Abstract
Purpose
To determine the feasibility of two magnetic resonance spectroscopy techniques in pediatric patients with diffuse intrinsic pontine gliomas (DIPGs) and evaluate the relationship of metabolic profiles determined by each technique. Utility of each technique for improving patient management is also discussed.
Methods and Materials
Children with DIPG (n=36) were evaluated using single voxel spectroscopy (SVS) and magnetic resonance spectroscopic imaging (MRSI) during the same imaging session. Patients were followed longitudinally (n=150 total studies). Technical feasibility was defined by sufficient water and lipid suppression for detection of metabolites. Correlation of metabolic data obtained by SVS and MRSI was determined using the Spearman rank method. Metabolite ratios, including Choline:N-acetyl-aspartate (Cho:NAA) and Cho:Creatine (Cho:Cr), were obtained from SVS and MRSI.
Results
SVS and MRSI acquisition were feasible in >90% of studies. Maximum Cho:NAA and Cho:Cr from MRSI analysis were strongly associated with Cho:NAA and Cho:Cr obtained by SVS (r=0.67 and 0.76, respectively). MRSI Cho:NAA values were more heterogeneous compared to Cho:Cr values within the same lesion, and a strong linear relationship between the range and maximum Cho:NAA values was observed.
Conclusions
SVS and MRSI acquisition was feasible, with a strong correlation in metabolic data. Both techniques may improve diagnostic evaluation and management of DIPG. SVS is recommended for global assessment of tumor metabolism before and after therapy. MRSI showed heterogeneous patterns of metabolic activity within these tumors and is recommended for planning and monitoring targeted therapies and evaluating nearby tissue for tumor invasion.
Keywords: Brainstem, glioma, pontine, children, tumor
Introduction
Diffuse intrinsic pontine gliomas (DIPGs) are invasive pediatric brainstem tumors which affect over 300 children in the United States each year (1). Outcome for these patients is poor, with 90% of patients surviving less than 2 years after diagnosis (2). Biopsy tissue is not routinely obtained in patients with DIPG, and radiographic presentation is the standard means of diagnosis. Radiation therapy is the mainstay of treatment, and magnetic resonance imaging (MRI) is used monitor disease status and treatment response. However, clinical management of DIPG based on MRI appearance is challenging, as there is a poor correlation between standard MRI characteristics and outcome in this population and optimal timing of MRI follow-up is unclear (3, 4). Treatment-related changes such as enhancement, mass effect and edema can mimic changes observed with tumor progression (5). Moderate tumor growth is difficult to detect and verify using current measurement standards (6). Advanced imaging techniques, including proton magnetic resonance spectroscopy (MRS), are under investigation to identify potential prognostic and surrogate markers of outcome in order to improve patient management and evaluate the underlying biology of these devastating tumors.
MRS is a noninvasive procedure that allows molecular imaging of tissue, and is frequently used to evaluate metabolic changes in primary brain tumors compared to normal tissue. The metabolic profile most frequently observed in brain tumors includes elevated choline (Cho), decreased N-acetyl-aspartate (NAA), decreased creatine (Cr) and the presence of lactate and/or lipids (7). Different MRS techniques, including single voxel spectroscopy (SVS) and multi-voxel magnetic resonance spectroscopic imaging (MRSI) are used, but acquisition parameters and processing techniques are not standardized (8). Both SVS and MRSI techniques have advantages and disadvantages. Acquisition of SVS is relatively quick (<5 minutes), and provides a global assessment of tissue within the selected volume. MRSI requires longer acquisition and processing time but provides metabolic data from multiple areas within the tumor and surrounding tissue. Both SVS and MRSI can be applied in a clinical setting and are currently under investigation to identify regions of tumor infiltration, distinguish metabolically active tissue from necrosis, assess treatment response, classify tumor type, identify tumor grade and predict patient outcome in children and adults with primary CNS tumors (7, 9). In adults, the majority of tumors are supratentorial and most MRS studies have focused on tumors in this region. In children, a large percentage of primary brain tumors are infratentorial (1). Performing MRS in this region is technically challenging due to proximity of pericranial lipids, air-tissue interfaces and small size of structures in the brainstem. SVS and MRSI techniques have been successfully used to study the posterior fossa of healthy volunteers (10, 11), but the optimal MRS technique for evaluating tumors in this region is not defined. Studies of posterior fossa tumors in children have typically employed single voxel techniques (12–14), with a limited number of studies using MRSI techniques (15–17). Metabolic data obtained from both SVS and MRSI in DIPG have been identified as potential biomarkers for predicting tumor progression (12, 13) and patient outcome (17). However, the relationship of the metabolic data acquired from each technique is unclear, making it difficult to interpret and compare the clinical utility of MRS techniques in patients with brain tumors. This study assesses the relationship and clinical utility of SVS and MRSI in a group of pediatric patients with DIPG, a posterior fossa tumor which is reliant on imaging for evaluation and therapy monitoring.
Methods and Materials
Patients
Pediatric patients (1 to < 21 years of age) were referred to our institution for evaluation of a newly diagnosed, progressive or refractory DIPG. Confirmation of diagnosis was made using standard radiographic and clinical criteria for DIPG—1) The epicenter of the lesion was in the pons, involved at least 50% of the pons, and appeared hyperintense on a T2-weighted image and 2) Patients presented with cranial nerve deficits, long tract signs, and/or ataxia at the time of diagnosis. Patients whose tumor had a significant exophytic component were excluded. Previous treatment was not an exclusion criterion, and patients were permitted to continue receiving therapy over the course of this study. Informed consent was obtained from patients or their legal guardian for enrollment to a protocol approved by the institutional review board, which included evaluation and follow-up with standard and advanced imaging techniques for the identification of potential in vivo biomarkers.
Magnetic Resonance Imaging
Imaging was performed on a 1.5T Signa HDx system (GE Medical Systems, Milwaukee, WI) using a standard quadrature head coil. The majority of scans were performed under sedation. Imaging was performed at multiple time points during each patient’s disease course, as outlined in the protocol or as clinically indicated. Clinical MRI sequences included axial pre- and post-contrast T1-weighted spin echo (TR/TE = 450/13 ms, FOV=22 × 22 cm, matrix=256 × 192, slice thickness=3 mm, no gap), pre-contrast fluid attenuated inversion recovery (FLAIR; TR/TE/TI = 10,000/140/2200 ms, FOV = 22 × 22 cm, matrix = 256 × 192, slice thickness = 3 mm, no gap) and pre-contrast T2-weighted fast spin echo (TR/TE = 3400/95 ms, FOV = 22 × 22 cm, matrix = 256 × 192, slice thickness = 3 mm, no gap) images. MRI results were reviewed by a neuroradiologist.
Single Voxel Spectroscopy
SVS and multi-slice MRSI data were obtained during the same imaging session prior to contrast administration. SVS was acquired using a point-resolved spin-echo sequence (PROBE-P, GE Medical Systems; TR/TE = 1500 ms/144 ms or 270 ms, averages = 128, FOV = 22 × 22 cm, slice thickness = 15 mm or 20 mm) with automated shim and water suppression. Voxels were positioned on a pre-contrast localizer image (TR/TE = 2000 ms/102 ms, FOV = 24 × 24cm, matrix = 256 × 128, 56 slices, slice thickness = 5 mm, no gap). Voxels were centered within the lesion, dorsal to the basilar artery on the slice with the largest axial diameter of the pons, and included as much of the MRI signal abnormality as possible, while avoiding regions of CSF, bone and air (minimum size = 3 cm3). Acquisition time was approximately 3.5 minutes. SVS data were processed using the scanner manufacturer’s automated processing routine with line width normalization and a Marquardt curve fitting of each major resonance in the spectrum (PROBE-P, GE Medical Systems), including choline (Cho, 3.20 ppm), creatine (Cr, 3.03 ppm) and N-acetylaspartate (NAA, 2.01 ppm). The signal intensity of each metabolite was expressed as the peak amplitude, proportional to the area under the curve. Results were available for immediate review with images of the reconstructed spectrum and voxel location. Peak amplitudes were used to calculate Cho:NAA and Cho:Cr ratios.
Multi-voxel magnetic resonance spectroscopic imaging
MRSI was acquired using a multi-slice, multi-voxel technique which applied a spin-echo sequence for slice selection (TR/TE = 2300ms/280 ms, FOV = 240 mm, matrix = 32 × 32, 4 slices, slice thickness = 15 mm, 3-mm gap), chemical shift saturation pulse for water suppression, and octagonal outer volume saturation (OVS) for lipid suppression (18). OVS bands were arranged more tightly in the inferior slices to optimize suppression of lipids in the posterior fossa (10). The center of one of the four slices was placed in the area of the pons with the largest tumor diameter, similar to SVS voxel placement. A 32 × 32 array of spectra (total = 1,024 voxels, nominal volume of each voxel = 0.84 cm3)was acquired for each MRSI slice. Acquisition time was approximately 25 minutes.
Raw MRSI data were transferred to a workstation for additional processing using a customized software package developed in IDL (ITT Visual Information Solutions, Boulder CO) (19). Processing took approximately 25 – 30 minutes per study. MRSI studies with inadequate water or lipid suppression throughout the 4 slices were excluded from further analysis. Signal intensity maps were generated for Cho, Cr, and NAA. The customized software was used to import and resample pre-contrast axial FLAIR images to match MRSI slice locations using the location and orientation information from the image headers. Regions of interest (ROIs) were manually prescribed on the axial FLAIR images and simultaneously applied to metabolite intensity maps. These ROIs were drawn to match the location and dimensions of the SVS volume of interest from the same imaging session. Voxels with apparent contributions from surrounding tissues, air, or CSF were excluded to reduce partial volume effects. The corresponding spectrum for each ROI voxel was evaluated using several automated quality control measures including Cr and Cho peak separation, width and shape of NAA peak, and baseline signal appearance as previously described (19). Relative concentrations for Cho, Cr and NAA, represented by the area under the signal intensity peak for each metabolite, were expressed as Cho:NAA and Cho:Cr ratios for each spectrum meeting quality control standards. Analysis results for each ROI, including the mean, median, range of values, and standard deviation were reported for both Cho:NAA and Cho:Cr. Maximum MRSI Cho:NAA and maximum Cho:Cr were identified for a “worst voxel” analysis of tumor metabolism. Metabolic heterogeneity of each tumor was estimated using the coefficient of variation (CV, defined as the standard deviation divided by the mean) of Cho:NAA and Cho:Cr and the range of Cho:NAA and Cho:Cr values within each ROI.
Statistical Analysis
We estimated 1) the feasibility of each technique and 2) the correlation between the SVS and MRSI Cho:NAA and Cho:Cr. Feasibility was defined as sufficient water and lipid suppression for detection of metabolites, determined by the quality of the metabolic signals obtained from each. Voxels with low-quality signals for the metabolites of interest were excluded to prevent under or over-estimation of metabolic concentrations. For SVS, scans were considered unacceptable if 1) the voxel included CSF, air, or bone 2) the spectrum showed significant baseline distortion and poor water or lipid suppression, resulting in overlap or interference with metabolite signals and calculation of metabolite concentrations or 3) multiple metabolite signals could not be detected. For MRSI, scans were considered unacceptable if 1) water and lipid signals were not suppressed properly and 2) metabolite signals could not be detected.
The relationship between MRSI maximum Cho:NAA and Cho:Cr and ratios from the corresponding SVS study was evaluated using the Spearman rank correlation method. A repeated measures analysis of variance was applied to account for patients with multiple scans. P ≤ 0.05 was considered statistically significant. Data were analyzed using SAS version 9.1.3 (SAS Institute Inc., Cary, NC).
Results
Patients
Thirty-six patients, ages 1.8 to 18.2 yr (median = 5.7 yr) were included in this study. Imaging studies were performed between May 2004 and March 2010. Both SVS and MRSI were obtained in 150 scans. All patients underwent imaging procedures without complication. Most patients completed a standard 6-week course of radiation therapy prior to study enrollment (median dose = 54.0 Gy, range = 50.4 – 61.2 Gy), with the exception of 2 patients whose families deferred radiation treatment. Patients were followed longitudinally, between 1 and 221 weeks following radiation therapy (median = 23 weeks). Multiple scans were obtained for most patients (median = 3 range = 1 – 21 scans), at a median interval of 8 weeks between scans (range = 1 – 112 weeks).
Feasibility and Quality of SVS and MRSI
Six (4%) SVS studies were excluded for inadequate water suppression. Thirteen (9%) MRSI studies had insufficient water or lipid suppression and were excluded from further analysis. The quality of spectra was evaluated in the remaining studies. Table 1 summarizes the results of the ROI analysis. Eleven SVS volumes of interest were excluded for failed detection of multiple metabolites (n = 6) and inclusion of CSF, air, bone or tissues outside the pons (n = 5). For MRSI, a total of 1205 voxels from 137 scans were evaluated, with an overall acceptance of 72% (871 voxels).
Table 1.
Results of ROI Analysis
| MRS Technique | Median (Range) | Mean (SD) |
|---|---|---|
| SVS | ||
| Voxel size | 9.0 cm3 (3.5 – 20.7 cm3) | 9.5 cm3 (3.5 cm3) |
| Cho:NAA | 2.0 (0.6 – 5.9) | 2.2 (1.1) |
| Cho:Cr | 2.0 (1.0 – 5.2) | 2.2 (0.9) |
| MRSI | ||
| ROI size | 10 voxels (4 – 19 voxels) | 10 voxels (3 voxels) |
| ROI voxel acceptance | 80% (11% – 100%) | 75% (27%) |
| Max Cho:NAA | 2.1 (0.7 – 7.8) | 2.3 (1.2) |
| Max Cho:Cr | 2.5 (1.3 – 4.9) | 2.7 (0.8) |
| ROI CV Cho:NAA | 27% (4% – 59%) | 27% (12%) |
| ROI CV Cho:Cr | 14% (3% – 51%) | 16% (9%) |
Relationship of SVS and MRSI data
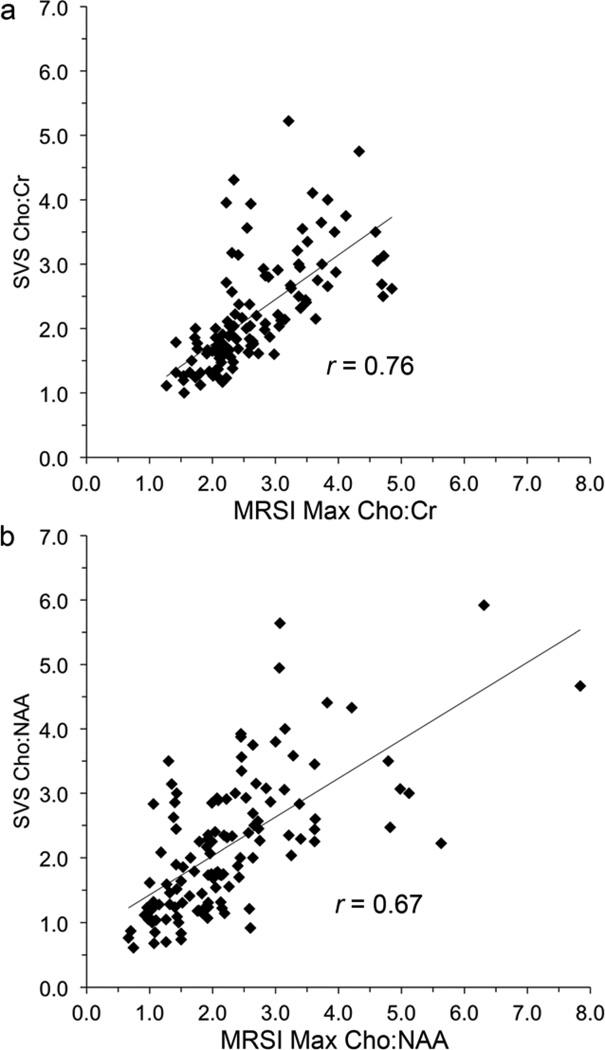
A strong linear association between SVS and MRSI data was identified (Figure 1). Correlations between MRSI maximum and SVS Cho:NAA and Cho:Cr were 0.67 and 0.76, respectively (p < 0.0001 for each). Figure 2 illustrates SVS and MRSI results for a single patient on study.
Figure 1.
Linear relationship between SVS and MRSI (a) Cho:Cr and (b) Cho:NAA.
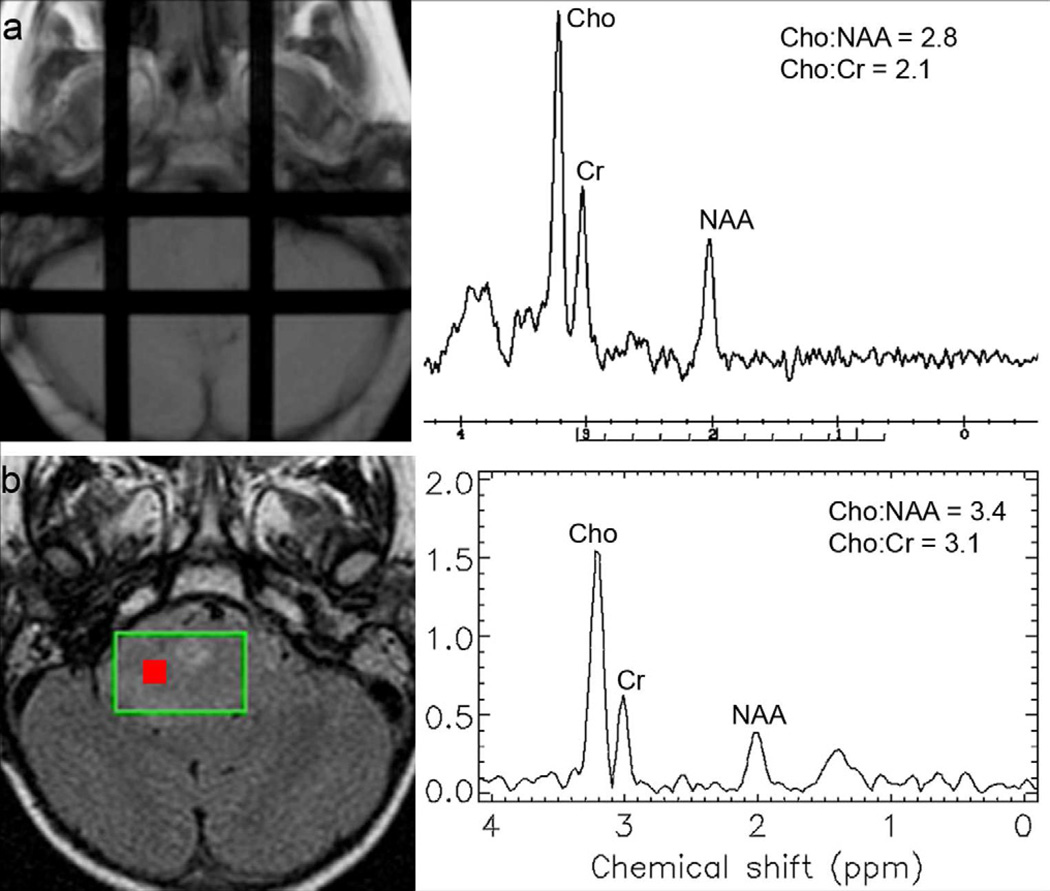
Figure 2.
(a) SVS and (b) MRSI results for a single patient on study. All voxels for the MRSI regions of interest (n = 15) yielded good quality data. MRSI spectral data from the voxel with the maximum Cho:NAA are shown (Voxel location indicated by red box).
Heterogeneity
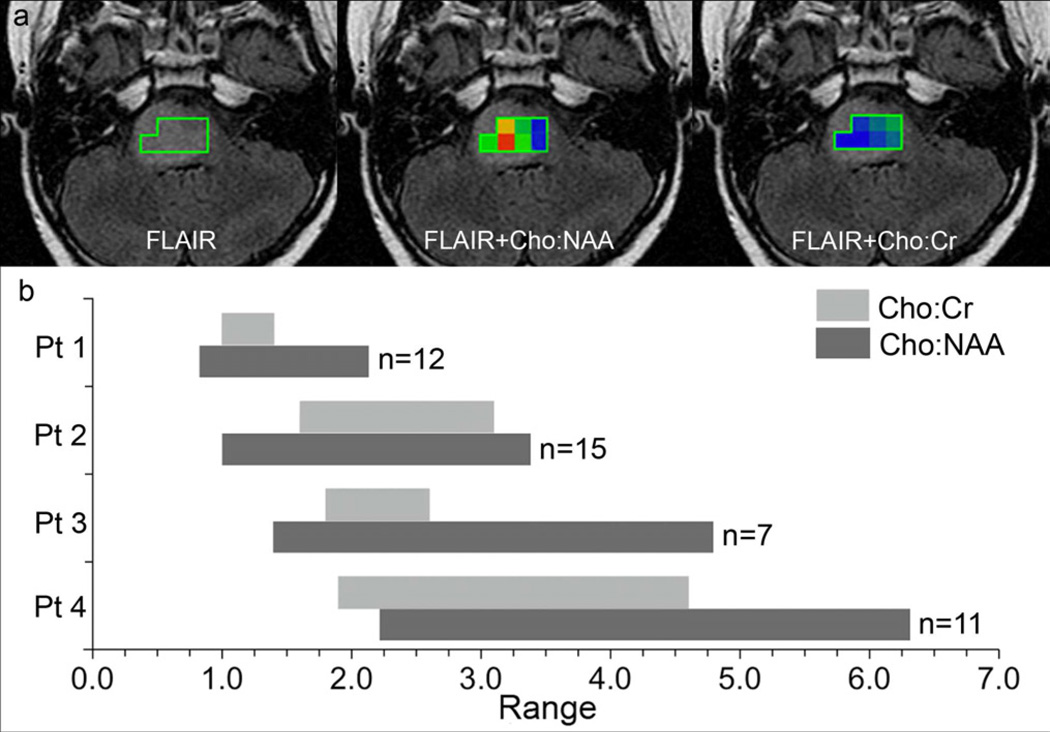
ROIs from MRSI were further evaluated to determine the metabolic heterogeneity of tumors using two measures—1) coefficient of variation, CV and 2) range in values for all MRSI measures. Results are summarized in Table 1. ROI CV for Cho:NAA was larger (i.e., more heterogeneous) than that of Cho:Cr in the majority (83%) of studies. Analysis of the range in MRSI values yielded similar results. The range in Cho:Cr values for each ROI was typically narrower (i.e., less heterogeneous) compared to that of Cho:NAA values. The maximum Cho:NAA was strongly related to the range in Cho:NAA ratio values. Patients with larger maximum Cho:NAA tended to have a broader range of Cho:NAA values throughout the tumor ROI (Figure 3).
Figure 3. Metabolic heterogeneity of DIPG.
(a) MRSI regions of interest for a single case (Patient 3 in Figure 3b). Color maps represent the range in values within the ROI (Possible values include 0.5 – 5.0; blue = minimum and red = maximum). (b) Ranges in Cho:NAA and Cho:Cr for 4 patients on study. The upper limit of each bar represents the maximum value of each range. Cases illustrate the linear relationship between the range and maximum Cho:NAA. Patient 1 (ROI = 12 voxels) had the lowest maximum Cho:NAA and the narrowest range in Cho:NAA values. Patient 4 (ROI = 11 voxels) had the highest maximum Cho:NAA with the broadest range in Cho:NAA values.
Discussion
MRS is increasingly used to noninvasively assess the metabolic activity of CNS tumors for clinical management, providing additional information to standard MRI. Although promising as a potential biomarker or surrogate endpoint for clinical trials, major drawbacks include the lack of standardization in the methods used to acquire and analyze data. Selection of the spectroscopic method is frequently operator- or institutional-dependent. The objectives of this study were to evaluate the feasibility and relationship of two spectroscopy methods in pediatric DIPG, a tumor that is highly dependent on imaging for diagnosis and evaluation. We found that SVS and MRSI are feasible and informative of tumor metabolism in this population, and identified a strong correlation between the two techniques.
Previous MRS studies of patients with DIPG used different techniques, sequence parameters, analysis methods and evaluated different regions within the lesions, which precludes an accurate comparison between studies (12, 13, 15–17). Our study evaluated the relationship between SVS and MRSI results obtained during the same scan and evaluating similar regions of interest for each patient. Though SVS and MRSI results were not equivalent, a strong linear relationship was identified.
The association between the maximum Cho:NAA and Cho:Cr obtained from MRSI and the corresponding ratios obtained using SVS was not unexpected, given the biologic implication of these metabolites. NAA and Cr are markers of normal brain tissue, and both are decreased in tumors. NAA is predominantly found in neurons and is considered a marker of neuronal integrity. Cr is a marker of normal cell energy storage. Both of these physiologic conditions are compromised in tumor (7). Cho is found in cell membranes and increased cell density would be expected throughout most of the pons in this infiltrative lesion.
Several recommendations can be made for using each MRS technique in the evaluation of pediatric DIPG. Multiple prognostic biomarkers have been identified using SVS techniques (12, 13, 17). Careful placement of voxels is critical for accurate results (20), but given its shorter acquisition and automated processing, we recommend using SVS for global evaluation of tumors before and after therapy or evaluating a specific area within a lesion (e.g., differentiating necrosis from viable tumor). Metabolic data from MRSI is also predictive of patient outcome (17), but MRSI has a longer acquisition time and requires additional analysis, typically performed by a trained spectroscopist. The use of MRSI techniques in the clinical setting without advanced technical assistance may be impractical until manufacturer software with semi-automated processing and analysis becomes more widely available (8). However, MRSI provides greater spatial resolution than SVS, identifies areas of highest metabolic activity, and also allows evaluation of the entire tumor and surrounding tissues. Results of the MRSI used in this study showed that DIPGs are metabolically heterogeneous tumors. Therefore, we recommend MRSI for planning and monitoring targeted therapies, such as convection enhanced delivery, identifying biopsy targets, and monitoring possible rostral or caudal tumor growth often observed in patients with DIPG.
Results from this study must be interpreted in the context of the following limitations. Although our goal was to compare SVS and MRSI for each individual patient at separate time points, patients evaluated on this study were heterogeneous in regards to their treatment and time of evaluation, preventing any correlation of MRS results with treatment related-changes. Standard clinical MRI sequences were used to determine voxel selection. Necrosis, edema, and hemorrhage could not be reliably distinguished using standard MRI, and regions with these pathological conditions may be included in the analysis. Regions of interest for MRSI studies were manually selected to cover the same volume as the corresponding SVS study. However, MRSI and SVS studies were not co-registered. MRSI studies were successfully obtained in the majority of scans, but the percentage of voxel acceptance for each ROI varied depending on the quality of spectra. In some MRSI spectra, NAA peaks were too small to distinguish from baseline noise and were excluded from analysis. As a result, the maximum Cho:NAA obtained by MRSI was not irrefutably representative of the worst voxel in the tumor, and histologic validation was not possible.
Proton spectroscopy continues to be evaluated as a potential biomarker and surrogate endpoint for patients with CNS tumors due to the inability to acquire repeat tissue samples. However, MRS techniques are not standardized and the relationship of data obtained by different techniques was unclear. This study compares data obtained from both MRSI and SVS in children with DIPG. Obtaining good quality spectra in the brainstem was feasible using the methods applied in this study. While there is a strong concordance between the maximum Cho:NAA from MRSI and Cho:NAA determined by SVS, the results are not equivalent, and each technique has a distinct application in radiographic follow-up of these patients. SVS is recommended for monitoring global changes in tumor metabolism before and after therapy. MRSI is recommended for identifying the area of greatest activity within the tumor for targeted treatment and monitoring surrounding tissue. Both techniques provide additional insight into the biologic behavior of DIPG and can be incorporated into the evaluation of these tumors, depending on the clinical indication being addressed.
Acknowledgements
The authors thank Dr. Jan Willem van der Veen for his contributions to the MRSI acquisition and Dr. Alan Barnett for the development of the MRSI registration and analysis software. Our research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The views expressed in this article are those of the authors and do not reflect the official policy of the National Institutes of Health, Department of Army, Department of Defense, or U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Meeting presentation line: This work was presented in part at the International Society of Pediatric Neuro-Oncology (ISPNO) Annual Meeting, Chicago 2008.
Conflict of Interest Statement: The authors declare no conflict of interest.
References
- 1.Central Brain Tumor Registry of the United States (CBTRUS) CBTRUS statistical report: primary and central nervous system tumors diagnosed in the United States in 2004–2006. Hindsdale, IL: 2010. pp. 10–25. [Google Scholar]
- 2.Hargrave D, Bartels U, Bouffet E. Diffuse brainstem glioma in children: critical review of clinical trials. Lancet Oncol. 2006;7:241–248. doi: 10.1016/S1470-2045(06)70615-5. [DOI] [PubMed] [Google Scholar]
- 3.Hargrave D, Chuang N, Bouffet E. Conventional MRI cannot predict survival in childhood diffuse intrinsic pontine glioma. J Neurooncol. 2008;86:313–319. doi: 10.1007/s11060-007-9473-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu AK, Brandon J, Foreman NK, et al. Conventional MRI at presentation does not predict clinical response to radiation therapy in children with diffuse pontine glioma. Pediatr Radiol. 2009;39:1317–1320. doi: 10.1007/s00247-009-1368-5. [DOI] [PubMed] [Google Scholar]
- 5.Yang I, Huh NG, Smith ZA, et al. Distinguishing Glioma Recurrence from Treatment Effect After Radiochemotherapy and Immunotherapy. Neurosurgery Clinics of North America. 2010;21:181–186. doi: 10.1016/j.nec.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Hayward RM, Patronas N, Baker EH, et al. Inter-observer variability in the measurement of diffuse intrinsic pontine gliomas. J Neurooncol. 2008;90:57–61. doi: 10.1007/s11060-008-9631-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soares DP, Law M. Magnetic resonance spectroscopy of the brain: review of metabolites and clinical applications. Clin Radiol. 2009;64:12–21. doi: 10.1016/j.crad.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Mandal PK. In vivo proton magnetic resonance spectroscopic signal processing for the absolute quantitation of brain metabolites. Eur J Radiol. 2011 doi: 10.1016/j.ejrad.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 9.Horska A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am. 2010;20:293–310. doi: 10.1016/j.nic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs MA, Horska A, van Zijl PC, et al. Quantitative proton MR spectroscopic imaging of normal human cerebellum and brain stem. Magn Reson Med. 2001;46:699–705. doi: 10.1002/mrm.1248. [DOI] [PubMed] [Google Scholar]
- 11.Galanaud D, Le Fur Y, Nicoli F, et al. Regional metabolite levels of the normal posterior fossa studied by proton chemical shift imaging. MAGMA. 2001;13:127–133. doi: 10.1007/BF02668161. [DOI] [PubMed] [Google Scholar]
- 12.Panigrahy A, Nelson MD, Jr, Finlay JL, et al. Metabolism of diffuse intrinsic brainstem gliomas in children. Neuro Oncol. 2008;10:32–44. doi: 10.1215/15228517-2007-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamasaki F, Kurisu K, Kajiwara Y, et al. Magnetic resonance spectroscopic detection of lactate is predictive of a poor prognosis in patients with diffuse intrinsic pontine glioma. Neuro Oncol. 2011;13:791–801. doi: 10.1093/neuonc/nor038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porto L, Hattingen E, Pilatus U, et al. Proton magnetic resonance spectroscopy in childhood brainstem lesions. Childs Nerv Syst. 2007;23:305–314. doi: 10.1007/s00381-006-0221-5. [DOI] [PubMed] [Google Scholar]
- 15.Laprie A, Pirzkall A, Haas-Kogan DA, et al. Longitudinal multivoxel MR spectroscopy study of pediatric diffuse brainstem gliomas treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2005;62:20–31. doi: 10.1016/j.ijrobp.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Thakur SB, Karimi S, Dunkel IJ, et al. Longitudinal MR spectroscopic imaging of pediatric diffuse pontine tumors to assess tumor aggression and progression. AJNR Am J Neuroradiol. 2006;27:806–809. [PMC free article] [PubMed] [Google Scholar]
- 17.Steffen-Smith EA, Shih JH, Hipp SJ, et al. Proton magnetic resonance spectroscopy predicts survival in children with diffuse intrinsic pontine glioma. J Neurooncol. 2011 doi: 10.1007/s11060-011-0601-x. [Epub ahead of print May 13 2011]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duyn JH, Gillen J, Sobering G, et al. Multisection proton MR spectroscopic imaging of the brain. Radiology. 1993;188:277–282. doi: 10.1148/radiology.188.1.8511313. [DOI] [PubMed] [Google Scholar]
- 19.Tedeschi G, Bertolino A, Campbell G, et al. Reproducibility of proton MR spectroscopic imaging findings. AJNR Am J Neuroradiol. 1996;17:1871–1879. [PMC free article] [PubMed] [Google Scholar]
- 20.Ricci PE, Pitt A, Keller PJ, et al. Effect of voxel position on single-voxel MR spectroscopy findings. AJNR Am J Neuroradiol. 2000;21:367–374. [PMC free article] [PubMed] [Google Scholar]