Abstract
Reporter bacteria are beneficial for the rapid and sensitive screening of cultures producing peptide antibiotics, which can be an addition or alternative to the established antibiotics. This study was carried out to validate the usability of specific reporter strains for the target mediated identification of antibiotics produced by native Bacillus spp. isolated from different food sources. During preliminary classification, cell wall stress causing Bacillus isolates were screened by using reporter strain Bacillus subtilis BSF2470. The isolates which induced cell wall stress were further characterized for their specific mode of action by using other B. subtilis reporter strains (TMB 488, TMB 299 and TMB 279). The isolate B.licheniformis N12 was found to produce bacitracin confirmed by the response to reporter strain B. subtilis TMB 279 and by putative identification of bacitracin biosynthetic loci. The other isolate B. subtilis EC1 also induced B. subtilis TMB 279, but does not possess the bacitracin gene cluster indicating that it can be a novel, bacitracin like antibiotic. The different but related subsets of peptide antibiotics that bind the pyrophosphate moiety of the lipid carrier of cell wall biosynthesis can be identified using this whole cell based reporter strains.
Keywords: Reporter strains, Mode of action, Bacitracin, Bacillus
Introduction
The emergence of multi-drug resistance in pathogens that are once susceptible to various drugs has promoted the search for novel antibacterial agents of microbial origin which leads to the development in genomics and post genomics technologies [1]. Antimicrobial peptides of Gram-positive bacteria are known to be promising candidates for treatment of diseases caused by several human pathogens including those associated with botulism and listeriosis [2]. A Gram-positive food-grade bacterium, Bacillussubtilis is known to produce over two dozen antibiotics, predominantly peptides that are either ribosomally synthesized and post-translationally modified, or non-ribosomally generated, as well as a couple of non-peptidic compounds such as polyketides, aminosugars and phospho-lipids [3]. Peptide antibiotics such as bacitracin and gramicidin that are synthesized in Bacillus by multienzyme complexes or sequential enzyme reactions have not yet achieved widespread application in the treatment of infectious diseases. However, studies of mersacidin and epidermin, ribosomal synthesized peptides of the lantibiotic class have suggested that they may be at least as effective as some currently used therapeutic agents for the treatment of staphylococcal infections in mice and acne in humans, respectively [4].
Conventional characterization of antibacterial peptides involves routine screening by agar well diffusion assay followed by purification and studying its properties. However, this growth inhibition assays requires high concentration of sample and longer incubation time and moreover these assays does not provide information on the molecular target of inhibitory compounds [5]. High throughput screening (HTS) with target based assays became the major choice for discovering novel drugs due to the progress in laboratory robotics together with the availability of large collections of chemical entities. These assay systems are based on cells that carry reporters such as β-galactosidase or luciferase genes fused to promoters that specifically respond to certain types of antibiotic stress [6, 7]. HTS of antibiotic producing microorganisms is commonly defined as automatic testing of potential candidates at a rate in excess of 10,000 compounds per week. The reporter strains can help in the search for novel antibacterial entities through a rapid, sensitive and cost effective screening method. Other importance of reporter strains is the pathway-based drug-screening applications. For this development, selection of marker genes is important and was achieved solely by analyzing the data generated in a large-scale gene expression profiling project. They can be used as mode of action (MOA) specific whole cell screening assays or as tools to assign a MOA to uncharacterized whole cell-active compounds [8].
Bacterial cell envelope and other essential metabolic pathway are the major targets for numerous antibiotics. The identification of antibiotic affecting cell wall biosynthesis requires efficient screening method that can tentatively identify a group of antibiotics. VanRS based reporter strain was developed in B. subtilis by Ulijasz et al. [9] and is able to differentiate cell wall inhibiting antibiotics. This reporter strain was used later by Pascale et al. [7] for HTS of cell wall inhibitors. Mascher et al. [10] developed a B. subtilis LiaRS (lipid II cycle interfering antibiotic response regulator and sensor) two component system that specifically senses and responds to the cell wall active antibiotics. Burkard and Stein [2] explored LiaRS system for characterizing the response of lantibiotics, subtilin, epidermin, gallidermin and cinnamycin. However, LiaRS do not differentiate further classes of antibiotics that affect other vital steps of peptidoglycan formation. The recently developed PbceA- and PpsdA- based Bacillus reporter strains are sensitive and more specific biosensor for lipid II binding peptide antibiotics, cationic lipid II binding lantibiotics and bacitracin, respectively [11]. There are no reports on the evaluation of these reporter strains for the characterization of antibiotics produced by natural isolates.
The aim of this study was to explore the newly developed reporter bacteria to characterize the MOA of the antibiotic of native Bacillus isolates. In this study, we report rapid and effective identification of the bacitracin and bacitracin-like antibacterial compound producing native Bacillus isolates using bacitracin specific biosensor.
Materials and Methods
Chemicals, Reagents and Bacteriological Media
The media used throughout the work, Luria–Bertani (LB) agar and broth was purchased from Hi-media (Mumbai, India). X-gal, ONPG, lysozyme, agarose, oligonucleotide primers, PCR reagents and bacteriocins such as bacitracin and nisin were obtained from Sigma (USA).
Bacterial Cultures and Maintenance
Native Bacillus cultures isolated previously in our laboratory from different food sources, exhibiting inhibitory activity against the indicator, Micrococcus luteus ATCC9341 were used for this experiment. The reporter strains used includes, B. subtilis BSF2470 (CU1065 lial::pMUTIN) [10], B. subtilis TMB 488, B. subtilis TMB 299 (W168 amyE::pER605), B. subtilis TMB 279 (W168 amyE::pER603) [12]. The known subtilin producer, B. subtilis ATCC6633 and B. subtilis W168 was used as positive and negative controls, respectively. The cultures used in this study were stored at −20°C in LB broth containing 16% (v/v) glycerol. All the cultures were propagated aerobically twice in fresh LB broth for 24 h at 37°C and 150 rpm, before use.
Chromogenic Plate Assay
Chromogenic plate assay for native isolates was carried out against the reporter strain B. subtilis BSF2470 using the method as described by Burkard et al. [13]. The overnight grown (16 ± 2 h) native Bacillus isolates were streaked against reporter strain B. subtilis BSF 2470 on the LB agar plate supplemented with 50 μg/ml of X-gal. The plates were incubated for 24–36 h at room temperature until a blue coloration is observed at the interjunction due to the induction of lacZ resulting in the production of β-galactosidase.
β-gal Induction Assay
For β-galactosidase activity, Bacillus isolates were grown for 24 h in LB broth at 37°C under shaking condition (150 rpm). The culture filtrate was collected by centrifugation at 10,000 rpm for 10 min and kept at −20°C, until further use. Freshly grown reporter strains of B. subtilis (TMB 488, TMB 299, TMB 279 and TMB 134) were inoculated into 50 ml of LB broth and allowed it to grow as conditions described above, until it reached an OD600 of ~0.5. Different concentrations (0, 10, and 30% v/v) of previously collected culture filtrate of the isolates was added in 2 ml each of the reporter strains and incubated for 30 min at 37°C. Cell pellet was collected after incubation and resuspended in 1 ml of working buffer (20 mM β-mercaptoethanol, 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCL and 1 mM MgSO4, pH 7.0) and assayed for β-galactosidase activity [14]. Each value represents the mean value ± SEM from three measurements. Bacitracin and nisin (1 mg/ml) were used as positive and negative control, respectively for B. subtilis TMB 279 and vice versa for B. subtilis TMB 299.
DNA isolation, Identification and Sequencing of Bacitracin Gene
Total genomic DNA of the Bacillus isolates was extracted from overnight grown cultures by phenol–chloroform method as described by Mora et al. [15]. The quality of isolated DNA was checked by agarose gel electrophoresis and used as a template for PCR analysis. The 23S rRNA gene of the isolates were amplified using the forward primer 5′TACCGAATTCAGTCAAACTCC3′ and reverse primer 5′GGCGTCCTACTCTCACAGG3′ at an annealing temperature of 55°C. PCR amplification of bacitracin biosynthetic gene was carried out using the primer as listed in Table 1. The following thermo cycling conditions for amplification of bacitracin biosynthetic gene were used, denaturation at 95°C for 5 min and 35 cycles at 95°C for 45 s, 50°C for 45 s, 72°C for 3 min, followed by one cycle of elongation for 15 min at 72°C. PCR was performed in the Thermocycler Gene AmpPCR system 9700 (Applied Biosystem, USA). The amplified product was sequenced (Eurofins MWG biotech, Germany) and sequence homology was identified using BLASTN search tool.
Table 1.
Characterization of bacitracin biosynthetic loci
| Gene/s | Sequence | Expected size | Bacillus sp. N12 | Bacillus sp. Ec1 | ||
|---|---|---|---|---|---|---|
| PCR product (kb) | Homology (%) | PCR product (kb) | Homology (%) | |||
| bacA | F-GCTAAACATTCATTAGAAAATGG | 1.5 | 1.5 | 99, Bacitracin synthase I | – | – |
| R-GAGCTTTTCTGTCGACTTTCC | ||||||
| bacB | F-TGACTTAAAACAGCAAATCAAGC | 1.5 | 1.5 | 97, Bacitracin synthase II | 1.0 | 98 (putative ABC transporter) |
| R-AGCTCTCGATATGTCAATTGC | ||||||
| bacC | F-GGAGAGCTGTATATCAGCGG | 1.7 | 1.7 | 97, BcrC | – | – |
| R-ACAGCCTTAACGCATCTTCTCC | ||||||
| bacRS | F-TCCGATCCTGATGCTGACC | 1.4 | 1.4 | 97, BacS | 1.2 | 98 (Aldehyde dehydrogenase) |
| R-TAGGCAGCGAAAAACCAAACG | ||||||
Results and Discussion
Screening of Bacillus Cultures Inducing LiaRS
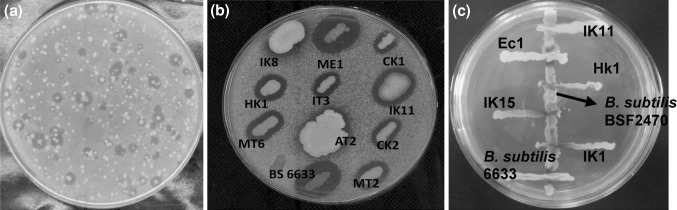
Genetically engineered recombinant reporter bacteria facilitate the easy screening of pathway based novel antibacterial entities with the presence of intact cells [7]. Among 78 zone producing Bacillus isolates tested against M. luteus (Fig. 1a, b), the cultures which produced cell wall stress inducing antimicrobial compounds were selected based on the cell biosensor, B. subtilis BSF2470 [10]. Result indicated that only 30% of the tested isolates produce antibiotics with MOA on cell wall (Fig. 1b). The MOA of the isolates which didn’t induce β-gal in B. subtilis BSF2470 could be other than cell wall for which other studies based on specific reporter strains were required. The isolates showing positive response to B. subtilis BSF2470 were selected to further differentiate the MOA of their antibiotics using other specific reporter strains. For the detection of lantibiotic subtilin, Burkard et al. [13] used the reporter strain B. subtilis ΔspaS amyE::PspaS–lacZ and found it as sensitive and unsusceptible method to identify subtilin producing B. subtilis wild type strains from both culture collections and soil samples. Similarly, Pascale et al. [7] used B. subtilis strain expressing the Enterococcus faecium vanRS genes and vanH–lacZ fusion genes and producing β-galactosidase activity in the presence of cell wall inhibitors to validate reporter gene based novel antibiotic discovery.
Fig. 1.
Representative plates showing initial screening. a Zone of growth inhibition of M. luteus by spore forming Bacillus from fermented potato, b Inhibitory activity of the isolates against Micrococcus luteus ATCC9341, c plate showing chromogenic assay
Specific Mode of Action by β-gal Activity
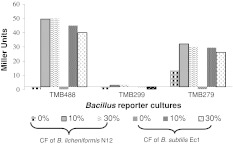
Bacitracin inhibits cell wall biosynthesis by binding undecaprenyl pyrophosphate and inhibiting its dephosphorylation, thereby blocking its recycling and cell wall biosynthesis [12]. PbceA (TMB 299) and PpsdA (TMB 299) based reporter strains are sensitive and more specific biosensor for lipid II-binding peptide antibiotics than any of the established cell wall antibiotic biosensors currently available, most notably, the PypuA and PliaI derived reporter strains [10, 16]. Thus, the Bacillus isolates which exhibited positive results against B. subtilis BSF 2470 were checked for induction of β-gal activity against reporter strain TMB 488, which differentiates cell-wall acting lantibiotics from other groups of antibiotics. Six isolates showing positive response included Pk5, B9, Me1, N12, Ec1 and C2. These isolates were tentatively identified: Pk5 as B. megaterium, B9 and Ec1 as B. subtilis, C2 as B. cereus, and Me1 and N12 as B. licheniformis. Among these cultures, only B. licheniformis N12 and B. subtilis Ec1 exhibited higher β-gal activity of about 40–50 Miller Units (Fig. 2). PpsdA is induced by lipid II binding lantibiotics such as nisin, subtilin, actagardine and gallidermin, whereas, PbecA can only be induced by lipid II lantibiotic, namely, bacitracin [11]. The six isolates were also checked with another set of reporter strains TMB 134 (PbcrC–lacZ), TMB 279 (PbecA–lacZ), TMB 299 (PpsdA–lacZ). Among them, B. megaterium Pk5 and B. cereus C2 responded to TMB 134, whereas TMB 299 got induced by antibiotic of B. licheniformis Me1 (Data not shown). The other three isolates (B. subtilis B9, B. subtilis Ec1, and B. licheniformis N12) were induced by TMB 279, a bacitracin specific reporter strain (Fig. 2). The β-gal activity of B. subtilis B9 was less than other two and thus, it was eliminated from further studies.
Fig. 2.
β-gal activity of cultures aB. licheniformis N12 and bB. subtilis Ec1 against bacitracin specific reporter strain
Characterization of Bacitracin Producers
The antibiotics produced by Bacillus cultures viz. B. licheniformis N12 and B. subtilis Ec1 was sensed by reporter strain TMB 279 were subsequently analyzed for the conserved bacitracin biosynthetic loci using gene specific primers. Nucleotide sequence of the amplified gene was determined. Homology search using BLAST program indicated that in B. licheniformis N12 culture, bceA has 99% homology, bceB—97% and bceRS—47% (Table 1). Even though B. subtilis Ec1 showed amplification with becB and becRS primers, the nucleotide sequence did not match with the bacitracin operon suggesting B. subtilis Ec1 lacking bacitracin biosynthetic gene cluster. However, it might be producing bacitracin-like antibiotics, since it was found to induce bacitracin specific reporter bacteria. Similarly in a previous study of subtilin auto-induction bioassay, employing sensor strain B. subtilis ATCC6633 ΔspaS PspaS- lacZ, identified the novel subtilin-like lantibiotic entianin from B. subtilis subsp. spizizenii DSM15029, which differs from subtilin with few amino acids [17] and other subtilin-isoforms such as ericin [13].
Thus we conclude this study that, application of HTS using cell biosensors facilitates the rapid characterization and identification of novel antibacterial compounds produced by natural isolates incl. Bacillus strains, as used in this study. With the help of specific MOA sensing bacterial cultures, we specifically identified a new bacitracin producer (B. licheniformis N12) and bacitracin like antibiotic producer (B. subtilis Ec1) that indeed induces a bacitracin specific promoter.
Further characterization of Ec1 culture is underway to investigate its MOA and biosynthetic loci associated with production of novel antibiotics.
Acknowledgments
The authors wish to acknowledge The Director, CFTRI, Mysore and Head, Food Microbiology Department, CFTRI for providing the facilities. Part of work has been carried out under CSIR-DAAD scientific exchange program 2009 (Courtesy Prof. Thorsten Mascher) at KIT, Karlsruhe, Germany. NV acknowledges CSIR for the fellowship.
References
- 1.Ji Y. The role of genomics in the discovery of novel targets for antibiotic therapy. Pharmacogenomics. 2002;3:315–323. doi: 10.1517/14622416.3.3.315. [DOI] [PubMed] [Google Scholar]
- 2.Burkard M, Stein T. Microtiter plate bioassay to monitor the interference of antibiotics with lipid II cycle essential for peptidoglycan synthesis. J Microbiol Methods. 2008;75:70–74. doi: 10.1016/j.mimet.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 4.Jack RW, Tagg JR, Ray B. Bacteriocins of Gram-positive bacteria. Microbiol Rev. 1995;59:171–200. doi: 10.1128/mr.59.2.171-200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro E, Baneyx F. Stress based identification and classification of antibacterial agents: second generation Escherichia coli reporter strains and optimization of detection. Antimicrob Agents Chemother. 2002;46:2490–2497. doi: 10.1128/AAC.46.8.2490-2497.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi AA, Baneyx F. Stress responses as a tool to detect and characterize the mode of action of antibacterial agents. Appl Environ Microbiol. 1999;65:5023–5027. doi: 10.1128/aem.65.11.5023-5027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascale GD, Grigoriadou C, Losi D, Ciciliato I, Sosio M, Donadio S. Validation for high throughput screening of a VanRS based reporter gene assay for bacterial cell wall inhibitors. J Appl Microbiol. 2007;103:133–140. doi: 10.1111/j.1365-2672.2006.03231.x. [DOI] [PubMed] [Google Scholar]
- 8.Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, Freiberg C, Ziegelbauer K, Rock CO, Ivanov I, Loferer H. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob Agents Chemother. 2004;48:2838–2844. doi: 10.1128/AAC.48.8.2838-2844.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulijasz AT, Grenader A, Weisblum B. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J Bacteriol. 1996;178:6305–6309. doi: 10.1128/jb.178.21.6305-6309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mascher T, Zimmer SL, Smith TA, Helmann JD. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother. 2004;48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staron A, Finkeisen DE, Mascher T. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob Agents Chemother. 2011;55:515–525. doi: 10.1128/AAC.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rietkotter E, Hoyer D, Mascher T. Bacitracin sensing in Bacillus subtilis. Mol Microbiol. 2008;68:768–785. doi: 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- 13.Burkard M, Entian KD, Stein T. Development and application of a microtiter plate-based autoinduction bioassay for detection of the lantibiotic subtilin. J Microbiol Method. 2007;70:179–185. doi: 10.1016/j.mimet.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
- 15.Mora D, Fortina MG, Parini C, Daffonchio D, Manachini PL. Genomic subpopulations within the species Pediococcus acidilactici detected by multilocus typing analysis: relationships between pediocin AcH/PA-1 producing and non-producing strains. Microbiology. 2000;146:2027–2038. doi: 10.1099/00221287-146-8-2027. [DOI] [PubMed] [Google Scholar]
- 16.Urban A, Eckermann S, Fast B, Metzger S, Gehling M, Zeigelbauer K, Weigmann RH, Freiberg C. Novel whole-cell antibiotic biosensors for compound discovery. Appl Environ Microbiol. 2007;73:6436–6443. doi: 10.1128/AEM.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs SW, Jaskolla TW, Bochmann S, Koetter P, Wichelhaus T, Karas M, Stein T, Entian KD. Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM15029T with high antimicrobial activity. Appl Environ Microbiol. 2011;77:698–1707. doi: 10.1128/AEM.01962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]