Abstract
The fatty acid desaturase (FADS) gene family at 11q12-13.1 includes FADS1 and FADS2, both known to mediate biosynthesis of omega-3 and omega-6 long-chain polyunsaturated fatty acids (LCPUFA). FADS3 is a putative desaturase due to its sequence similarity with FADS1 and FADS2, but its function is unknown. We have previously described 7 FADS3 alternative transcripts (AT) and 1 FADS2 AT conserved across multiple species. This study examined the effect of dietary LCPUFA levels on liver FADS gene expression in vivo and in vitro, evaluated by qRT-PCR. Fourteen baboon neonates were randomized to three diet groups for their first 12 weeks of life: C: Control, no LCPUFA; L: 0.33% docosahexaenoic acid (DHA)/ 0.67% arachidonic acid (ARA) (w/w); and L3: 1.00% DHA/ 0.67% ARA (w/w). Liver FADS1 and both FADS2 transcripts were downregulated by at least 50% in the L3 group compared to controls. In contrast, FADS3 AT were upregulated (L3>C), with four transcripts significantly upregulated by 40% or more. However, there was no evidence for a shift in liver fatty acids to coincide with increased FADS3 expression. Significant upregulation of FADS3 AT was also observed in human liver-derived HepG2 cells after DHA or ARA treatment. The PPARγ antagonist GW9662 prevented FADS3 upregulation, while downregulation of FADS1 and FADS2 was unaffected. Thus, FADS3 AT were directly upregulated by LCPUFA by a PPARγ-dependent mechanism unrelated to regulation of other desaturases. This opposing pattern and mechanism of regulation suggests a dissimilar function for FADS3 AT compared to other FADS gene products.
Keywords: docosahexaenoic acid, arachidonic acid, polyunsaturated fatty acids, fatty acid desaturase, FADS3, alternative splicing
Introduction
Biosynthesis of long-chain polyunsaturated fatty acids (LCPUFA) requires introduction of cis double bonds by the Δ5 and Δ6 desaturases, encoded by the FADS1 and FADS2 genes, respectively. FADS1 and FADS2 span a 100kb cluster on the long arm of chromosome 11 (11q12-13.1), together with a third member of the gene family, designated FADS3. FADS3 is a putative fatty acid desaturase gene due to its high degree of sequence homology with FADS2 (62%) and FADS1 (52%), but no function for FADS3 has been demonstrated experimentally [1].
Although its exact function is unknown, genetic evidence suggests FADS3 plays an important role in lipid metabolism and diseases. For example, single nucleotide polymorphisms in FADS3 have been associated with plasma sphingolipids and triglyceride levels, and with risk of myocardial infarction [2, 3]. Expression of FADS3 is altered in familial combined hyperlipidemia [3], and FADS3 is one of the six most highly expressed genes at the implantation site in mice at the initiation of pregnancy [4].
Early attempts in our laboratory to characterize FADS3 expression resulted in the discovery of seven alternative transcripts (AT) of FADS3 with distinctive patterns of expression in primate tissues [5]. In addition, we recently reported an alternative splice variant for FADS2 [6]. FADS2 AT1 and at least five of the FADS3 AT were conserved from chickens to humans [7]. Despite this evidence of crucial roles in essential processes, functions of the splice variants remain unclear.
Patterns of regulation can often provide clues to function; we reasoned that if the FADS3 AT encoded functional fatty acid desaturases, they were likely to be regulated similarly to the classical desaturase genes, FADS1 and FADS2. These two genes encode desaturases required for biosynthesis of the omega-3 and omega-6 LCPUFA docosahexaenoic acid (DHA, 22:6n-3) and arachidonic acid (ARA, 20:4n-6). These two products of the biosynthetic pathway have been shown to decrease the classical transcripts of FADS1 and FADS2 [8]. DHA and ARA are both known to bind members of the peroxisome proliferator-activated receptor (PPAR) family of transcription factors (especially PPARα and PPARγ), which form heterodimers with the retinoid X receptor (RXR) and influence gene expression [9, 10]. The effect of dietary LCPUFA on FADS1 and FADS2 gene expression has been shown to occur via PPARα and the sterol response element binding protein, SREBP-1c [11]. Nutrients, hormones, and drugs regulating FADS1 and FADS2 are known to regulate both in concert, with the same directionality of change [12], as would be expected for genes functioning in the same biosynthetic pathway.
Here we asked whether dietary LCPUFA affect expression of FADS3 AT and FADS2 AT1 similarly to classical FADS1 and FADS2, both in vivo and in vitro. Neonatal baboons were fed infant formula with varying levels of DHA and ARA for 12 weeks, and liver fatty acids and FADS gene expression examined. In vitro, human liver-derived HepG2 cells were studied to determine whether the observed effects were reproducible in human cells, and if it was a direct response to a fatty acid or an endocrine response.
Experimental Procedures
Animals and diets
All baboon work was carried out at the Southwest Foundation for Biomedical Research (SFBR) in San Antonio, TX. Animal protocols were approved by the SFBR and Cornell University Institutional Animal Care and Use Committee (IACUC, protocol # 02–105.) Diets and feeding protocols were described in detail previously [13]. Briefly, fourteen baboon neonates were randomized to one of three diet groups with varying concentrations of ARA and DHA, as described in Table 1. The infant formulas used for C and L groups correspond to the human infant formulas Enfamil and Enfamil LIPIL, respectively, and the L3 group was targeted to have three-fold higher DHA concentration, corresponding with the upper end of DHA levels found in human breast milk worldwide [14]. These diets were identical to a subset of those used in recently reported human studies [15, 16]. As shown in Table 1, analysis of the prepared diets showed that the actual concentrations used were slightly higher than target values, since the diets were prepared within tolerances designed to account for losses and variation during manufacturing and storage. Infant baboons consumed the experimental diets until 12 weeks of life, when tissues were harvested for lipid and RNA extraction.
Table 1.
Characteristics of experimental groups and diets
| C | L | L3 | |
|---|---|---|---|
| Number of animals (n) | 5 | 4 | 5 |
| Female | 4 | 3 | 3 |
| Male | 1 | 1 | 2 |
| DHA (% w/w) | 0 | 0.42 ± 0.02 [0.33] | 1.13 ± 0.04 [1.00] |
| ARA (% w/w) | 0 | 0.77 ± 0.02 [0.67] | 0.71 ± 0.01 [0.67] |
Analyzed concentrations are shown as mean ± SD; target values are shown in brackets.
Quantitative real-time PCR
Baboon liver RNA was extracted as described previously [5]. For HepG2 cells, RNA was extracted using the RNeasy kit (Qiagen), and RNA quality was checked by agarose gel electrophoresis to verify RNA integrity and by 260/280 nm ratios on a NanoDrop 2000 (Thermo Scientific). cDNA was prepared using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative real-time PCR was carried out using SYBR Green Master Mix (Roche) on a LightCycler 480 instrument (Roche). Human and baboon PCR primers were obtained from Integrated DNA Technologies (sequences available upon request), except for 18S, which was obtained from Qiagen as a QuantiTect Primer Assay. PCR primers designed for FADS splice variants were validated by cloning and sequencing PCR products. PCR reaction efficiency was calculated from standard curves, and reactions were assessed by both melting curves and by running on agarose gels to verify reaction products and the absence of primer-dimers. Quantitative cycle (Cq) values were determined using LightCycler 480 SW1.5.0SP3 software, version 1.5.0.39 (Roche). Relative quantification was carried out using the method of Pfaffl [17], taking into account reaction efficiency and using multiple reference genes for greater accuracy (β-actin and GAPDH for baboon experiments; β-actin, GAPDH, and 18S for HepG2 cell experiments).
Fatty acid analysis
Lipids from baboon liver samples were extracted and fatty acids analyzed by covalent adduct chemical ionization tandem mass spectrometry as described in detail previously [13]. Percent conversion of substrates (S) to products (P) was calculated as: [(P) / (S + P)] * 100, and normalized to the control group.
Cell treatments
For all experiments, human HepG2 hepatocellular carcinoma cells were maintained within ten passages of the original passage received from the ATCC. HepG2 cells were grown in MEM with 10% FBS (media and serum obtained from HyClone) in a humidified environment at 37°C with 5% CO2. For fatty acid treatment, free fatty acids were first non-covalently bound to fatty-acid free bovine serum albumin (BSA). Fatty acid sodium salts were suspended in PBS, then mixed with fatty-acid free bovine serum albumin (U.S. Biologicals) in a 3:1 ratio of fatty acid to albumin, and incubated for 5 hours at 37°C. Fatty acids conjugated to albumin were sterilized by passage through a syringe filter before cell treatments. Cells were treated with 100 µM of DHA-BSA, ARA-BSA, palmitic-BSA, and/or 2 µM GW9662 (Sigma) for 78 hours in media containing 0.5% FBS.
Statistical methods
Data are presented as mean ± standard deviation. Bootstrapping and randomization techniques were used in REST 2009 software (Qiagen) to calculate significance of fold changes in expression for qRT-PCR experiments. Statistical analysis of changes in fatty acid conversion was conducted using Student’s t-test to compare LCPUFA supplementation with control. Linear regression analysis of fatty acid data was carried out in SAS v.9.2 (SAS Institute, Cary NC).
Results
FADS expression in baboon liver
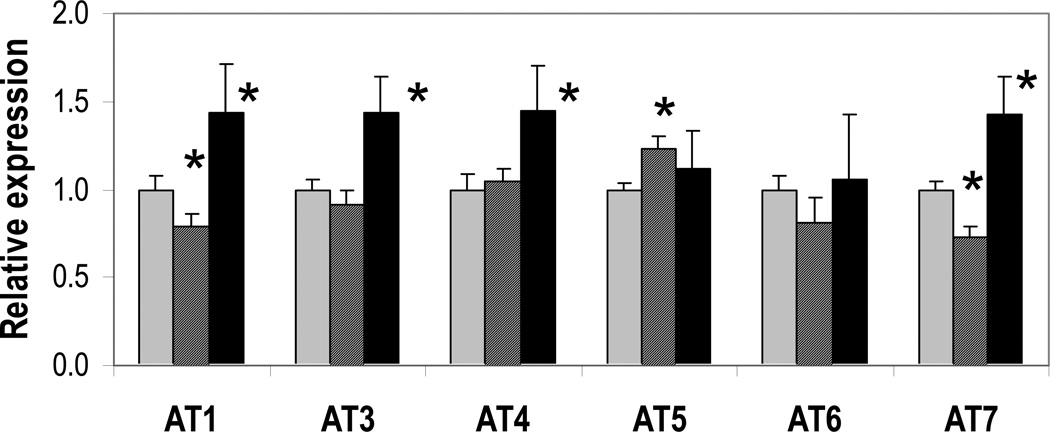
The splicing and expression patterns of the seven alternative transcripts (AT) of FADS3 and the one splice variant for FADS2 have been described in detail previously [5, 6]. Because of shared sequences across transcripts, it was not possible to design PCR primers unique to the full-length FADS3 classical transcript. However, the splice variants could each be assessed by quantitative real-time PCR (qRT-PCR), with the exception of FADS3 AT2, which was present at levels too low to quantify accurately. Thus, to assess the effect of dietary LCPUFA on FADS3 expression, six of the alternative transcripts were evaluated by qRT-PCR in samples obtained from baboon livers after 12 weeks on diets L3, L, or C (described in Materials and Methods). As shown in Figure 1, FADS3 AT1, AT3, AT4, and AT7 were about 40% upregulated in the highest LCPUFA group, L3, relative to control. FADS3 AT5 and AT6 also had apparently elevated mean expression in L3, but it was not statistically significant. However, FADS3 AT5 was significantly upregulated in the intermediate group L compared to control. Interestingly, AT1 and AT7 had a significant U-shaped expression response, with lower expression in L compared to control, but higher expression in L3.
Figure 1. FADS3 isoform expression is altered by dietary LCPUFA.
FADS3 splice variant expression in livers from baboons fed diet C (gray bars), L (striped bars), or L3 (black bars) was measured by qRT-PCR, with GAPDH and beta-actin as reference genes. Most isoforms were significantly upregulated in the L3 diet relative to C (* p < 0.05 compared to C). Downregulated AT1 and AT7, and upregulated AT5, were observed for the intermediate diet, L.
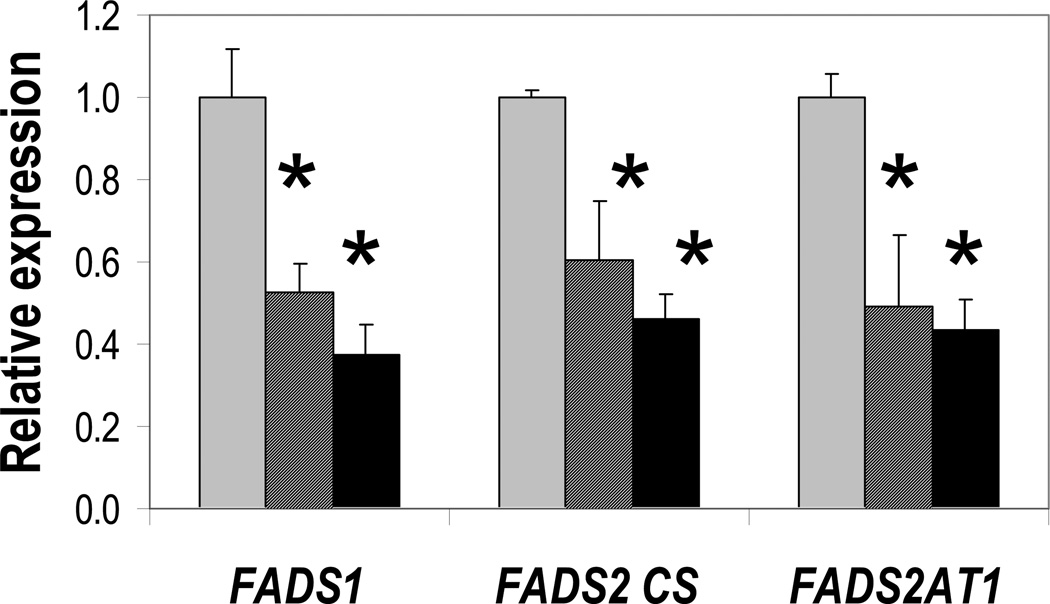
As shown in Figure 2, the other two members of the FADS gene cluster had an entirely different pattern of regulation. FADS1, FADS2, and the alternative transcript FADS2 AT1 were all downregulated in both the L and the L3 groups relative to control. The magnitude of expression change was similar for both genes; FADS1 and total FADS2 transcripts were reduced by at least 50% for both L and L3. These data are consistent with previous studies showing lower appearance of labeled 22:6n-3 from labeled 18:3n-3 in liver and blood pools of animals fed with 22:6n-3 in a diet with DHA-ARA similar to the present L diet compared to the C diet [18].
Figure 2. FADS1 and both FADS2 isoforms are downregulated by dietary LCPUFA.
Expression in baboon liver from animals fed diet C (gray bars), L (striped bars), or L3 (black bars) was measured by qRT-PCR, with GAPDH and beta-actin as reference genes. * p < 0.05 compared to C group.
Liver PUFA substrate-product ratios
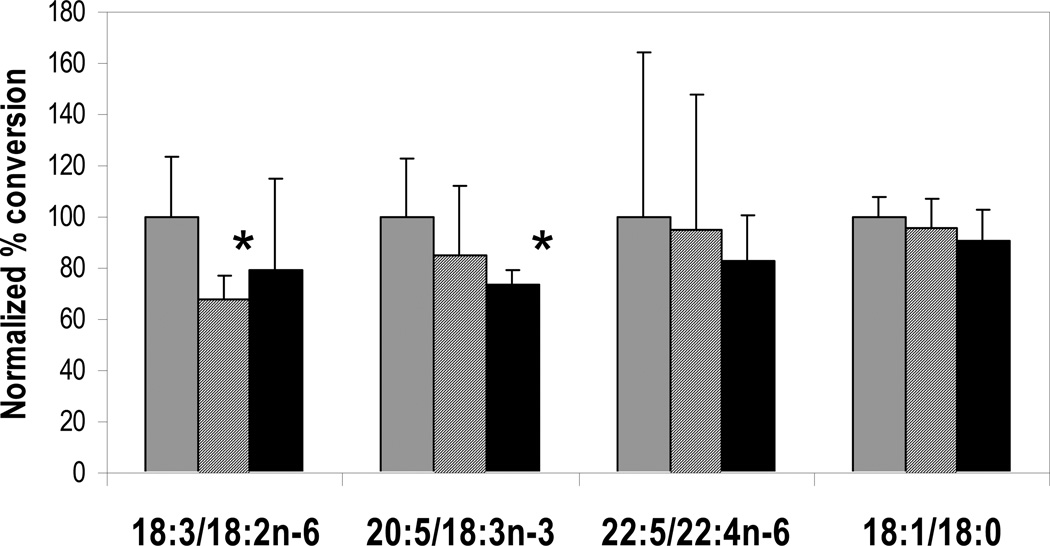
To investigate whether FADS3 upregulation affected desaturase reactions, levels of fatty acid substrates and products in baboon liver were evaluated as indicators of apparent desaturase activity. DHA and ARA were not included in ratios because their concentrations reflected both biosynthesis and incorporation pre-formed from the experimental diets. Instead, several other substrate/product pairings were used to infer activity, as shown in Figure 3. FADS2-encoded Δ6-desaturase activity catalyzes the desaturation of 18:2n-6 to 18:3n-6. The percent conversion to the 18:3n-6 product was significantly decreased in L compared to C, but L3 was not significantly different from control, though the L and L3 means were similar. Diet 20:4n-6 is zero in the C treatment and 0.64 % (w/w) in both the L and L3 treatments, consistent with the concept that ARA downregulated both L and L3.
Figure 3. Apparent desaturase activity is consistent with downregulation.
Percent conversion of fatty acid substrates into desaturated products (ratios shown, calculated as product/[substrate + product]) was measured in baboon liver and normalized to C group (gray bars). Group L is shown as striped bars, and L3 as black bars. * p < 0.05 compared to C group.
Conversion of 22:4n-6 to 22:5n-6 is presumed to proceed via FADS2-mediated Δ6-desaturation. A linear regression against diet DHA yielded a significant slope (p<0.05) and thus a significant downward trend for decreasing percent conversion with increasing LCPUFA. The pairing of 18:3n-3 with the downstream product 20:5n-3 was used as an aggregate measure of total desaturase activity, since both FADS1 and FADS2 gene products are required for 20:5n-3 synthesis. Substrate conversion resulting in 20:5n-3 was significantly decreased in L3 compared to C. Finally, a reaction catalyzed by a desaturase outside the FADS cluster, the conversion of 18:0 to 18:1 by stearoyl-coA desaturase, was also examined since this conversion is sensitive to overall dietary unsaturation. The downward trend in mean conversion with increasing LCPUFA was not significant. Thus, significant differences in pairwise comparisons and trends associated with more unsaturation implied steadily decreasing desaturation activity with increasing dietary LCPUFA. No substrate-product pairs were significantly increased by dietary LCPUFA, leaving no obvious candidates for FADS3 substrates.
FADS expression in response to LCPUFA supplementation in HepG2 cells
To investigate the mechanism for the effect of LCPUFA on FADS3 transcripts, human liver-derived HepG2 cells were grown in low-serum media supplemented with albumin-bound fatty acids. Cells were treated either with docosahexaenoic acid (DHA, 22:6n-3) alone, arachidonic acid (ARA, 20:4n-6) alone, or palmitic acid (16:0) as a control. As summarized in Table 2, DHA alone significantly upregulated all FADS3 transcripts except AT7. ARA alone significantly upregulated FADS3 AT1 and AT3, while FADS3 AT4, AT6, and AT7 increases were comparable to those observed with DHA treatment, but were not statistically significant. As in baboon livers, both fatty acids also significantly downregulated FADS1 and both FADS2 transcripts.
Table 2.
Fold changes in FADS gene expression in HepG2 cells for LCPUFA treatments with or without the PPARγ antagonist GW9662, shown relative to control (palmitic acid).
| Gene | DHA | DHA + GW9662 |
ARA | ARA + GW9662 |
|---|---|---|---|---|
| FADS3 AT1 | 1.2** | 1.1 | 1.3** | 1.1 |
| FADS3 AT3 | 1.2** | 1.0 | 1.2** | 1.1 |
| FADS3 AT4 | 1.2* | 0.93 | 1.2 | 1.1 |
| FADS3 AT5 | 1.4* | 0.92 | 0.91 | 1.1 |
| FADS3 AT6 | 1.6** | 0.85 | 1.4 | 0.92 |
| FADS3 AT7 | 1.2 | 0.90 | 1.2 | 0.93 |
| FADS2 | 0.59** | 0.37** | 0.47** | 0.22** |
| FADS2 AT1 | 0.47** | 0.34** | 0.32** | 0.31** |
| FADS1 | 0.81** | 0.50** | 0.65** | 0.49** |
p < 0.05,
p < 0.01 for being different from 1 (control = palmitic acid)
To understand the role of transcription factors in mediating these expression changes, the effect of co-incubation of fatty acids with the PPARγ antagonist GW9662 was evaluated. GW9662 treatment completely blocked the upregulation of FADS3 transcripts observed with both DHA and ARA treatment, but had no effect on the downregulation of FADS1 or FADS2 transcripts.
Discussion
We have evaluated transcriptional changes in FADS genes in infant baboon liver in response to diets corresponding to the physiological range of LCPUFA found in human breastmilk worldwide. FADS3 transcripts followed a pattern of regulation opposite to the other members of the FADS gene cluster, FADS1 and FADS2. FADS3 transcripts were upregulated by approximately 40% in livers of animals fed the highest LCPUFA diet, L3, compared to control. In contrast, the same diet downregulated FADS1, FADS2, and the alternative transcript FADS2 AT1, by at least 50%. The similarity of the downregulation of FADS2 and FADS2 AT1 suggests transcription-level control rather than any change in splicing regulation.
The expression changes observed in baboon liver were reproduced in human liver-derived HepG2 cells treated with either DHA or ARA. DHA produced statistically significant fold changes in FADS3 isoforms, while ARA treatment produced similar magnitude fold-changes, though most were not statistically significant. Thus, there was no obvious difference in sensitivity of the FADS3 response to DHA or ARA, but the DHA response was of greater precision in this experiment. Moreover, because the cell treatments did not differ in caloric or fat content from the control (palmitic acid treatment), the response was a specific, direct response to DHA or ARA rather than an endocrine mechanism or a general response to energy density of the diet/media. Both DHA and ARA have previously been shown to bind and activate the transcription factor PPARγ [19]. Co-incubation with the PPARγ antagonist GW9662 prevented FADS3 upregulation by DHA or ARA, suggesting that the fatty acids acted by a PPARγ-dependent mechanism. In contrast, GW9662 did not prevent the downregulation of FADS1 and both FADS2 isoforms by DHA or ARA. Thus, FADS3 expression is regulated in opposite sense to the other FADS genes, and occurs by a different mechanism. Among the FADS3 AT, we note that the expression pattern for FADS3AT5 is unique because it is responsive to the L diet but not to the higher DHA level in L3. FADS3AT5 is the only AT that we have detected to retain an intron (between exons 8 and 9), though we cannot yet speculate on putative functions.
Extensive substrate screening in our laboratory has so far failed to uncover a substrate for any FADS3 isoforms (unpublished data). Investigation of baboon liver fatty acids as evidence of desaturase enzyme activity produced evidence for downregulation of FADS1 and FADS2, but no evidence for increased desaturation of any substrate/product pair to correspond with the upregulation of FADS3 isoforms. This observation could be explained if FADS3 isoforms function as desaturases on non-LCPUFA substrates. Alternatively, they may act as non-functional dominant negative inhibitors by binding non-productively to LCPUFA substrates, and removing them from availability for desaturase reactions. In favor of the latter theory, Δ5-desaturase, Δ6-desaturase, and stearoyl-coA desaturases are all downregulated by dietary unsaturated fatty acids [20, 21], probably as a mechanism to maintain the unsaturation index of cell membranes within certain limits. It is difficult to reconcile the opposite pattern of regulation of FADS3 with a putative function as a desaturase. Moreover, dominant negative inhibition is a common mode of action for splice variants of other enzymes [22], and binding specificity for different fatty acids could explain the large number of splice variants for FADS3. Further studies are required to test this hypothesis.
We have demonstrated that FADS3 is regulated by a different mechanism from other members of the FADS gene cluster, and expression is upregulated when other desaturases are downregulated. These results suggest that, despite a high degree of sequence similarity, FADS3 isoforms have a function quite distinct from FADS1 and FADS2, and may not encode functional desaturases at all. FADS3 has been implicated in cardiovascular conditions of enormous public health import, so determining the true function of FADS3 and its alternative transcripts should be a high priority. Further work characterizing regulation of FADS3 transcription and alternative mRNA splicing may yield more clues to functional roles.
Acknowledgments
H.R. was supported by a Ruth L. Kirchstein-NRSA predoctoral training fellowship in reproductive sciences and genomics (Grant Number T32HD052471) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NICHD or the NIH.
Funding
This work was funded by Mead Johnson Nutrition, Evansville, Indiana.
Abbreviations
- ARA
arachidonic acid 20:4n-6
- AT
alternative transcripts
- DHA
docosahexaenoic acid 22:6n-3
- LCPUFA
long-chain polyunsaturated fatty acids
- PPAR
peroxisome proliferator-activated receptor
- qRT-PCR
quantitative real-time polymerase chain reaction
- RXR
retinoid X receptor
- SREBP-1c
sterol response element binding protein 1c
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors report no conflicts of interest.
References
- 1.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 2.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, Jonasson I, Zorkoltseva IV, Pattaro C, Hayward C, Isaacs A, Hengstenberg C, Campbell S, Gnewuch C, Janssens AC, Kirichenko AV, Konig IR, Marroni F, Polasek O, Demirkan A, Kolcic I, Schwienbacher C, Igl W, Biloglav Z, Witteman JC, Pichler I, Zaboli G, Axenovich TI, Peters A, Schreiber S, Wichmann HE, Schunkert H, Hastie N, Oostra BA, Wild SH, Meitinger T, Gyllensten U, van Duijn CM, Wilson JF, Wright A, Schmitz G, Campbell H. Genetic determinants of circulating sphingolipid concentrations in european populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plaisier CL, Horvath S, Huertas-Vazquez A, Cruz-Bautista I, Herrera MF, Tusie-Luna T, Aguilar-Salinas C, Pajukanta P. A systems genetics approach implicates usf1, fads3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 2009;5:e1000642. doi: 10.1371/journal.pgen.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma XH, Hu SJ, Ni H, Zhao YC, Tian Z, Liu JL, Ren G, Liang XH, Yu H, Wan P, Yang ZM. Serial analysis of gene expression in mouse uterus at the implantation site. J Biol Chem. 2006;281:9351–9360. doi: 10.1074/jbc.M511512200. [DOI] [PubMed] [Google Scholar]
- 5.Park WJ, Kothapalli KS, Reardon HT, Kim LY, Brenna JT. Novel fatty acid desaturase 3 (fads3) transcripts generated by alternative splicing. Gene. 2009;446:28–34. doi: 10.1016/j.gene.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park WJ, Reardon HT, Tyburczy C, Kothapalli KS, Brenna JT. Alternative splicing generates a novel fads2 alternative transcript in baboons. Mol Biol Rep. 2009;37:2403–2406. doi: 10.1007/s11033-009-9750-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenna JT, Kothapalli KS, Park WJ. Alternative transcripts of fatty acid desaturase (fads) genes. Prostaglandins Leukot Essent Fatty Acids. 2010;82:281–285. doi: 10.1016/j.plefa.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 9.Keller H, Mahfoudi A, Dreyer C, Hihi AK, Medin J, Ozato K, Wahli W. Peroxisome proliferator-activated receptors and lipid metabolism. Ann N Y Acad Sci. 1993;684:157–173. doi: 10.1111/j.1749-6632.1993.tb32279.x. [DOI] [PubMed] [Google Scholar]
- 10.Kliewer SA, Sundseth SS, Jones SA, Brown PJ, Wisely GB, Koble CS, Devchand P, Wahli W, Willson TM, Lenhard JM, Lehmann JM. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc Natl Acad Sci U S A. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 12.Brenner RR. Hormonal modulation of delta6 and delta5 desaturases: Case of diabetes. Prostaglandins Leukot Essent Fatty Acids. 2003;68:151–162. doi: 10.1016/s0952-3278(02)00265-x. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh AT, Anthony JC, Diersen-Schade DA, Rumsey SC, Lawrence P, Li C, Nathanielsz PW, Brenna JT. The influence of moderate and high dietary long chain polyunsaturated fatty acids (lcpufa) on baboon neonate tissue fatty acids. Pediatr Res. 2007;61:537–545. doi: 10.1203/pdr.0b013e318045bec9. [DOI] [PubMed] [Google Scholar]
- 14.Brenna JT, Varamini B, Jensen RG, Diersen-Schade DA, Boettcher JA, Arterburn LM. Docosahexaenoic and arachidonic acid concentrations in human breast milk worldwide. Am J Clin Nutr. 2007;85:1457–1464. doi: 10.1093/ajcn/85.6.1457. [DOI] [PubMed] [Google Scholar]
- 15.Birch EE, Carlson SE, Hoffman DR, Fitzgerald-Gustafson KM, Fu VL, Drover JR, Castaneda YS, Minns L, Wheaton DK, Mundy D, Marunycz J, Diersen-Schade DA. The diamond (dha intake and measurement of neural development) study: A double-masked, randomized controlled clinical trial of the maturation of infant visual acuity as a function of the dietary level of docosahexaenoic acid. Am J Clin Nutr. 2010;91:848–859. doi: 10.3945/ajcn.2009.28557. [DOI] [PubMed] [Google Scholar]
- 16.Drover JR, Hoffman DR, Castaneda YS, Morale SE, Garfield S, Wheaton DH, Birch EE. Cognitive function in 18-month-old term infants of the diamond study: A randomized, controlled clinical trial with multiple dietary levels of docosahexaenoic acid. Early Hum Dev. 2011;87:223–230. doi: 10.1016/j.earlhumdev.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 17.Pfaffl MW. A new mathematical model for relative quantification in real-time rt-pcr. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkadi-Nagy E, Wijendran V, Diau GY, Chao AC, Hsieh AT, Turpeinen A, Lawrence P, Nathanielsz PW, Brenna JT. Formula feeding potentiates docosahexaenoic and arachidonic acid biosynthesis in term and preterm baboon neonates. J Lipid Res. 2004;45:71–80. doi: 10.1194/jlr.M300106-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Yu YH, Lin EC, Wu SC, Cheng WT, Mersmann HJ, Wang PH, Ding ST. Docosahexaenoic acid regulates adipogenic genes in myoblasts via porcine peroxisome proliferator-activated receptor gamma. J Anim Sci. 2008;86:3385–3392. doi: 10.2527/jas.2008-1051. [DOI] [PubMed] [Google Scholar]
- 20.Ntambi JM. Regulation of stearoyl-coa desaturase by polyunsaturated fatty acids and cholesterol. J Lipid Res. 1999;40:1549–1558. [PubMed] [Google Scholar]
- 21.Matsuzaka T, Shimano H, Yahagi N, Amemiya-Kudo M, Yoshikawa T, Hasty AH, Tamura Y, Osuga J, Okazaki H, Iizuka Y, Takahashi A, Sone H, Gotoda T, Ishibashi S, Yamada N. Dual regulation of mouse delta(5)- and delta(6)- desaturase gene expression by srebp-1 and pparalpha. J Lipid Res. 2002;43:107–114. [PubMed] [Google Scholar]
- 22.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]