05, bP < 0.05 vs control (CTRL) and mock.
Downregulation of CLIC1 expression induces cell cycle arrest in gastric cancer cells
The cell cycle assay indicated that the inhibition of CLIC1 expression by specific CLIC1 siRNA3 significantly changed the proportions of the G0/G1, S and G2/M phases at 48 h after transfection with CLIC1 siRNA3. The percentage of the G2/M phase proportion increased notably in after transfection with CLIC1 siRNA3 in SGC-7901 (2.66-fold) and MGC-803 (1.28-fold), respectively, while there was decrease in the proportion of G0/G1 and S phases (Figure 4).
Figure 4.
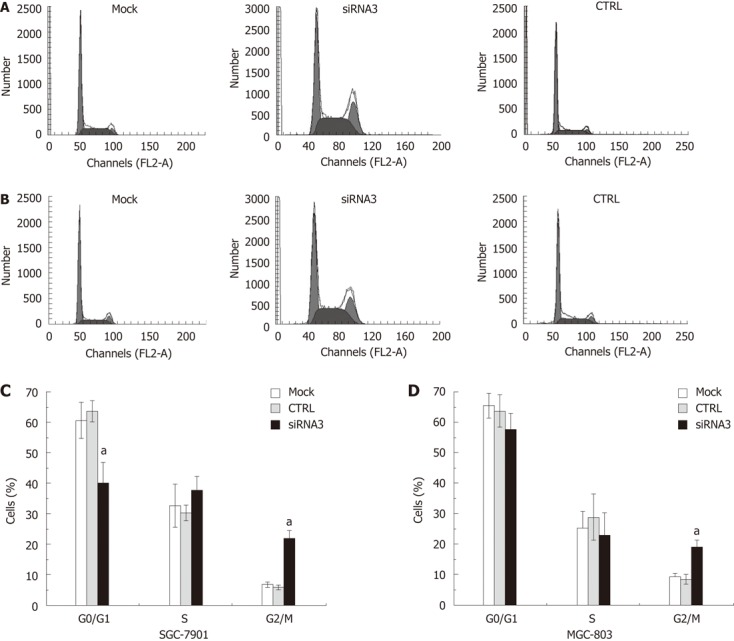
Measurement of the cell-cycle distribution by flow cytometry. Downregulation of chloride intracellular channel 1 (CLIC1) expression by small interference RNA (siRNA) 3 transfection changed cell cycle distribution in SGC-7901 and MGC-803 cells. At 48 h post-transfection, the cell cycle distribution of these cells was analyzed using propidium iodide staining on flow cytometry. The experiment was done in triplicate. The figure represented one of the 3 experiments performed in duplicate. A: The figure was obtained from SGC-7901 cells; B: The figure was obtained from MGC-803; C: The data was obtained from SGC-7901 cells; D: The data was obtained from MGC-803 cells. aP < 0.05 vs control (CTRL) and mock.
Downregulation of CLIC1 expression inhibits gastric cancer cell apoptosis in vitro
We investigated whether CLIC1 could induce human gastric cancer SGC-7901 and MGC-803 cell apoptosis. Cells were harvested and disposed by the Annexin V assay with indicated treatment at 48 h after transfection in SGC-7901 and MGC-803 cells. Then apoptosis was examined by FCM. The rates of apoptosis were obviously decreased in both SGC-7901 and MGC-803 cells (Figure 5). The CLICI siRNA3 showed the least apoptosis rate, suggesting that CLIC1 expression can increase apoptosis of gastric cancer cells in vitro.
Figure 5.
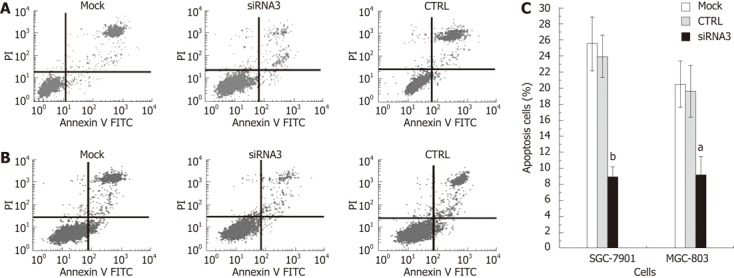
Chloride intracellular channel 1 knockdown reduces apoptosis in gastric cancer cell lines. Apoptosis detection by flow cytometry. For each sample, 10 000 cells were counted, and the apoptotic cells were summarized in the right panel. A: Figure was obtained from SGC-7901 cells; B: Figure was obtained from MGC-803 cells; C: The apoptotic rate of the chloride intracellular channel 1 (CLIC1) specific small interference RNA (siRNA) 3 group obviously decreased in both SGC-7901 cells (62.24%, P = 0.000) and MGC-803 cells (52.67%, P = 0.000), compared with the control (CTRL) group; while there were no significant difference of cell apoptosis rate between the negative control group cells and the mock group cells. The experiment was done in triplicate. The figure represented one of the 3 experiments performed in duplicate. aP < 0.05, bP < 0.05 vs CTRL and mock.
Downregulation of CLIC1 expression inhibits gastric cancer cell migration and invasion in vitro
We determined the effect of decreased CLIC1 expression on gastric cancer cell invasion, and examined the role of CLIC1 in gastric cancer cell migration. There was a significant decrease in CLIC1 siRNA3 transfected cells in the Matrigel invasion assay (Figure 6A and B). Decreased CLIC1 expression led to the inhibition of invasion by 54.31% (P = 0.000) in SGC-7901 and 40.74 % (P = 0.000) in MGC-803 cells. And cancer cell migration was also significantly reduced by CLIC1 siRNA3 in the migration assay (Figure 6C and D). Decreased CLIC1 expression inhibited the cell migration by 33.62% (P = 0.001) in SGC- 7901 and 29.26% (P = 0.002) in MGC-803. Taken together, these results clearly indicate that suppression of CLIC1 inhibits the invasion and migration ability of SGC-7901 and MGC-803 cells.
Figure 6.
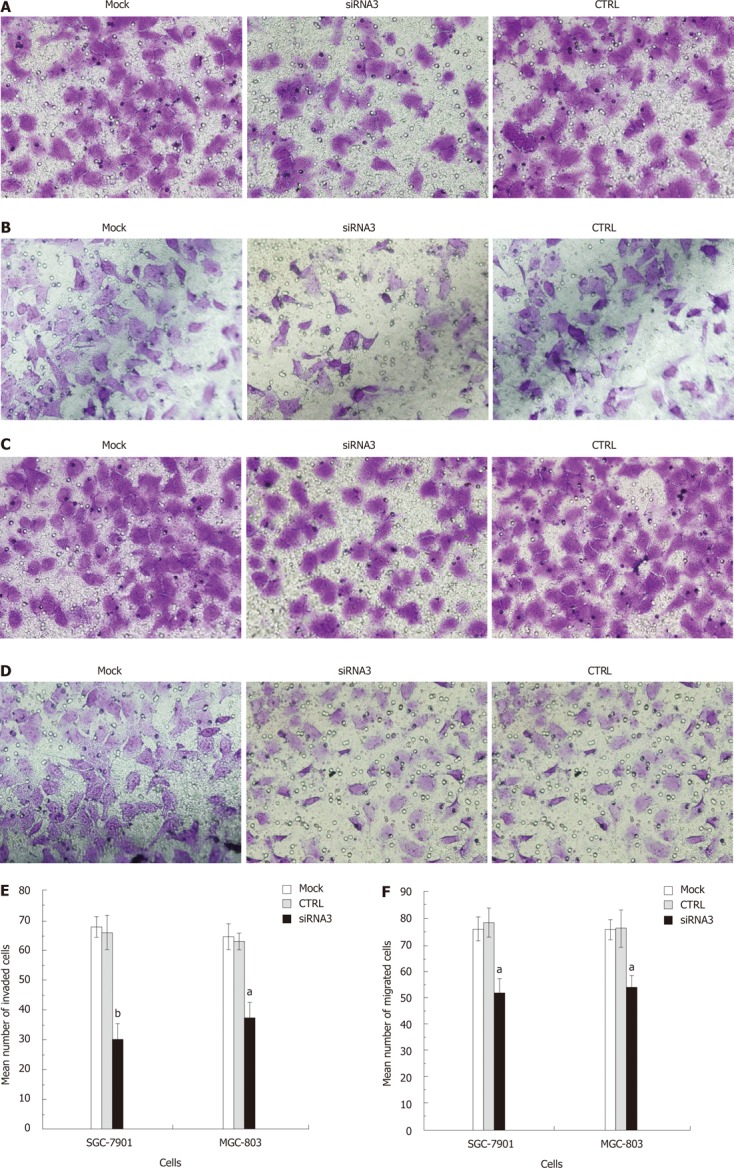
Effects of chloride intracellular channel 1 knockdown on cell migration and invasion in SGC-7901 and MGC-803 cell lines at 48 h. A: The figure was obtained from the invasion assay of SGC-7901 cells; B: The figure was obtained from the invasion assay of MGC-803 cell; C: The figure was obtained from the migration assay of SGC-7901 cells; D: The figure was obtained from the invasion assay of MGC-803 cell; E: The data was obtained from the invasion assay; F: The data was obtained from the migration assay. The experiment was done in triplicate. The figure represented one of the 3 experiments performed in duplicate. Cell invasion and migration were evaluated after two days SGC-7901 and MGC-803 cells were transfected in vitro. Representative microscopic images were presented in the upper panel of each assay graph. The number of cells that invaded or migrated through the filter was counted. Data are expressed as average number of migrated or invaded cells. The cells transfected with chloride intracellular channel 1 (CLIC1) specific small interference RNA (siRNA) 3 were significantly less able to invade or migrate than the mock cells or the negative control cells in both SGC-7901 and MGC-803 cells. aP < 0.05, bP < 0.05 vs control (CTRL) and mock.
DISCUSSION
Gastric cancer is a highly aggressive and lethal malignancy. It accounts for 8.6% of all new cancer cases worldwide, and is the second leading cause of cancer-related deaths[15]. Surgery is the standard treatment for localized gastric cancer, but advanced patients with distant metastasis or recurrence remain incurable[16]. Identification of key regulatory molecules in metastasis is crucial for understanding the mechanism for tumor dissemination as well as development of novel interventions. However, molecular events promoting invasiveness of gastric carcinoma cells remain unknown and routine biomarkers for metastastic gastric carcinoma are not yet available[17].
CLIC1 (also called nuclear chloride channel-27 or NCC27), the first identified human member of a growing family of organelle ion channels, is a transmembrane protein sufficient to form a functional ion channel as a tetrameric assembly of subunits. Both human and murine CLIC1 genes are located within the major histocompatibility complex class III region, one of the most conserved and important regions in the genome. In humans, it is located at 6p21.3[18]. The structure of the soluble form of CLIC1 is a typical soluble glutathione S-transferase superfamily protein, but contains a glutaredoxin-like active site[19]. Some recent studies have suggested that CLIC1 plays an important role in tumor metastasis[10,20].
Chen et al[9] reported that CLIC1 expression was significantly up-regulated in 67.9% of gastric cancer patients. The CLIC1 expression in tumor tissues increased 1.95-fold (range: 0.01-6.19-fold) compared with that expressed by adjacent noncancerous mucosa. Elevated CLIC1 expression was strongly correlated with lymph node metastasis, lymphatic and perineural invasion, pathological staging and poor survival. Additionally, the 5-year survival rate of the low CLIC1 expression group was higher than that of the high CLIC1 expression group. But further investigations are needed to observe this phenomenon in vitro and elucidate the mechanisms of CLIC1’s involvement in tumor metastasis. In this study, we used RNAi technology to inhibit the expression of CLIC1 in gastric cancer cell lines SGC-7901 and MGC-803, and explored the possible mechanism by observing the biological behavior changes (such as invasion, migration, etc.) after inhibition.
We designed four pairs of siRNA against the CLIC1 gene in this study to guarantee that there was at least one CLIC1 siRNA that could inhibit gene expression effectively. We used the four CLIC1 siRNAs to downregulate the expression of CLIC1 in human gastric cancer cell lines SGC-7901 and MGC-803. Quantitative real-time PCR and Western blotting assay showed that CLIC1 mRNA and protein expression were down-regulated by CLIC1 siRNA. Generally, the inhibition rate of CLIC1 siRNA3 was higher than siRNA1, siRNA2 and siRNA4, but there were no significant differences among siRNA5 group, mock group and CTRL group cells at all the time points in both cell lines (P > 0.05). And 24, 48 and 72 h after transfection with siRNA3, the expression of mRNA and protein was down-regulated significantly in both SGC-7901 ( inhibition rates of mRNA: 98.35%, 94.15% and 95.38%; inhibition rates of protein: 70.00%, 76.39% and 87.90%) and MGC-803 cells (inhibition rates of mRNA: 78.33%, 86.81% and 84.65%; inhibition rates of protein: 72.63%, 81.97% and 64.06%). These results showed that CLIC1 siRNA3 can efficiently inhibit CLIC1 expression. Therefore, CLIC1 siRNA3 was chosen for the subsequent experiments in vitro. However, the decreased level of mRNA and protein expressions after RNAi are inconsistent in SGC-7901 but in MGC-803, the specific mechanism needs to be further explored. In addition, we noted that the inhibition rates in the two cell lines were different, i.e., the inhibition rate was higher in SGC-7901 than in MGC-803 cells. The possible reasons for the difference were different cell growth characteristics and tissue source between the two cell lines. SGC-7901 originates from metastatic lymph node of gastric carcinoma, but MGC-803 comes from poorly differentiated adenocarcinoma of gastric mucous cells. Wang et al[21] found that the ability of cell growth, proliferation, migration and invasion of MGC-803 was much stronger than SGC-7901. In addition, MGC-803 has the characteristic of short growth cycle, unstable adhesion and high sensitivity to external stimuli. After successfully transfected with siRNA, MGC-803 exfoliated easily due to its vulnerable characteristics. Therefore, the inhibition rate of MGC-803 was lower than SGC-7901 under the same transfection conditions.
In this study, the A value increased significantly by the silencing of CLIC1 in the cell growth assay, suggesting that downregulation of CLIC1 by siRNA3 promoted the proliferation of gastric cancer cells. That is to say, CLIC1 gene itself could inhibit the proliferation of gastric cancer cells. Some scholars observed the same phenomenon in their studies[20,22,23].
It has been found that CLIC1 expression in the cell membrane only occurred at G2/M phase, and located in the cytoplasm in G1/S phase. Some researchers utilized CLIC1 blocker and found Chinese hamster ovary (CHO) cell cycle arrest of CHO cell was in the G2/M phase, suggesting that CLIC1 was involved in cell cycle[24]. Unlike typical membrane protein, the soluble CLICl presented firstly in the cytoplasm after synthesized, and then transferred to the plasma membrane to play a role of an ion channel[25]. Using the technology of RNAi, we inhibited the expression of CLIC1 in gastric cancer cell lines SGC-7901 and MGC-803, and detected the cells with the FCM. As a result, we found that cell cycle arrest in the G2/M phase was increased notably. Along with previous studies, we speculated that the expression of CLIC1 in this phase was enhanced significantly, and then played a role of an ion channel, such as promotion of cell growth. The findings proved that CLIC1 participated in regulation of cell cycle in the G2/M phase, and played an important role in the process of cell division. In addition, the apoptosis rate was decreased markedly in the CLIC1 siRNA3 group. Compared with control group, the apoptosis rates in SCC-7901 and MGC-803 were reduced by 62.24% and 52.67%, respectively. This suggests that the gene itself can promote gastric cancer cell apoptosis. And some studies also found this function in colon cancer, mouth cavity squamous cell carcinoma, melanoma and mouse hepatocellular carcinoma[20,23,26].
Some scholars[20,27,28] found that overexpression of CLIC1 significantly increased cell motility, and knock-down of CLIC1 markedly inhibited cell migration and invasion. But the role of CLIC1 in gastric cancer metastasis remains largely unclear. We studied the CLIC1 functions with a Transwell assay in gastric cancer cells, and the results demonstrated that down-regulation of CLIC1 expression by siRNA3 obviously suppressed the invasion and migratory capacity of SGC-7901 and MGC-803 cells in vitro. These results were consistent with previous experiments. A recent study to explain how CLIC1 promotes the movement of cancer cells found that CLIC1 has been implicated in modifying cell adhesion[24,29]. Tung et al[27] reported that reduced CLIC1 expression caused a reduction in endothelial migration, branching morphogenesis, capillary-like network formation, and capillary-like sprouting. FACS analysis showed that reducing CLIC1 expression increased integrins of β1, α3 and αvβ3 expression, but decreased the αvβ5 expression. Integrins are essential for invasion and metastasis of carcinoma cells and are a major family of cell adhesion molecules,which mediate cell-cell adhesion or cell-extracellular matrix adhesion and affect signal transduction, cell proliferation, differentiation, survival and apoptosis[30]. By increasing the surface expression of β1, α3 and αvβ3, downregulation of CLIC1 expression can increase endothelial cell adhesion to the extracellular matrix and inhibit motility by preventing the cell from breaking its contact with the extracellular matrix. These shifts in integrin expression also provide a possible explanation for the cell growth and viability defects[31]. Additionally, studies confirmed that invasion and migration of gastric cancer was closely associated with integrins[32-34], therefore, we speculated that invasion and migration inhibition of gastric cancer cells after knock-down of CLIC1 was associated with the up-regulation of β1, α3 and αvβ3. However, the specific mechanism needs to be further studied.
In summary, our findings show that CLIC1 siRNA3 can efficiently inhibit CLIC1 expression, and suppress gastric cancer migration and invasion in vitro. However, downregulation of CLIC1 expression promotes the proliferation of gastric cancer and reduces gastric cancer cellular apoptosis. The molecular mechanism remains unclear. Future studies need to focus on the precise mechanism of tumorigenesis. In addition, animal models need to be established. In short, CLIC1 plays an important role in regulating the biological behavior of gastric cancer cells. Our research lays a solid foundation for future studies.
COMMENTS
Background
Gastric cancer is a highly aggressive and lethal malignancy. Metastasis or recurrence is the main obstacle to improvement of the treatment efficacy of gastric cancer. Identification of key regulatory molecules in metastasis is crucial for understanding tumor dissemination and for development of novel interventions. However, molecular events promoting invasiveness of gastric carcinoma cells are still hardly known and routine biomarkers for metastastic gastric carcinoma are not yet available.
Research frontiers
Some researchers found that chloride intracellular channel 1 (CLIC1) was overexpressed in gastric cancer and was a potential prognostic marker, and elevated CLIC1 expression was strongly correlated with lymph node metastasis, lymphatic invasion, perineural invasion, and pathological staging. However, its role in gastric cancer deserves further studies.
Innovations and breakthroughs
Previous studies mainly focused on the phenomenon that the level of CLIC1 expression was associated with clinicopathologic features of gastric cancer in vivo. However, whether it is consistent with the phenomenon in vitro, and the mechanisms of CLIC1’s involvement in tumor metastasis remain unclear. In this study, the authors used RNA interference (RNAi) technology to inhibit the expression of CLIC1 in gastric cancer cell line SGC-7901 and MGC-803, and explored the possible mechanism by observing the biological behavior changes (cell proliferation, apoptosis, migration and invasion) after inhibition.
Applications
High CLIC1 expression can efficiently inhibit proliferation and enhance apoptosis, migration and invasion of gastric cancer cells in vitro. CLIC1 can be used as a promising target for the treatment of gastric cancer.
Terminology
CLIC1 is the first cloned human member of the CLIC family, and is a 241 amino acid ion channel protein. Like other members of the family, CLIC1 is highly conserved across a wide range of species and its relatives can be identified in the genome of all vertebrates so far sequenced. CLIC1 was initially found to localize to the cell nucleus and intracellular vesicles. RNAi is the highly specific, homology dependent suppression of gene expression by small double-stranded RNA (dsRNA). Long dsRNAs are cleaved by the endoribonuclease Dicer into short dsRNA duplexes or small interference RNA (siRNA). siRNA are loaded onto RNA-induced silencing complex (RISC). RISC contains argonaute 2 (Ago-2) which cleaves and releases one strand from the dsRNA, resulting in an activated form of RISC with a single-strand RNA (guide siRNA) that directs the specificity of the target mRNA recognition through complementary base pairing. Ago-2 then cleaves the target mRNA between bases 10 and 11 related to the 5’ end of the siRNA antisense strand, thereby causing mRNA degradation and gene silencing, which occurs at a post-transcriptional level.
Peer review
This is a good study in which the authors employed CLIC1 siRNA to knockdown the expression of CLIC1, investigated its effects on gastric cancer cell lines SGC-7901 and MGC-803, and then explored the possible mechanisms. The manuscript is well prepared, study design is reasonable, statistical methods are appropriate, and conclusions are based on the convincing statistical analysis.
Footnotes
Supported by The National Natural Science Foundation of China, No. 30560151; the Key Research Project of Guangxi Municipal Health Bureau, No. 200824; the Research Project of Guangxi Educational Department, No. 201012MS062 and No. 2011105981002M204; and the Natural Science Foundation of Guangxi, No. 0832113
Peer reviewer: Haruhiko Sugimura, MD, PhD, Professor, Department of Pathology, Hamamatsu University School of Medicine, 1-20-1 Handayama, Higashi-ku, Hamamatsu 431-3192, Japan
S- Editor Lv S L- Editor Ma JY E- Editor Xiong L
References
- 1.Nishiyama M. Chemotherapy for gastric cancer in Japan. Int J Clin Oncol. 2008;13:191–192. doi: 10.1007/s10147-008-0790-2. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467–477. doi: 10.1007/978-1-60327-492-0_23. [DOI] [PubMed] [Google Scholar]
- 3.Kunzelmann K. Ion channels and cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 4.Schönherr R. Clinical relevance of ion channels for diagnosis and therapy of cancer. J Membr Biol. 2005;205:175–184. doi: 10.1007/s00232-005-0782-3. [DOI] [PubMed] [Google Scholar]
- 5.Yang JY, Jung JY, Cho SW, Choi HJ, Kim SW, Kim SY, Kim HJ, Jang CH, Lee MG, Han J, et al. Chloride intracellular channel 1 regulates osteoblast differentiation. Bone. 2009;45:1175–1185. doi: 10.1016/j.bone.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, Valenzuela SM, Mazzanti M, Brown LJ, Breit SN, et al. The enigma of the CLIC proteins: Ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010;584:2093–2101. doi: 10.1016/j.febslet.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 7.Valenzuela J. [Role of nitric oxide in gastrointestinal physiology and in the pathogenesis of digestive diseases] Rev Med Chil. 1997;125:1408–1411. [PubMed] [Google Scholar]
- 8.Ulmasov B, Bruno J, Woost PG, Edwards JC. Tissue and subcellular distribution of CLIC1. BMC Cell Biol. 2007;8:8. doi: 10.1186/1471-2121-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CD, Wang CS, Huang YH, Chien KY, Liang Y, Chen WJ, Lin KH. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7:155–167. doi: 10.1002/pmic.200600663. [DOI] [PubMed] [Google Scholar]
- 10.Pan X, Thompson R, Meng X, Wu D, Xu L. Tumor-targeted RNA-interference: functional non-viral nanovectors. Am J Cancer Res. 2011;1:25–42. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Lu Z, Wientjes MG, Au JL. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Grimm D. Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev. 2009;61:672–703. doi: 10.1016/j.addr.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Sun M, Zhang A, Lv C, De W, Wang Z. Adenovirus-mediated siRNA targeting Bcl-xL inhibits proliferation, reduces invasion and enhances radiosensitivity of human colorectal cancer cells. World J Surg Oncol. 2011;9:117. doi: 10.1186/1477-7819-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern M, et al. MicroRNAs as a potential prognostic factor in gastric cancer. World J Gastroenterol. 2011;17:3976–3985. doi: 10.3748/wjg.v17.i35.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HJ, Eun JY, Jeon YW, Yun J, Kim KH, Kim SH, Kim HJ, Lee SC, Bae SB, Kim CK, et al. Efficacy and safety of oxaliplatin, 5-Fluorouracil, and folinic Acid combination chemotherapy as first-line treatment in metastatic or recurrent gastric cancer. Cancer Res Treat. 2011;43:154–159. doi: 10.4143/crt.2011.43.3.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y, He L, Wang J, Nie Y, Shi Y, et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS One. 2011;6:e26401. doi: 10.1371/journal.pone.0026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu MR, Jiang L, Matthaei KI, Schoenwaelder SM, Kuffner T, Mangin P, Joseph JE, Low J, Connor D, Valenzuela SM, et al. Generation and characterization of mice with null mutation of the chloride intracellular channel 1 gene. Genesis. 2010;48:127–136. doi: 10.1002/dvg.20590. [DOI] [PubMed] [Google Scholar]
- 19.Littler DR, Harrop SJ, Fairlie WD, Brown LJ, Pankhurst GJ, Pankhurst S, DeMaere MZ, Campbell TJ, Bauskin AR, Tonini R, et al. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J Biol Chem. 2004;279:9298–9305. doi: 10.1074/jbc.M308444200. [DOI] [PubMed] [Google Scholar]
- 20.Li RK, Tang JW, Zhang J, Wang SQ, Wang M, Wang B, Zhang YH. [Effects of silencing chloride intracellular channel 1 gene expression on the proliferation and invasion of mouse hepatocellular carcinoma cell lines] Zhonghua Ganzangbing Zazhi. 2010;18:131–135. doi: 10.3760/cma.j.issn.1007-3418.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 21.Wang YQ, Han M, Hou XL. [Comparison of proliferation and invasion among four human gastric cancer cell lines in vitro] Linchuang he Shiyan Yixue Zazhi. 2006;5:1486–1489. [Google Scholar]
- 22.Nawarak J, Huang-Liu R, Kao SH, Liao HH, Sinchaikul S, Chen ST, Cheng SL. Proteomics analysis of A375 human malignant melanoma cells in response to arbutin treatment. Biochim Biophys Acta. 2009;1794:159–167. doi: 10.1016/j.bbapap.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Scheper MA, Shirtliff ME, Meiller TF, Peters BM, Jabra-Rizk MA. Farnesol, a fungal quorum-sensing molecule triggers apoptosis in human oral squamous carcinoma cells. Neoplasia. 2008;10:954–963. doi: 10.1593/neo.08444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Warton K, Musgrove EA, Campbell TJ, Breit SN. The nuclear chloride ion channel NCC27 is involved in regulation of the cell cycle. J Physiol. 2000;529 Pt 3:541–552. doi: 10.1111/j.1469-7793.2000.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song MY, Tang JW, Sun MZ, Liu SQ, Wang B. [Localization and expression of CLIC1 in hepatocarcinoma ascites cell lines with high or low potentials of lymphatic spread] Zhonghua Binglixue Zazhi. 2010;39:463–466. [PubMed] [Google Scholar]
- 26.Skvortsov S, Skvortsova I, Sarg B, Loeffler-Ragg J, Lindner H, Lukas P, Tabernero J, Zwierzina H. Irreversible pan-ErbB tyrosine kinase inhibitor CI-1033 induces caspase-independent apoptosis in colorectal cancer DiFi cell line. Apoptosis. 2005;10:1175–1186. doi: 10.1007/s10495-005-1322-4. [DOI] [PubMed] [Google Scholar]
- 27.Tung JJ, Kitajewski J. Chloride intracellular channel 1 functions in endothelial cell growth and migration. J Angiogenes Res. 2010;2:23. doi: 10.1186/2040-2384-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang JW, Peng SY, Li JT, Wang Y, Zhang ZP, Cheng Y, Cheng DQ, Weng WH, Wu XS, Fei XZ, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71–81. doi: 10.1016/j.canlet.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Suh KS, Mutoh M, Gerdes M, Crutchley JM, Mutoh T, Edwards LE, Dumont RA, Sodha P, Cheng C, Glick A, et al. Antisense suppression of the chloride intracellular channel family induces apoptosis, enhances tumor necrosis factor {alpha}-induced apoptosis, and inhibits tumor growth. Cancer Res. 2005;65:562–571. [PubMed] [Google Scholar]
- 30.Singh H, Cousin MA, Ashley RH. Functional reconstitution of mammalian ‘chloride intracellular channels’ CLIC1, CLIC4 and CLIC5 reveals differential regulation by cytoskeletal actin. FEBS J. 2007;274:6306–6316. doi: 10.1111/j.1742-4658.2007.06145.x. [DOI] [PubMed] [Google Scholar]
- 31.Stupack DG, Puente XS, Boutsaboualoy S, Storgard CM, Cheresh DA. Apoptosis of adherent cells by recruitment of caspase-8 to unligated integrins. J Cell Biol. 2001;155:459–470. doi: 10.1083/jcb.200106070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao GT, Zhao XL, Li LM, Ji CX, Xiao P, Zhang Q, Zhang JX. Study of the correlation of integrin β1 and VEGF with invasion and metastasis of gastric carcinoma. Zhongliu Yanjiu Yu Linchuang. 2009;21:101–103. [Google Scholar]
- 33.Takatsuki H, Komatsu S, Sano R, Takada Y, Tsuji T. Adhesion of gastric carcinoma cells to peritoneum mediated by alpha3beta1 integrin (VLA-3) Cancer Res. 2004;64:6065–6070. doi: 10.1158/0008-5472.CAN-04-0321. [DOI] [PubMed] [Google Scholar]
- 34.Song G, Ming Y, Mao Y, Bao S, Ouyang G. Osteopontin prevents curcumin-induced apoptosis and promotes survival through Akt activation via alpha v beta 3 integrins in human gastric cancer cells. Exp Biol Med (Maywood) 2008;233:1537–1545. doi: 10.3181/0805-RM-164. [DOI] [PubMed] [Google Scholar]


