Not so long ago, the idea of deriving functional models from just looking at large subcellular structures in different functional states seemed unrealistic. In the meantime, the visualization of such structures by three-dimensional reconstructions from cryoelectron microscopic images has made enormous progress. Studies on ribosomes, from both prokaryotes and eukaryotes, have been particularly rewarding. The structure of the Escherichia coli ribosome was determined at ≈25-Å resolution in 1995 by the groups of Frank (1) and van Heel (2) and was improved to 18 Å recently (3). Likewise, reconstructions of eukaryotic ribosomes have been obtained recently (4–6). Regarding functional complexes, it was possible to localize tRNA molecules in three binding sites (A, P, and E) on the E. coli ribosome (7) and to determine their respective pre- and post-translocation positions (8). The complex of elongation factor (EF) Tu with Phe-tRNA was visualized in the ribosomal A site at 18-Å resolution (3), and, by fitting the crystal structure of the complex (9), the structure could be interpreted in terms of specific interactions with the ribosome. Another recent reconstruction visualized at the protein exit site of the eukaryotic ribosome a protein complex, Sec 61, that is involved in cotranslational protein translocation through the membrane of the endoplasmic reticulum (4).
The paper by Agrawal et al. (10) in a recent issue of the Proceedings reports the structure of another important functional ribosomal complex that is a complex of EF-G with E. coli ribosomes. The structure represents a late state of the translocation reaction in the ribosomal elongation cycle. Translocation is catalyzed by EF-G at the expense of GTP and consists of the coordinated movement of two mRNA-bound tRNAs from their pretranslocational positions to their post-translocation positions. The present reconstruction has a resolution of 20 Å and clearly shows the factor, a five-domain, 77-kDa protein of elongated shape spanning the intersubunit space and making contacts with both 30S and 50S ribosomal subunits. By fitting the crystal structure of EF-G-GDP (11, 12) to the density attributable to the factor—allowing some structural rearrangement of the factor for optimal fit—the factor could be placed in a well defined position. The arrangement is remarkably similar to that proposed recently on the basis of hydroxyl radical probing of the RNAs of the small and large subunit ribosomal directed from a large variety of surface positions of the five domains of EF-G (13). The ribosome-EF-G complexes used in the two studies are comparable biochemically [except for the presence (13) and absence (10) of tRNA] in that they were prepared in the presence of an antibiotic, fusidic acid, which binds to EF-G-GDP on the ribosome and stabilizes the complex.
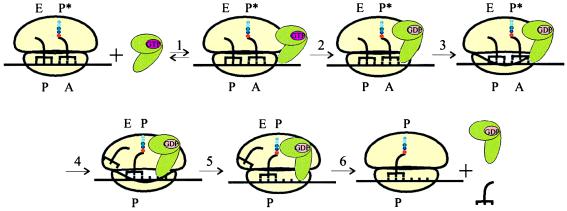
Before addressing potential functional implications of the topographical information, the partial reactions of translocation are summarized briefly (Fig. 1) ; hypothetical structural changes are discussed below. In the pretranslocation state, deacylated tRNA resides in the P site with the 3′ end toward or in the E site [P/E hybrid state (14)], and peptidyl-tRNA resides in the A site with the peptidyl end toward the P site (A/P* hybrid state, P* denoting a puromycin-unreactive state). Immediately after the binding of EF-G-GTP (Step 1), GTP is hydrolyzed [Step 2 (15)]. Most likely, GTP hydrolysis or subsequent release of Pi causes a conformational change of EF-G, which, in turn, induces the formation of the transition state of the ribosome (Step 3). In the transition state, the movement of the tRNA–mRNA complex takes place (Step 4). This step comprises (i) the movement of the 3′ ends of the two tRNAs on the 50S subunit into their respective post-translocation positions [i.e., the E site for the deacylated tRNA and the puromycin-reactive position of the peptidyl end of the peptidyl-tRNA in the P site (A/P state)]; and (ii) the movement of the anticodon arms of both tRNA molecules together with the mRNA to their immediate post-translocation positions on the 30S subunit. In step 5, the ribosome returns to the ground state and EF-G assumes the GDP-bound conformation; this step is inhibited by fusidic acid. Step 6 then describes the dissociation of both EF-G-GDP and the deacylated tRNA (the order of dissociation is not known) to reach the final post-translocation state of the ribosome with peptidyl-tRNA in the P site and a free A site.
Figure 1.
Reaction scheme of translocation as discussed in the text. EF-G (green) is depicted in three conformations: the GTP-bound form, an intermediate GDP-bound form on the ribosome, and the GDP-bound form that dissociates from the ribosome (step 6); the latter conformation probably is represented in the crystal structures of EF-G-GDP and nucleotide-free EF-G (11, 12). Pi is omitted because it is not known when after GTP hydrolysis it is released. The transition state of the ribosome, formed in step 3, is symbolized by an altered conformation of the small ribosomal subunit; of course, alterations of the large subunit or an altered arrangement of the subunits relative to each other also may be involved. A, P (P*), and E denote the tRNA binding sites on the two subunits and are indicated when occupied.
Previous studies using immunoelectron microscopy (16), crosslinking (17), chemical footprinting (18), and functional assays (19) have defined several EF-G-ribosome interaction sites, mainly on the 50S ribosomal subunit. The factor binding site has been mapped to the base of the L7/12 stalk of the 50S subunit, and in 23S RNA the sarcin stem-loop (around residue 2660) as well as the thiostrepton binding site (around residue 1070) have been identified as presumed contact sites (20). In the electron microscopic reconstruction of Agrawal et al. (10), domain 5 of EF-G appears to contact the base of the stalk, which could account for the contacts mentioned, whereas the G domain (domain 1), presumably through the G′ subdomain, is involved in an interaction with a structural element extending from the L7/12 stalk; the latter contact is strikingly reminiscent of a similar connection seen with EF-Tu (3). The GTPase activity of EF-G, as the one of EF-Tu, is activated when the factor binds to the ribosome. By analogy to other GTPases, the stimulation of the GTPase activity probably requires that a ribosomal structure contacts the G domain in the vicinity of the nucleotide binding site. L7/12 has been implicated in factor function on the ribosome repeatedly, and the contact may well reflect an interaction involved in GTPase activation, although other interactions on the 50S subunit, as discussed above, may be involved as well.
Interactions of EF-G with the small ribosomal subunit were not reported previously, except for a crosslink to protein S19 (17), and, therefore, mainly the 50S subunit was discussed as target for EF-G action in translocation. Only when the remarkable similarities of the tertiary structures of EF-G-GDP (11, 12) and the EF-Tu-5′-guanylylimidodiphosphate-Phe-tRNA (9) complex were discovered, it was suggested that domain 4 of EF-G, which strikingly matches the tRNA anticodon domain of the ternary complex, acts on the 30S subunit. Indeed, the deletion of domain 4 strongly impairs the activity of EF-G in promoting translocation (15). An important functional role of domain 4 also is suggested by mutational studies on the homologous eukaryotic EF-2 in yeast (21) and by the fact that the inactivation of mammalian EF-2 by diphtheria toxin is caused by ADP ribosylation at the tip of domain 4 (22). The present positioning of EF-G (10), with domain 4 reaching into the cleft of the 30S subunit at or near the decoding center, which is corroborated by chemical probing data (13), is fully consistent with the functional studies and supports the contention of a crucial role of domain 4 in translocation catalysis by EF-G that is exerted on the small subunit of the ribosome.
Much less is known about domain 2, through which the protein appears to contact the small ribosomal subunit in the S4/S12 region, in keeping with hydroxyl radical hits from domain 2 in the vicinity of the S4-binding region of 16S RNA (13). A similar orientation of domain 2, albeit without binding contact, was observed in the ribosome complex with EF-Tu-Phe-tRNA (3). The S4 region of 16S RNA, in particular the 530 loop, has been implicated as an important target for EF-Tu in influencing the function of the decoding center (23), and mutants of protein S4 were among the first described to affect ribosomal accuracy. Thus, the contact of domain 2 with the S4 region could be the means by which the factor(s) affect the decoding center, thereby influencing tRNA-ribosome contacts.
Translocation is an exergonic process because the spontaneous EF-G-independent reaction can be observed in vitro (24). Thus, the potential for the molecular movements involved in translocation resides in the structure of the ribosome. However, there is a large activation barrier to be overcome for translocation to proceed fast enough to sustain the rate of protein elongation in vivo (10 to 20 s−1 per amino acid incorporated in E. coli). The activation barrier is lowered by binding of EF-G-GTP to the ribosome in the pretranslocation state (studied with nonhydrolyzable GTP analogues) and by GTP hydrolysis (15). The latter probably induces conformational strain in EF-G, which, by establishing binding interactions with the ribosome, is coupled to conformational changes of the ribosome that allow, or promote, tRNA movement.
Does the present structure explain how energy coupling may be accomplished in terms of structure? The two tRNAs in A and P sites are bound to their respective sites by interactions not only with the mRNA but also with the ribosome, notably with 16S RNA and 23S RNA. As has been pointed out in a recent review by Wilson and Noller (25), these interactions are different in the two sites, and it is difficult to imagine that they are all disrupted at once before, and reformed after, the movement. Thus, a direct action of EF-G on the tRNA by way of a “push” mechanism (26) seems unlikely. It is more likely that EF-G, by binding interactions with the ribosome that are reinforced by conformational changes brought about by GTP hydrolysis, induces conformational rearrangements of the tRNA binding sites. The structural changes destabilize some of those tRNA-ribosome interactions and/or promote the formation of alternative interactions that stabilize the transition and, eventually, post-translocation states. There is direct evidence that supports such a mechanism for the movement of the 3′ end of P site-bound tRNA into the E site on the 50S subunit (27), and the same may apply for interactions in the decoding center. Additionally, it has been shown that the anticodon regions of the two tRNAs move together during translocation (28), which is also difficult to reconcile with a simple push mechanism.
Conformational changes of EF-G, induced by GTP hydrolysis and Pi release, that have been implicated in the above discussion are largely hypothetical at present because the structure of EF-G in the GTP-bound form is not known. It is, therefore, interesting to note that Agrawal et al. (10), to optimize the fit in modeling the observed density, had to introduce a conformation of EF-G that differs from the crystal structure of EF-G-GDP by a rotation of domains 4 and 5 relative to the domains 1–3. By analogy to other GTPases, in particular to EF-Tu, for which the conformational differences between the GTP and GDP-bound forms are well characterized (29, 30), one may expect that the conformations of EF-G-GDP and EF-G-GTP differ with respect to the interactions of the G domain with neighboring domains 2 and 5 and that the modulation of these interactions is functionally important. In the present structure, a state after the transition state and preceding EF-G release is frozen by the binding of fusidic acid to the factor. Inhibition of EF-G release also results from deletion of domain 4 (15), indicating that conformational coupling between the factor and the ribosome is required to release binding interactions of the transition state. Of interest, a frozen EF-G-GDP-ribosome complex also is observed in the presence of thiostrepton (unpublished results), an antibiotic that binds to the 50S subunit and may inhibit the conformational relaxation of the complex by blocking transitions of the ribosome.
It should be stressed that the scenario developed above at present remains largely hypothetical. Because the structure of EF-G-GTP is not known, we can only speculate about conformational changes in EF-G in response to GTP hydrolysis, which may lead to the known GDP-bound structure and would make EF-G a motor protein (15). The structure reported by Agrawal et al. (10) shows the position of EF-G and the structure of the EF-G-ribosome complex in a state equivalent to the state after the movement of the tRNAs has taken place (if there were tRNAs in the complex), and we do not know how closely this state resembles the transition state. Thus, one would like to look at the transition state complex, in particular to see whether conformational changes of the ribosome, as implied in the above discussion, can be demonstrated. We also do not know to what extent the interactions between EF-G and the ribosome seen in the present structure are indicative of the interactions that induce translocation. To resolve this issue, the structure of the pretranslocation complex with EF-G bound to it has to be determined. The paper of Agrawal et al. (10) is an excellent example demonstrating how functional information can emerge from looking at a structure.
Footnotes
The companion to this commentary is published on pages 6134–6138 in issue 11.
References
- 1.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata R K, Agrawal R K. Nature (London) 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 2.Stark H, Mueller F, Orlova E V, Schatz M, Dube P, Erdemir T, Zemlin F, Brimacombe R, van Heel M. Structure (London) 1995;3:815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- 3.Stark H, Rodnina M V, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. Nature (London) 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- 4.Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 5.Verschoor A, Warner J R, Srivastava S, Grassucci R A, Frank J. Nucleic Acids Res. 1998;26:655–661. doi: 10.1093/nar/26.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dube P, Wieske M, Stark H, Schatz M, Stahl J, Zemlin F, Lutsch G, van Heel M. Structure (London) 1998;6:389–399. doi: 10.1016/s0969-2126(98)00040-9. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal R K, Penczek P, Grassucci R A, Li Y, Leith A, Nierhaus K H, Frank J. Science. 1996;271:1000–1002. doi: 10.1126/science.271.5251.1000. [DOI] [PubMed] [Google Scholar]
- 8.Stark H, Orlova E V, Rinke-Appel J, Junke N, Mueller F, Rodnina M, Wintermeyer W, Brimacombe R, van Heel M. Cell. 1997;88:19–28. doi: 10.1016/s0092-8674(00)81854-1. [DOI] [PubMed] [Google Scholar]
- 9.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal R K, Penczek P, Grassucci R A, Frank J. Proc Natl Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czworkowski J, Wang J, Steitz T A, Moore P B. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ævarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson L A, Liljas A. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson K S, Noller H F. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- 14.Moazed D, Noller H F. Nature (London) 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 15.Rodnina M V, Savelsbergh A, Katunin V I, Wintermeyer W. Nature (London) 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 16.Girshovich A S, Bochkareva E S, Gudkov A T. FEBS Lett. 1982;150:99–102. doi: 10.1016/0014-5793(82)81312-4. [DOI] [PubMed] [Google Scholar]
- 17.Sköld S E. Nucleic Acids Res. 1983;11:4923–4932. doi: 10.1093/nar/11.14.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moazed D, Robertson J M, Noller H F. Nature (London) 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 19.Hausner T P, Atmadja J, Nierhaus K H. Biochimie (Paris) 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 20.Munishkin A, Wool I G. Proc Natl Acad Sci USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimata Y, Kohno K. J Biol Chem. 1994;269:13497–13501. [PubMed] [Google Scholar]
- 22.Foley B T, Moering J M, Moering T J. J Biol Chem. 1995;270:23218–23225. doi: 10.1074/jbc.270.39.23218. [DOI] [PubMed] [Google Scholar]
- 23.Powers T, Noller H F. Trends Genet. 1994;10:27–31. doi: 10.1016/0168-9525(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 24.Spirin A S. Prog Nucleic Acid Res Mol Biol. 1985;32:75–114. doi: 10.1016/s0079-6603(08)60346-3. [DOI] [PubMed] [Google Scholar]
- 25.Wilson K S, Noller H F. Cell. 1998;92:337–349. doi: 10.1016/s0092-8674(00)80927-7. [DOI] [PubMed] [Google Scholar]
- 26.Abel K, Jurnak F. Structure (London) 1996;4:229–238. doi: 10.1016/S0969-2126(96)00027-5. [DOI] [PubMed] [Google Scholar]
- 27.Lill R, Robertson J M, Wintermeyer W. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen H, Wintermeyer W. Biochemistry. 1986;25:2749–2756. doi: 10.1021/bi00358a002. [DOI] [PubMed] [Google Scholar]
- 29.Abel K, Yoder M D, Hilgenfeld R, Jurnak F. Structure (London) 1996;4:1153–1159. doi: 10.1016/s0969-2126(96)00123-2. [DOI] [PubMed] [Google Scholar]
- 30.Polekhina G, Thirup S, Kjeldgaard M, Nissen P, Lippmann C, Nyborg J. Structure (London) 1996;4:1141–1151. doi: 10.1016/s0969-2126(96)00122-0. [DOI] [PubMed] [Google Scholar]