Abstract
Yersinia pestis, the causative agent of plague, is unique among the enteric group of Gram-negative bacteria in relying on a blood-feeding insect for transmission. The Yersinia-flea interactions that enable plague transmission cycles have had profound historical consequences as manifested by human plague pandemics. The arthropod-borne transmission route was a radical ecologic change from the food- and water-borne transmission route of Yersinia pseudotuberculosis, from which Y. pestis diverged only within the last 20,000 years. Thus, the interactions of Y. pestis with its flea vector that lead to colonization and successful transmission are the result of a recent evolutionary adaptation that required relatively few genetic changes. These changes from the Y. pseudotuberculosis progenitor included loss of insecticidal activity, increased resistance to antibacterial factors in the flea midgut, and extending Yersinia biofilm-forming ability to the flea host environment.
Introduction
The genus Yersinia consists of seventeen species of Gram-negative rod-shaped bacteria in the family Enterobacteriaceae of the gammaproteobacteria. Most of them are widely distributed in the environment and two of them, Yersinia enterocolitica and Yersinia pseudotuberculosis, cause relatively mild food- and water-borne enteric diseases. A third pathogenic species, Yersinia pestis, stands out in several respects. Y. pestis, the cause of plague, is one of the most virulent bacterial pathogens responsible for three devastating pandemics in human history. Plague is primarily a disease of rodents, however, and Y. pestis is transmitted between them primarily via flea vectors. Maintenance of Y. pestis in nature depends on these rodent-flea transmission cycles, and the ecology of plague is complex, involving many different rodent and flea species. Today, Y. pestis exists in permanently entrenched enzootic foci in many parts of the world. Thus, unlike all other Yersinia species, Y. pestis no longer needs to survive in the environment but instead alternates between life in two different eukaryotic hosts-insects and mammals. Remarkably, this radical change in ecology is a recent evolutionary adaptation, because Y. pestis is essentially a clonal variant that diverged from the closely related Y. pseudotuberculosis only within the last 1,500 to 20,000 years [1,2].
As with most bacteria of public health importance, Yersinia research has focused on bacterial interactions with the mammalian host that lead to disease. The interactions of Y. pestis with fleas that lead to transmission have received comparatively little attention. In this review we examine the mechanisms by which Y. pestis is able to colonize the flea digestive tract and to produce a transmissible infection, and the evolutionary pathway that led to arthropod-borne transmission in the genus Yersinia.
The flea host environment
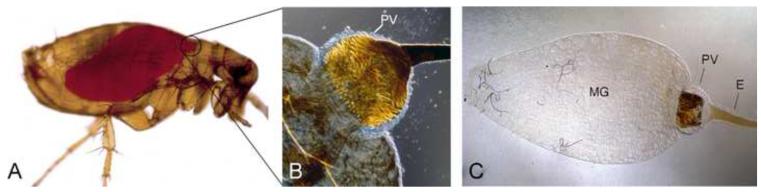
Fleas are wingless insects of the order Siphonaptera, most closely related to the Diptera and Mecoptera orders. Over 2,000 species have been described, and adult fleas are obligate blood-feeding ectoparasites. Most live in close association with their hosts and depend on small but frequent blood meals for survival. Unlike mosquitoes, blood meal storage, digestion, and adsorption all take place in a simple, non-compartmentalized midgut (Fig. 1); hemolysis and complete liquefaction of the blood meal occur within a few hours and digestion is fully completed within a few days.
Figure 1.
The flea digestive tract. (A) Uninfected X. cheopis flea immediately after taking a blood meal, with the midgut filled with fresh blood. (B, C) Digestive tracts dissected from uninfected fleas showing the anatomy of the proventriculus and the simple midgut.
Little is known about the biochemical and physiological environment of the flea digestive tract. The intense digestive milieu of the flea midgut and continual disturbance due to peristalsis and excretion would appear to be a somewhat hostile environment, because fleas have limited (but as yet poorly defined) normal midgut flora, and few pathogens are transmitted by fleas. Nevertheless, rickettsia-like endosymbionts are common in flea tissue, protists occur in the hindgut and Malpighian tubules, and Gram-positive and Gram-negative bacteria can also be detected in fleas [3-6]. How this normal flora might influence the flea gut environment or flea-Yersinia interactions and vector competency is unknown. The flea immune response to infection with Y. pestis has not been characterized either, but likely will resemble the immune response of mosquitoes and other insects to ingested foreign bacteria. Both Y. pestis and Y. pseudotuberculosis are constitutively resistant to cationic antimicrobial peptides, common elements of insect immunity, especially at low growth temperatures that match the flea environment [7-9].
Yersinia infection of the flea digestive tract
Y. pestis infection of the flea is confined to the digestive tract, entering the midgut as individual, planktonic bacteria when a flea feeds on an infected rodent. The bacteria do not adhere to or invade the midgut epithelium and are initially quite susceptible to elimination in flea feces-up to half of fleas completely clear themselves of infection in this way even after feeding on blood containing >108 Y. pestis per milliliter [10,11]. Thus, plague transmission cycles depend on the ability of Y. pestis to produce a high-density bacteremia in the mammalian host. Y. pestis can achieve levels of 109 per ml in the peripheral blood of susceptible rodents, and this overwhelming sepsis accounts for the extreme virulence and high mortality rate of plague.
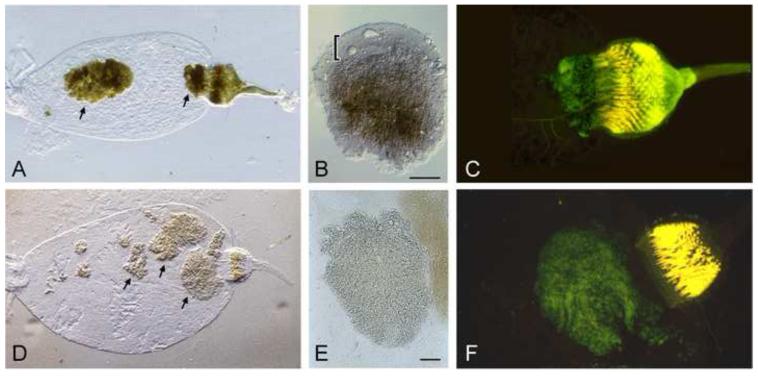
Persistence in the flea gut depends on the ability of Y. pestis to form multicellular aggregates that are too large to be passed in the feces. Clumps of bacteria begin to form within the first few days after infection and grow and consolidate as the bacteria multiply during the first week (Fig. 2). Y. pestis numbers in a successfully colonized flea reach 105 to >106 by the second week after infection, and then plateau [12].
Figure 2.
The Y. pestis life stage in the flea. (A) Digestive tract from an X. cheopis flea dissected two weeks after infection with wild-type Y. pestis. Arrows indicate large aggregates of bacteria, one of which fills the proventriculus. (B) Wild-type Y. pestis aggregate dissected from the midgut. The dense mass of bacteria is brown-pigmented due to hmsHFRS-dependent sequestration of hemin derived from the flea blood meal and is enveloped by a viscous layer (indicated by the bracket) composed of biofilm ECM and lipid components, also derived from the blood meal [16]. (C) Fluorescence micrograph of the digestive tract from a flea blocked with wild-type Y. pestis expressing green fluorescent protein (GFP). The PV is swollen due to the large bacterial mass that fills it. (D) Digestive tract from a flea dissected two weeks after infection with hmsHFRS− Y. pestis. Multicellular aggregates of bacteria are present in the midgut but the proventriculus is uninfected. (E) Aggregate of hmsHFRS− Y. pestis dissected from the midgut. The bacterial aggregate is not pigmented and not surrounded by ECM. (F) Fluorescence micrograph of the digestive tract from a flea infected with hmsHFRS− Y. pestis expressing GFP. The infection is confined to the midgut and does not involve the proventriculus. Dissected digestive tracts in both Fig. 1 and Fig. 2 were mounted in H20, which clears red blood cell material from previous blood meals but not the bacterial aggregates. MG, midgut; PV, proventriculus; E, esophagus; ECM, extracellular matrix. Scale bars = 0.05 mm.
Survival of Y. pestis in the flea midgut depends on the activity of a phospholipase D (PLD) enzyme, originally characterized as a toxin for mice and rats, that is encoded by the ymt (Yersinia murine toxin) gene carried on a Y. pestis-specific plasmid [13]. Ymt is a member of a large, phylogenetically widespread family of PLDs characterized by signature HKD motifs in the catalytic site of the enzyme. Y. pestis mutants lacking functional Ymt assume an aberrant spherical shape, indicative of loss of outer membrane integrity, and rapidly disappear from the flea midgut within a day after infection. Presumably, Ymt protects the bacteria against a bacteriolytic agent that is generated in the flea midgut during digestion of the blood meal, but the agent and mechanism by which PLD activity protects against it remain to be identified. Interestingly, transformation of Y. pseudotuberculosis and E. coli with the Y. pestis ymt gene greatly increase their ability to infect the flea midgut also [13].
Role of bacterial biofilm in producing a transmissible infection of fleas
Colonization of the flea midgut is not sufficient to produce a transmissible infection. Unlike many other arthropod-borne pathogens, which are transmitted via vector saliva after dissemination to the salivary glands, Y. pestis is transmitted by regurgitation from the digestive tract. Regurgitative transmission is possible when the bacteria infect the proventriculus, a valve in the flea foregut that connects the esophagus and the midgut (Fig. 1, 2). The proventriculus opens and closes rhythmically during feeding, but otherwise is tightly closed to seal the blood meal in the midgut. The interior of the proventriculus is arrayed with stiff spines that are coated with cuticle, the same hard acellular material that makes up the exoskeleton.
Y. pestis infection of the proventriculus depends on its ability to grow in the form of a biofilm on the surface of the proventricular spines [14,15]. As the biofilm grows and consolidates, it can impede and eventually completely block normal blood feeding, a process that typically requires one to two weeks after the infectious blood meal. Complete blockage of the proventriculus results in gradual starvation, persistent feeding attempts, and eventual death of the flea; but infected, unblocked fleas do not experience increased morbidity or mortality. Interference with proventricular valve function by the Y. pestis biofilm (which does not require complete blockage) potentiates regurgitation of Y. pestis from the biofilm into the flea bite site [16]. The polysaccharide extracellular matrix (ECM) of the biofilm is synthesized and exported by the Y. pestis hmsHFRS genes and is similar or identical in composition to the poly-β-1,6-N-acetyl-d-glucosamine ECM of E. coli and staphylococcal biofilms [17,18]. Y. pestis hmsHFRS mutants are able to colonize the flea midgut in the form of multicellular aggregates nearly as well as wild-type bacteria, but are devoid of ECM and associated material and completely unable to colonize the proventriculus (Fig. 2) [12].
A second mechanism of regurgitative transmission can occur the first time a flea feeds again on a naïve animal in the week following an infectious blood meal. The mechanism of this early-phase transmission is not known, but it does not require biofilm formation [19]. We have proposed an ingestion-salivation mechanism for early-phase transmission, similar to the mechanism by which aphids transmit plant viruses [20]. According to this model, a residue of infected blood remains on the interior surface of the flea mouthparts and distal foregut following an infectious blood meal. Upon the next feeding, salivation washes residual bacteria present in the salivary groove into the bite site to effect transmission [21].
Most research on Yersinia-flea interactions has been with the rat flea Xenopsylla cheopis. Proventricular blockage and transmission rates are higher in this species than in others, but the physiological basis for these differences is unknown. Since bacterial adherence to insect cuticle appears to be critical for a transmissible infection, subtle differences in the biophysical properties of the proventricular or mouthpart epicuticle of different flea species could be important. Bacterial adhesion and biofilm formation are highly dependent on surface charge and other properties of the substrate [22].
Molecular mechanisms of Yersinia-flea interactions
Upon entering a flea in a blood meal, Y. pestis experiences a rapid drop in temperature from 37°C to < 26°C. This temperature switch is a major environmental cue that Y. pestis uses to regulate gene expression appropriately in order to adapt to the insect host. Many Y. pestis phenotypes, including biofilm formation, are temperature-dependent [23]. The transcriptomes of Y. pestis in infected fleas and in the lymph node of rats with bubonic plague, as well as the transcriptomes of Y. pestis grown in liquid culture media at 21° and 37°C, have been characterized and compared to identify adaptational gene expression responses associated with successful colonization and blockage of the flea [24,25].
Regulation of biofilm formation in the flea
The bacterial biofilm phenotype, critical to the regurgitative transmission mechanism of Y. pestis, involves a complex, multistage developmental process. Although this process has a common end point– a dense surface-attached multicellular bacterial aggregate embedded in a self-synthesized ECM– the responsible genetic pathways differ among different bacteria and environmental conditions [26]. A few common elements are evident, however. Synthesis and transport of the biofilm ECM is essential, and in many bacteria this is under the control of the bacterial second messenger c-di-GMP [27]. Intracellular levels of c-di-GMP depend on the opposing activities of GGDEF-domain diguanylate cyclase (DGC) enzymes that synthesize c-di-GMP and EAL-domain phosphodiesterase (PDE) enzymes that degrade it. Y. pestis has two functional DGC genes, hmsT and y3730; and a single functional PDE gene, hmsP [28,29]. Interestingly, the Y3730 DGC has the major role in biofilm formation in the flea but virtually no role in biofilm formation in vitro, which depends almost entirely on HmsT [29]. The two DGC genes are transcribed equally in both environments, suggesting that Y3730 activity is specifically modulated by an environmental signal detected only in the flea. Y3730, but not HmsT, has a molecular architecture common to many signal transduction proteins in which a HAMP signal converter domain separates an extracellular sensor domain and a cytoplasmic output domain. This pattern suggests that a flea-specific environmental signal sensed by Y3730 is transduced via the HAMP signaling domain to activate the GGDEF catalytic domain, resulting in c-di-GMP production and the appropriate physiological response– biofilm formation [29].
The genetic pathways and molecular mechanisms leading from c-di-GMP to ECM synthesis and biofilm formation in Y. pestis have yet to be delineated. Like the c-di-GMP metabolizing genes, expression of the Y. pestis hmsHFRS operon responsible for the synthesis and transport of the biofilm ECM is not significantly upregulated in the flea or during growth at low temperatures [24]; temperature-dependent ECM production is controlled post-transcriptionally [30]. The phosphoheptose isomerase gene gmhA, required for synthesis of the inner core of the bacterial lipopolysaccharide (LPS), is also required for biofilm formation and proventricular blockage in the flea, but is not required for midgut colonization [31] and is not differentially expressed in the flea gut environment [24].
Physiologic adaptations to the flea gut environment
In the flea gut, many Y. pestis genes involved in the uptake and catabolism of dipeptides, oligopeptides, and the L-glutamate group of amino acids (Gln, His, Arg, Pro) are significantly upregulated. In contrast, genes for hexose carbohydrate uptake, glycolysis, and fermentation are differentially underexpressed in the flea compared to the other environments. This would seem to match the available nutrients (mammalian blood and its digestion byproducts), which contains comparatively little carbohydrate. Blood also contains substantial lipid, but Y. pestis fatty acid uptake or catabolism genes were not upregulated in the flea. Thus, Y. pestis appears to rely on oligopeptides and select amino acids as its major carbon and energy source during persistent infection of the flea gut.
LPS, the major component of the Gram-negative outer membrane, also varies with the temperature shift that accompanies infection of the flea gut. At 37°C, the lipid A moiety of Y. pestis LPS is tetra-acylated whereas at < 26°C hexa-acylated lipid A predominates [8,32]. However, a Y. pestis mutant able to synthesize only tetra-acylated lipid A was able to infect and block fleas normally [33]. Exposure to the flea antibacterial response may induce expression in Y. pestis of genes that protect against both the insect and the mammalian innate immune response. For example, the Y. pestis PhoP-PhoQ gene regulatory system, which modifies LPS and other outer membrane components to confer protection against cationic antimicrobial peptides, is upregulated in the flea [24].
Insecticidal-like toxin complex (Tc) genes of Yersinia
Most Yersinia species encode homologs of the toxin complex (Tc) family of insect toxins of Photorhabdus and other bacteria that are associated with insects and other invertebrates. The Y. pestis Tc genes are strikingly upregulated during infection of the flea [24,34], but their role, if any, in the Yersinia-flea interaction is not obvious. Y. pestis Tc mutants show no defect in their ability to infect and block fleas. Although the Tc proteins of several Yersinia species have some oral toxicity to Manduca sexta larvae, there is no evidence that they are orally toxic to fleas [34-36]. The recently described Yersinia entomophaga, the first true insect pathogen in the genus, encodes a more complex array of potent insecticidal Tc toxins [37].
Evolution of flea-borne transmission in Yersinia
Comparing the interactions of Y. pestis and Y. pseudotuberculosis with fleas identifies some of the important adaptive changes that were necessary. To begin with, Y. pseudotuberculosis, unlike Y. pestis, is orally toxic to fleas– when fleas take a blood meal containing Y. pseudotuberculosis they experience acute diarrhea and significant mortality [34]. The responsible enterotoxic factor(s) have not been identified but the Tc insecticidal-like proteins have been ruled out. Toxicity subsides within 24 hours after the infectious blood meal, and most serotypes of Y. pseudotuberculosis are able to persist in the flea gut, although their numbers decrease with time unless they are transformed with the Y. pestis ymt gene [13,38]. Notably, Y. pseudotuberculosis does not form biofilm in the flea or colonize the proventriculus, but exists in the flea gut in a planktonic, single-celled state [38].
Some of the key genetic changes in Y. pestis that enabled flea-borne transmission have now been identified, and both gene gain and gene loss were important (Table). Y. pestis acquired two new plasmids since it diverged from Y. pseudotuberculosis, and each contains a gene important for transmission. One (ymt) is highly similar to the Photorhabdus luminscens ymt gene, suggesting that it was acquired by horizontal gene transfer from that species recently. A plasminogen activator gene (pla) on the smaller Y. pestis-specific plasmid is irrelevant to the flea interaction but is highly expressed in the flea and greatly enhances Y. pestis dissemination from the flea bite site following transmission [39,40]. Pla is an omptin-family outer surface protease that is catalytically active at 37°C but not at the flea temperature. The combined activities of Pla result in fibrinolysis and damage to the basement membrane and extracellular matrix of host tissues at the site of infection, disrupting normal barrier functions and enabling systemic spread [41].
Table 1.
Yersinia genes important for interaction with fleas and for transmission
| Role | Genea | Functionb | Location in the genome |
Present in : |
Differential expression in the fleac |
|
|---|---|---|---|---|---|---|
| Y. pestis | Y. pseudotuberculosis | |||||
| Survival in the flea midgut | ymt | phospholipase D | Y. pestis-specific plasmid (pFra) |
+ | − | yes |
| Proventricular biofilm formation/regulation |
hmsHFRS | ECM synthesis/transport |
chromosome | + | + | no |
| hmsT, y3730 | DGC; c-di-GMP synthesis |
chromosome | + | + | no | |
| hmsP | PDE; c-di-GMP degradation |
chromosome | + | + | no | |
| rcsA | transcriptional regulation |
chromosome | pseudogene | + | unknown | |
| nghA | glycosyl hydrolase; ECM degradation |
chromosome | pseudogene | + | unknown | |
| Dissemination from the flea bite site |
pla | plasminogen activator (surface protease) |
Y. pestis-specific plasmid (pPla) |
+ | − | no |
| Other | Toxin complex (Tc) genes |
antiphagocytic | chromosome | + | + | yes |
| unidentified | flea enterotoxin | unknown | − | + | unknown | |
Gene differences between Y. pseudotuberculosis and Y. pestis are indicated in bold type.
ECM, extracellular matrix of biofilm ; DGC, diguanylate cyclase ; PDE, phosphodiesterase
See [24] for details
Although Y. pseudotuberculosis forms biofilm in some environments, it does not do so in fleas [38]. The Y. pestis orthologs of >200 Y. pseudotuberculosis genes are nonfunctional pseudogenes [42,43], and selective loss of gene function has been important in extending biofilm-forming ability to the flea gut environment. For example, rcsA, a negative regulator of biofilms, is a functional gene in Y. pseudotuberculosis but not in Y. pestis. Replacement of the Y. pestis pseudogene with the functional Y. pseudotuberculosis version of rcsA results in greatly reduced proventricular biofilm and blockage of fleas [44]. Similarly, pseudogene degradation in Y. pestis of nghA, which in Y. pseudotuberculosis encodes a glycosyl hydrolase that degrades the ECM of Yersinia biofilms, may have led to enhanced stability of Y. pestis biofilms [18]. Two functional PDE genes of Y. pseudotuberculosis are pseudogenes in Y. pestis [28], and it is possible that this gene loss also contributed to biofilm-forming ability in the flea [29].
Conclusions
Like other environmental Yersinia, the Y. pseudotuberculosis progenitor of Y. pestis would have been subject to ingestion by invertebrates. Based on flea infections, the insect digestive tract does not appear to be a favored niche for Y. pseudotuberculosis– it induces diarrhea and does not form biofilm in fleas, both of which lead to rapid elimination from the insect back into the environment. In contrast, the new Y. pestis life cycle depended on establishing a stable infection in the flea. This ecological about-face evolved recently and required only a few key genetic changes, including some that served to extend the preexisting biofilm-forming ability to the flea gut. The Y. pestis ancestor probably had the advantage of already being constitutively resistant to insect antimicrobial defenses because the genus Yersinia appears to be more closely related to insect- and invertebrate-associated genera of the Enterobacteriaceae (Photorhabdus, Serratia, Sodalis) than to vertebrate-associated genera (Escherichia, Salmonella) [45,46]. Perhaps reflective of an as yet brief coevolutionary history of Y. pestis-flea interactions, vector infectivity and transmission efficiency are low compared to other arthropod-borne disease systems [11]. The rather poor vector competence of fleas would have imposed positive selection pressure for Y. pestis strains able to produce severe septicemia in the mammal. Thus, the evolution of flea-borne transmission and increased virulence of Y. pestis probably went hand-in-hand and were were mutually reinforcing.
Highlights.
Flea-borne transmission of Y. pestis is a recent evolutionary phenomenon
Y. pestis biofilm formation in the flea gut is important for transmission
Y. pestis biofilm requires hms genes and enzymes controlling c-di-GMP levels
Y. pestis regulates its gene expression to specifically adapt to the flea vector
Gene gain and gene loss contributed to the evolution of flea-borne transmission
Acknowledgements
This work was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci U S A. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard CB, Butler JF, Hall DW. Prevalence and biology of endosymbionts of fleas (Siphonaptera: Pulicidae) from dogs and cats in Alachua County. Florida. JMed Entomol. 1990;27:1050–1061. doi: 10.1093/jmedent/27.6.1050. [DOI] [PubMed] [Google Scholar]
- 4.Pornwiroon W, Kearney MT, Husseneder C, Foil LD, Macaluso KR. Comparative microbiota of Rickettsia felis-uninfected and -infected colonized cat fleas, Ctenocephalides felis. ISME J. 2007;1:394–402. doi: 10.1038/ismej.2007.38. [DOI] [PubMed] [Google Scholar]
- 5.Jones RT, McCormick KF, Martin AP. Bacterial communitites of Bartonella-positive fleas: diversity and community assembly patterns. Appl. Environ. Microbiol. 2008;74:1667–1670. doi: 10.1128/AEM.02090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson DL, Anderson NE, Cromar LM, Jolley A. Bacterial communities associated with flea vectors of plague. J Med Entomol. 2009;46:1532–1536. doi: 10.1603/033.046.0642. [DOI] [PubMed] [Google Scholar]
- 7.Bengoechea JA, Lindner B, Seydel U, Diaz R, Moriyon I. Yersinia pseudotuberculosis and Yersinia pestis are more resistant to bactericidal cationic peptides than Yersinia enterocolitica. Microbiology. 1998;144(Pt 6):1509–1515. doi: 10.1099/00221287-144-6-1509. [DOI] [PubMed] [Google Scholar]
- 8.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 9.Anisimov AP, Dentovskaya SV, Titareva GM, Bakhteeva IV, Shaikhutdinova RZ, Balakhonov SV, Lindner B, Kocharova NA, Senchenkova SN, Holst O, et al. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect Immun. 2005;73:7324–7331. doi: 10.1128/IAI.73.11.7324-7331.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelthaler DM, Hinnebusch BJ, Rittner CM, Gage KL. Quantitative competitive PCR as a technique for exploring flea-Yersina pestis dynamics. Am. J. Trop. Med. Hyg. 2000;62:552–560. doi: 10.4269/ajtmh.2000.62.552. [DOI] [PubMed] [Google Scholar]
- 11.Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J. Inf. Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- 12.Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- 13.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg Å . Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- 14.Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- 15.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Inf. Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 16.Bacot AW. Further notes on the mechanism of the transmission of plague by fleas. J. Hygiene Plague. 1915;14(Suppl. 4):774–776. [PMC free article] [PubMed] [Google Scholar]
- 17.Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10:1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 18.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J Bacteriol. 2008;190:8163–8170. doi: 10.1128/JB.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Vetter SM, Eisen RJ, Schotthoefer AM, Montenieri JA, Holmes JL, Bobrov AG, Bearden SW, Perry RD, Gage KL. Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology. 2010;156:2216–2225. doi: 10.1099/mic.0.037952-0. This study showed that early-phase transmission of Y. pestis by fleas occurs by a different mechanism than the proventricular biofilm-dependent regurgitative transmission mechanism.
- 20.Martin B, Collar JL, Tjallingii WF, Fereres A. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 1997;78:2701–2705. doi: 10.1099/0022-1317-78-10-2701. [DOI] [PubMed] [Google Scholar]
- 21.Hinnebusch BJ. Biofilm-dependent and biofilm-independent mechanisms of transmission of Yersinia pestis by fleas. Adv. Exp. Med. Biol. 2012 doi: 10.1007/978-1-4614-3561-7_30. in press. [DOI] [PubMed] [Google Scholar]
- 22.Beloin C, Da Re S, Ghigo J-M. Colonization of abiotic surfaces. In: Böck A, Curtis R III, Kaper JB, Neidhardt FC, Nyström K, Rudd E, Squires CL, editors. EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. Vol. 2005. ASM Press; Washington, D.C.: 2005. [Google Scholar]
- 23.Perry RD, Fetherston JD. Yersinia pestis-etiologic agent of plague. Clin. Microbiol. Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **24.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathogens. 2010;6:e10000783. doi: 10.1371/journal.ppat.1000783. The in vivo transcriptome of Y. pestis in infected fleas was characterized to gain insight into adaptation to life in the insect vector. In addition to genes associated with physiological adaptation to the flea gut, a number of Y. pestis virulence factors important for mammalian infection were upregulated in the flea, suggesting that infection of the insect host primes Y. pestis for successful infection of the mammal.
- 25.Hinnebusch BJ, Sebbane F, Vadyvaloo V. Transcriptional profiling of the Yersinia pestis life cycle. In: Carniel E, Hinnebusch BJ, editors. Yersinia: Systems Biology and Control. Horizon Scientific Press; 2012. [Google Scholar]
- 26.Beloin C, Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. doi: 10.1016/j.tim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Hennge R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- *28.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 2011;79:533–551. doi: 10.1111/j.1365-2958.2010.07470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Sun Y-C, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One. 2011;6:e19267. doi: 10.1371/journal.pone.0019267. These two studies presented genetic and biochemical proof that Y. pestis encodes only three functional c-di-GMP metabolizing enzymes. One of the two enzymes capable of c-di-GMP synthesis primarily affects biofilm formation in vitro, while the other primarily affects biofilm formation in the flea.
- 30.Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 2004;186:1638–1647. doi: 10.1128/JB.186.6.1638-1647.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darby C, Ananth SL, Tan L, Hinnebusch BJ. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 2005;73:7236–7242. doi: 10.1128/IAI.73.11.7236-7242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 2002;70:4092–4098. doi: 10.1128/IAI.70.8.4092-4098.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebeil R, Ernst RK, Jarrett CO, Adams KN, Miller SI, Hinnebusch BJ. Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J. Bacteriol. 2006;188:1381–1388. doi: 10.1128/JB.188.4.1381-1388.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, ffrench-Constant RH, Hinnebusch BJ. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell. Microbiol. 2007;9:2658–2666. doi: 10.1111/j.1462-5822.2007.00986.x. [DOI] [PubMed] [Google Scholar]
- *35.Fuchs TM, Bresolin G, Marcinowski L, Schachtner J, Scherer S. Insecticidal genes of Yersinia spp.: taxonomical distribution, contribution to toxicity towards Manduca sexta and Galleria mellonella, and evolution. BMC Microbiol. 2008;8:214. doi: 10.1186/1471-2180-8-214. A comparative genetic analysis of the insecticidal-like Toxin complex (Tc) loci of thirteen Yersinia species and an evaluation of the toxicity of Yersinia Tc proteins to lepidopteran larvae.
- 36.Pinheiro VB, Ellar DJ. Expression and insecticidal activity of Yersinia pseudotuberculosis and Photorhabdus luminescens toxin complex proteins. Cell. Microbiol. 2007;9:2372–2380. doi: 10.1111/j.1462-5822.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- 37.Hurst MR, Jones SA, Binglin T, Harper LA, Jackson TA, Glare TR. The main virulence determinant of Yersinia entomophaga MH96 is a broad-host-range toxin complex active against insects. J Bacteriol. 2011;193:1966–1980. doi: 10.1128/JB.01044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J. Bacteriol. 2006;188:1113–1119. doi: 10.1128/JB.188.3.1113-1119.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinnebusch BJ, Fischer ER, Schwan TG. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Inf. Dis. 1998;178:1406–1415. doi: 10.1086/314456. [DOI] [PubMed] [Google Scholar]
- 40.Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc. Natl. Acad. Sci. USA. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lähteenmäki K, Edelman S, Korhonen TK. Bacterial metastasis: the host plasminogen system in bacterial invasion. Trends Microbiol. 2005;13:79–85. doi: 10.1016/j.tim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MTG, Prentice MB, Sebhaihia M, James KD, Churcher C, Mungall KL, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 43.Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Sun Y-C, Hinnebusch BJ, Darby C. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. USA. 2008;105:8097–8101. doi: 10.1073/pnas.0803525105. This study showed that gene loss during the evolution of Y. pestis resulted in increased ability to produce a transmissible infection in the flea vector, and hypothesized that such gene loss was subject to positive Darwinian selection rather than neutral selection.
- 45.Qi M, Sun F-J, Caetano-Anollés G, Zhao Y. Comparative genomic and phylogenetic analyses reveal the evolution of the core two-component signal transduction systems in Enterobacteria. J. Mol. Evol. 2010;70:167–180. doi: 10.1007/s00239-009-9318-2. [DOI] [PubMed] [Google Scholar]
- 46.Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, Dickerman AW. Phylogeny of gammaproteobacteria. J. Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]