Abstract
Trichoderma, soil-borne filamentous fungi, are capable of parasitising several plant pathogenic fungi. Twelve isolates of Trichoderma spp. isolated from different locations of South Andaman were characterized for their cultural, morphological and antagonistic activity against soil borne and foliar borne pathogens. The sequencing of these isolates showed seven different species. The isolates revealed differential reaction patterns against the test pathogens viz., Sclerotium rolfsii, Colletotrichum gloeosporioides and C. capsici. However, the isolates, TND1, TWN1, TWC1, TGD1 and TSD1 were most effective in percentage inhibition of mycelial growth of test pathogens. Significant chitinase and β-1,3-glucanase activities of all Trichoderma isolates has been recorded in growth medium. T. viride was found with highest chitinase whereas T. harzianum was recorded with highest β-1,3-glucanase activities.
Keywords: Trichoderma, Spice, Biocontrol, Andaman and Nicobar Islands
Introduction
The anamorphic fungal genus Trichoderma (Hypocreales, Ascomycota) is a cosmopolitan soil-borne fungi frequently found on decaying wood [1, 2] of which some are economically important producers of industrial enzymes (Trichodermaressei) [3], antibiotics [4] and have been used as biocontrol (i.e., T. harzianum, T. atroviride and T. asperellum) agents against plant pathogens [5]. Trichoderma spp. are among the most frequently isolated soil fungi and present in plant root systems [6]. However, there is still considerable interest in finding more efficient mycoparasitic fungi especially within Trichoderma spp., which differ considerably with respect to their biocontrol effectiveness. It is important to isolate Trichoderma spp. having potentially higher antagonistic efficiency by the selection of isolates with high potential to secret extra cellular lytic enzymes chitinase and β-1,3-glucanase. The lytic enzymes break down cell wall polysaccharides into short oligomers and by this way facilitate the hyperparasite to penetrate into the cytoplasm of the target fungi [7]. The aim of this study was screening of Trichoderma spp. for their antagonistic ability by dual cultures as well as their capability of producing lytic enzymes against the test pathogens.
Materials and Methods
Soil Samples and Isolation
Soil samples were collected from different ecological habitat of spice crops of district, South Andaman, Andaman and Nicobar Islands (India) for the isolation of Trichoderma spp. (Table 1). Samples were brought to laboratory and stored at 4°C until used. Five-fold serial dilutions of each soil samples were prepared in sterilized distilled water and 0.5 ml diluted sample was poured on the surface of Trichoderma Specific Medium (TSM) [8]. Plates were incubated at 28 ± 2°C for 96 h. Morphologically different colonies appearing on the plates were purified in the Potato Dextrose Agar (PDA) (HiMedia, India). The purified isolates were preserved at 4°C and used during the course of study.
Table 1.
Trichoderma isolates studied, their identification and host
| S. No. | Isolate designationa | Crop nameb | Place of collection | Species identified | Accession number |
|---|---|---|---|---|---|
| 1 | TSD1 | Cinnamon | Sippighat | Trichoderma inhamatum | GQ426033 |
| 2 | TWN1 | Nutmeg | Wandoor | T. harzianum | GQ426034 |
| 3 | TGN1 | Nutmeg | Guptapara | T. harzianum | GQ426035 |
| 4 | TWD1 | Cinnamon | Wandoor | T. asperellum | GQ426036 |
| 5 | TND1 | Cinnamon | New Mangulton | T. erinaceum | GQ426037 |
| 6 | TJP1 | Pepper | Jirkatang | T. harzianum | GQ426038 |
| 7 | TWC2 | Clove | Wandoor | T. harzianum | GQ426039 |
| 8 | TMC1 | Clove | Manjery | T. longibrachiatum | GQ426040 |
| 9 | TWP1 | Pepper | Wandoor | Trichoderma sp. | GQ426041 |
| 10 | TCC1 | Cinnamon | Calicut | T. ovalisporum | GQ426042 |
| 11 | TGD1 | Cinnamon | Guptapara | T. viride | GQ426043 |
| 12 | TBC1 | Clove | Brichgunj | T. brevicompactum | GQ426044 |
aCoding system of isolates TTrichoderma, S place of collection and D crop name
bRhizosphere soil of various spice crops
Phenotype Characters of the Trichoderma Isolates
The morphological and cultural characteristics of 12 isolates of Trichoderma were studied in four different media viz., OMA, CMD, PDA and TSM following the protocol of Samules et al. [9]. Mycelial discs (6 mm) of young growing culture of respective isolates of Trichoderma was inoculated in the periphery of the Petri plates containing above said media and incubated at 28 ± 2°C for one week. Colony radius was measured at 24, 48 and 72 h. each growth rate experiment was repeated three times in triplicate and the results were averaged for each isolate. Additional characters include presence of pigments, green conidia, odor and colony appearance were also noted.
Morphological observations were recorded from cultures grown on PDA plates. The following characters were measured; Phialide width at the widest pint, phialide length, conidium length and width and presence of chlamydospores. Each character was measured from water after initial wetting in 3% KOH for each isolates.
Fungal Growth Conditions and DNA Extraction
Cultures were maintained on PDA at 25°C was grown in potato dextrose broth for 3 days. Mycelial mat was collected on filter paper, washed with distilled water for 2–3 times, frozen and were used for DNA extraction. Genomic DNA was extracted using method of Raeder and Broda [10]. DNA resuspended in 50 μl of TE buffer and quantified by use of ethidium bromide fluorescence.
PCR Amplification and Sequencing of Amplification Products
Primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) described by White et al. [11] were used to amplify a fragment of rDNA including ITS1 and ITS2 and the 5.8S rDNA gene. PCR amplifications were performed in a total volume of 50 μl by mixing 20 ng of the template DNA with 2.5 mM concentrations of each deoxynucleotide triphosphate, 1 μm concentrations of each primer and 3 U of Taq DNA polymerase in 10× Taq buffer A (GeNeI). These reactions were subjected to initial denaturations of 1 min at 95°C, followed by 35 cycles of 1 min at 95°C, 30 s at 55°C and 1.5 min at 72°C, with a final extension of 10 min at 72°C using GeneAmp® PCR system 9700 (Applied Biosystems). The PCR products were resolved using a 1% agarose gel. Sequencing of purified PCR product was undertaken by SAP services, GeneI, Banglore using ITS1 (forward primer).
Phylogenetic Analysis
ITS gene sequences of the isolates were compared with ITS sequences available by the BLAST search in the NCBI, GenBank database (http://www.ncbi.nlm.nih.gov). Multiple sequence alignment was performed using Clustal X (1.1). The method of Jukes and Cantor [12] was used to calculate evolutionary distances. Phylogenetic dendogram was constructed by the neighbour-joining method and tree topologies were evaluated by performing bootstrap analysis of 100 data sets using MEGA 3.1 (Molecular Evolutionary Genetic Analysis). The sequences were submitted to GenBank under the following accession numbers GQ426033–GQ426044 (12 sequences).
Antagonistic Activity of Trichoderma Isolates
The dual culture technique described by Morton and Stroube [13] was used to test the antagonistic ability of Trichoderma spp. against soil borne pathogen S. rolfsii and foliar borne pathogens viz., C. capsici and C. gloeosporioides. The pathogens and Trichoderma were grown on PDA for a week at room temperature (28 ± 2°C). Small blocks of the target fungi (C. capsici and C. gloeosporioides) cut from the periphery were transferred to the Petri dish previously poured with PDA. After 2 days of Colletotrichum spp. growth, the Trichoderma spp. was transferred aseptically in the same plate of opposite end and were incubated at room temperature with alternate light and darkness for 7 days and observed periodically. For S. rolfsii both pathogen and Trichoderma were inoculated at the same time. The experiment was replicated thrice and percent growth inhibition was calculated by the formula of I = (C−T)/C × 100, where C is mycelial growth in control plate, T is mycelial growth in test organisms inoculated plate and I is inhibition of mycelial growth.
Assay for Extra Cellular Enzyme Activity
Twelve isolates of Trichoderma were separately inoculated into 100 ml broth media with four combinations viz., Czapek Dox Broth alone, CDB + mycelial powder of S. rolfsii (10 g), CDB + mycelial powder of C. gloeosporioides (10 g) and CDB + mycelial powder of C. capsici (10 g) in Erlenmeyer flasks and incubated at 28 ± 1°C for 7 days with intermittent shaking at 125 rpm twice a day. The culture filtrate each isolate was harvested, filtered through the Whatman Filter Paper 42, centrifuged and assayed for chitinase and β-1,3-glucanase enzyme activities immediately.
β-1,3-Glucanase Activity
For assay of β-1,3-glucanase enzyme, 0.5 ml laminarin, 1.0 ml of 0.05 M citrate buffer (pH 4.8) and 0.5 ml culture filtrate was mixed and incubated at 40°C for 60 min. An equal volume of dinitrosalicyclic acid reagent was added to the reaction mixture and warmed in boiling water for 15 min. The absorbance of reaction mixture was measured at 575 nm in a spectrophotometer and compared with standard graph drawn by following the same procedure but using different concentrations of glucose instead of culture filtrate. The quantity of reducing sugar was calculated from the glucose standards used in the assay and activity of β-1,3-glucanase was expressed in nkat/ml. One nkatal corresponds to the release of 1 nmol glucose equivalent per second.
Chitinase Activity
A mixture of 0.5 ml culture filtrate, 0.5 ml suspension of colloidal chitin and 1.0 ml of Mcllvaines buffer (pH 4.0) was mixed and incubated at 37°C for 2 h in a water bath with constant shaking. The reaction was stopped by boiling 3 min in heated water bath. 3 ml potassium ferricyanide reagent was added and warmed in boiling water for 15 min. The amount of N-acetyl glucosamine (NAG) released was estimated following the methods of Reissing et al. [14]. The absorbance of reaction mixture was measured in a spectrophotometer at 420 nm. The amount of reducing sugar released was calculated from standard curves for NAG and chitinase enzyme activity was expressed in pkat (pmol/s) per millitre.
Statistical Analysis
Statistical analysis were performed with the Agres and Agdata using completely randomized analyses of variances (ANOVA) was used to compare the biocontrol efficacy of Trichoderma isolates and means separated by Fisher’s protected least significant difference (LSD). The significance of effects of Trichoderma on growth characteristics was determined by the magnitude of the F value (P = 0.05).
Results and Discussion
Morphological Characterization
Based on the observation of the conidia, phialides, colony texture, chlamydospore, conidiophore morphology the isolates were grouped into section Pachybasium B (clade Lixii/catoptron), Trichoderma (Rufa), Trichoderma (Pachybasium A), Longibrachiatum and Pachybasium B (Lutea). Section Pachybasium consists of the isolates TSD1, TWN1, TGN1, TJP1 and TWC1 where the condiophores were much branched, which arise in distinct and continues ring like zones. The main branches mostly in groups of 2–3 and stand at right angle to the bearer and their length increased with the distance from the tip of the main branch which gave a conical or pyramidal appearance. In Trichoderma section (Rufa) (TWD1, TGD1 and TCC1) the hyphae, branched, smooth walled and colourless. The condiophores were observed as less complicated and formed aerial hyphae. They produced smaller branches and ultimately a conifer-like branching system is formed. In Pachybasium B (Lutea) (TBC1) the conidiophores are smooth walled, verticiliately branched in the Pachybasium type pattern. Trichoderma (section of Pachybasium A) consists of T. asperellum seen the conidiophores have terminating 2 or more philides and primary branching arising at nearly 90 degrees to the main axis. In section Longibrachiatum (H. orientalis), the conidiophores were complicated and progressively longer, often paired, secondary branches. Phialides arising directly from secondary branches, typically not in whorls. Using the key of Rifai [15] in the earlier most of the strains isolated in these regions were identified as T. harzianum. T. hamatum and T. viride [16]. In fact, apart from T. harzianum, which, nevertheless accounted for the majority of isolates, a very interesting finding of this study was the coexistence of several species of Trichoderma (e.g. T. harzianum, T. erinaceum, T. asperellum, T. ovalisporum, T. viride and T. brevicompactum) in the Bay Island ecosystem.
Molecular Identification of Trichoderma Species
Sequence analysis of twelve isolates was done to confirm species identity, which initially has been done based solely on morphological parameters. Comparison of oligonucleotide fragments of rDNA sequences, which included the 5.8S gene and the flanking ITS1 and ITS2 regions, with reference sequences from public databases, showed that they were very similar.
Phylogenetic Analysis
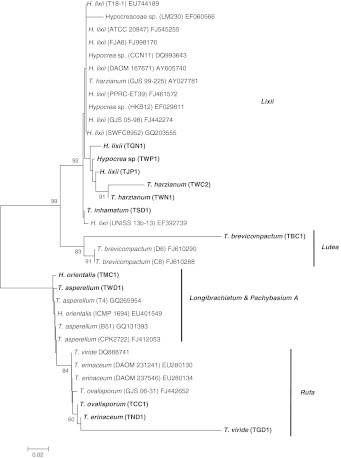
The phylogenetic tree obtained by sequence analysis of ITS1 and ITS4 of 12 of our isolate sequences isolates and the sequence of 22 Trichoderma spp. obtained from NCBI, GenBank is represented in Fig. 1. The ITS sequence was chosen for this analysis because it has been showed to be more informative with various sections of the genus Trichoderma [17–19]. Bootstrap analysis with 100 bootstrap replications demonstrated three major branches. On the basis of the bootstrap values, 34 Trichoderma spp. could be divided into 5 groups. Group 1, the T. harzianum complex includes 6 of our isolates. This group is supported by bootstrap value of 92% contains 3 subgroups supported by bootstrap values more than 90%. Group II (Lutea) includes 3 strains representing T. brevicompactum. This cluster is supported by a bootstrap value of 83%. Group IV (Rufa) includes 3 different isolates includes T. viride, T. ovalisporum and T. erinaceum, this cluster is supported by more than 60% of bootstrap values. In the case of group III cluster which, consists of section Longibrachiatum (H. orientalis) and Pachybasium A (T. asperellum). In this analysis no grouping among the isolates was observed, and did not show any bootstrap supports suggests instability of the clade.
Fig. 1.
Phylogram based on the ITS region of the genomic rRNA gene of 12 isolates and 22 representative strains of Trichoderma with isolate number indicated in parenthesis. The phylogram was generated after DNA distance based and neighbor-joining analysis of the data. Bootstrap 50% majority-rule consensus tree (1,000 replications), indicated above the nodes, was generated using MEGA 3.1
Antagonistic Activity and Enzyme Production of Trichoderma Isolates
Results from the dual culture test showed that all isolates of Trichoderma inhibited mycelial growth of S. rolfsii more than 50%. Two isolates TGD1 and TWN1 (T. viride and T. harzianum) showed statistically significant inhibition of mycelial growth (76.3%) over control against S. rolfsii (Table 2). The isolates TND1, TBC1, TCC1, TGN1 and TWC2 (T. erinaceum, T. brevicompactum, T. ovalisopum, H. lixii and T. harzianum) were next best isolates in inhibiting the growth of pathogen i.e., 73.3, 70.8, 72.5, 70.8 and 72.1 per cent over the control, respectively. Isolates TSD1 (T. inhamatum), TJP1 (H. lixii) and TND1 (T. erinaceum) showed statistically significant inhibition of mycelial growth of C. gloeosporioides in dual culture (Table 2). The growth inhibitions of these isolates were 45.6, 43.3 and 42.2% respectively over the control. Similarly in the case of C. capsici, isolates TWC2 (T. harzianum) and TSD1 (T. inhamatum) showed significant inhibition of mycelial growth.
Table 2.
Antagonistic potential of Trichoderma isolates against S. rolfsii, C.gloeosporioides and C. capsici
| S. No. | Isolate name | Mycelial growth inhibition* (%) | ||
|---|---|---|---|---|
| S. rolfsii | C. gloeosporioides | C. capsici | ||
| 1 | T. inhamatum | 69.6bc | 45.6a | 48.9ab |
| 2 | T. longibrachiatum | 67.1 cd | 35.6 cd | 42.2de |
| 3 | T. brevicompactum | 70.8ab | 38.9bc | 36.7e |
| 4 | T. ovalisporum | 72.5abc | 27.8d | 43.3 cd |
| 5 | T. viride | 76.3a | 30.0 cd | 46.7bc |
| 6 | T. erinaceum | 73.3ab | 42.2ab | 42.2cd |
| 7 | T. harzianum | 70.8ab | 35.6 cd | 46.7bc |
| 8 | T. asperellum | 69.6bc | 40.0bc | 38.9de |
| 9 | Trichoderma sp. | 62.1d | 28.9d | 42.2de |
| 10 | T. harzianum | 76.3a | 34.4 cd | 45.6bc |
| 11 | T. harzianum | 72.1ab | 27.8d | 50.0a |
| 12 | T. harzianum | 56.3e | 43.3ab | 46.7bc |
| Control (mm) | 80 | 30 | 30 | |
| CD (0.05) | 0.4387 | 0.3025 | 0.1946 | |
| SEd | 0.2126 | 0.1466 | 0.0943 | |
Data were analyzed using one-way analysis of variance (values with different lower case letters are significantly different; P ≤ 0.05%)
* Values are mean of three replications
The production of chitinase and β-1,3-glucanase are summarized in Tables 3 and 4. All isolates showed significantly higher chitinase and β-1,3-glucanase activities with supplement of different carbon sources as substrates in the basal media. The highest chitinase enzyme activity was recorded with T. viride (42.0 U) in CDB + mycelial powder of S. rolfsii, followed by T. harzianum (40.3 U), T. erinaceum (37.1 U), H. lixii (36.4 U), T. brevicompactum (35.1 U), T. ovalisporum (34.2 U) and Trichoderma sp. (33.2 U). Chitinase activity of test isolates of Trichoderma was relatively less when the basal medium was added with mycelial powder of C. gloeosporioides and C. capsici as compare to S. rolfsii whereas highest chitinase activity was recorded with T. harzianum. The partial substitution of sucrose by same carbon sources in the basal medium has profound effect as the β-1,3-glucanase activity as compared to basal media (CDB). Highest β-1,3-glucanase enzyme activity was recorded with T. harzianum (22.6, 31.5, 28.6 and 30.2 U) in CDB, CDB + S. rolsii, CDB + C. gloeosporioides and CDB + C. capsici followed by T. viride, T. erinaceum and T. inhamatum. Among all carbon sources as amendment of media, mycelial powder of S. rolsii induced higher β-1,3-glucanase and chitinase activity than C. gloeosporioides and C. capsici.
Table 3.
Chitinase enzyme activity of Trichoderma isolates from Andaman and Nicobar Islands (pkat/ml)
| Isolate | CDB | CDB + SR | CDB + CG | CDB + CC |
|---|---|---|---|---|
| T. inhamatum | 22.3 | 32.5 | 28.4 | 30.1 |
| T. longibrachiatum | 19.2 | 30.2 | 25.3 | 28.1 |
| T. brevicompactum | 23.9 | 35.1 | 26.4 | 23.5 |
| T. ovalisporum | 22.7 | 34.2 | 22.3 | 19.6 |
| T. viride | 28.3 | 42.0 | 24.5 | 27.8 |
| T. erinaceum | 23.5 | 37.1 | 28.0 | 28.6 |
| T. asperellum | 21.0 | 22.0 | 28.0 | 26.5 |
| T. harzianum | 25.1 | 36.4 | 26.4 | 24.8 |
| Trichoderma sp. | 20.6 | 33.2 | 19.6 | 21.3 |
| T. harzianum | 32.2 | 40.3 | 30.2 | 32.6 |
| T. harzianum | 30.5 | 38.6 | 29.4 | 31.2 |
| T. harzianum | 20.0 | 29.6 | 22.7 | 20.5 |
| SEd | 0.403 | 0.321 | 0.369 | 0.426 |
| CD (0.05) | 0.833 | 0.878 | 0.763 | 0.878 |
Means of four replications; CDB Czapek Dox Broth, S substrate, SRS. rolfsii, CGC. gloeosporiodes, CCC. capsici
Table 4.
β-1,3-Glucanase enzyme activity of Trichoderma isolates from Andaman and Nicobar Islands (nkat/ml)
| Isolate | CDB | CDB + SR | CDB + CG | CDB + CC |
|---|---|---|---|---|
| T. inhamatum | 15.2 | 22.3 | 24.9 | 22.5 |
| T. longibrachiatum | 17.4 | 23.5 | 19.4 | 21.0 |
| T. brevicompactum | 14.8 | 15.0 | 17.5 | 18.6 |
| T. ovalisporum | 9.5 | 18.1 | 21.2 | 19.6 |
| T. viride | 20.5 | 24.2 | 25.5 | 28.3 |
| T. erinaceum | 11.3 | 29.8 | 22.3 | 24.0 |
| T. asperellum | 11.5 | 12.8 | 16.5 | 20.0 |
| T. harzianum | 13.4 | 19.3 | 24.3 | 25.0 |
| Trichoderma sp. | 11.2 | 16.4 | 17.6 | 15.3 |
| T. harzianum | 22.6 | 31.5 | 28.6 | 30.2 |
| T. harzianum | 19.4 | 28.1 | 26.8 | 27.7 |
| T. harzianum | 9.3 | 17.5 | 18.7 | 21.0 |
| SEd | 0.372 | 0.320 | 0.408 | 0.406 |
| CD (0.05) | 0.768 | 0.654 | 0.843 | 0.837 |
Means of four replications; CDB Czapek Dox Broth, SRS. rolfsii, CGC. gloeosporiodes, CCC. capsici
The application of Trichoderma as biocontrol agent has been focused mainly in controlling root and soil-borne diseases while there are few reports on their application in controlling foliar diseases [20]. This stems from the fact that Trichoderma sp. are native (residents) in the soil but transients in the phylloplane [21]. The antagonism by Trichoderma spp. against many soils borne plant pathogen has been established [6, 22, 23]. Strong antagonism by Trichoderma spp. against a range of soil borne plant pathogens has been reported [6, 23]. It is well known that Trichoderma spp. have the potential to produce cell wall degrading enzymes by using the materials that are present in the growth medium [6]. Production of hydrolytic enzymes such as β-1,3-glucanase, chitinase, cellulase and proteinase increased significantly when Trichoderma spp. were grown in media supplemented with either autoclaved mycelium or purified host fungal cell walls [7, 24]. Ulhoa and Peberdy [25] found that products of chitin degradation also regulate the chitinase synthesis in T. harzianum. Kumar and Gupta [2] reported that cell wall of M. phaseolina and S. rolfsii is known to have glucan and chitin that should have resulted in the induction of glucanase and chitinase in mycelial mat amended media. High β-1,3-glucanase and chitinase activities were detected in dual culture when T. harzianum parasitized R. solani and S. rolfsii compared with low levels of substrates or in absence of pathogen [26]. In present investigation, it was found that chitinase and β-1,3-glucanase enzyme activities was increased with the substitution of specific carbon source at 1% concentration. These findings is in accordance with Ulhoa and Peberdy [25] where they suggested that chitinase activity was substrate’s concentration dependent above 0.5% (w/v) chitin there was no further synthesis of chitinase in the growth medium by T. harzianum was increased up to 1% concentration, whereas β-1,3-glucanase enzyme production was increase up to 1% concentration of laminarin but decreased at higher concentrations. This may be due to the fact that at higher concentration of sugar activity of this enzyme was inhibited. The degree of mycelial inhibition of target pathogen by T. erinaceum is notable example as biocontrol against pathogens tested.
Acknowledgments
This research was supported by grants from the project “Application of Microorganisms in Agriculture and Allied Sectors” of the NBAIM-ICAR, UP, Mau, India. We thank the Head Division of Field Crops, CARI for constant support and encouragements during the course of study.
Contributor Information
Krishna Kumar, Email: krishnaksingh2000@yahoo.co.in.
N. Amaresan, Email: amarcool82@yahoo.co.in
S. Bhagat, Email: sombhagat73@rediffmail.com
K. Madhuri, Email: madhurikbiotech@gmail.com
R. C. Srivastava, Email: ramesh_cari@yahoo.co.in
References
- 1.Samuels GJ. Trichoderma: a review of biology and systematics of the genus. Mycol Res. 1996;100:923–935. doi: 10.1016/S0953-7562(96)80043-8. [DOI] [Google Scholar]
- 2.Kumar A, Gupta JP. Variation in enzyme activity of tubeconazole tolerant biotypes of Trichoderma viride. Ind Phytopathol. 1999;52:263–266. [Google Scholar]
- 3.Kubicek CP, Penttila ME. Regulation of production of plant polysaccharide degrading enzymes by Trichoderma. In: Harman GE, Kubicek CP, editors. Trichoderma and Gliocladium enzymes biological control and commercial applications. London: Taylor and Francis Ltd; 1998. pp. 49–71. [Google Scholar]
- 4.Sivasithamparam K, Ghisalberti EL. Seconday metabolism in Trichoderma and Gliocladium. In: Kubicek CP, Harman GE, editors. Trichoderma and Gliocladium basic biology taxonomy and genetics. London: Taylor and Francis Ltd; 1998. pp. 139–191. [Google Scholar]
- 5.Harman GE. Myth and dogmas of biocontrol; changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis. 2000;84:377–393. doi: 10.1094/PDIS.2000.84.4.377. [DOI] [PubMed] [Google Scholar]
- 6.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 7.Cruz L, Pinter-Toro JA, Benitez T, Llobell A. Purification and characterization of an endo-β-1,3-glucanase from Trichoderma harzianum that is related to its mycoparasitism. J Bacteriol. 1995;177:1864–1877. doi: 10.1128/jb.177.7.1864-1871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elad Y, Chet I, Henis Y. Degradation of plant pathogenic fungi by Trichodermaharzianum. Can J Microbiol. 1982;28:719–725. doi: 10.1139/m82-110. [DOI] [Google Scholar]
- 9.Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia. 2002;94:146–170. doi: 10.2307/3761854. [DOI] [PubMed] [Google Scholar]
- 10.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x. [DOI] [Google Scholar]
- 11.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal DNA for phylogenetics. In: Innes MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic; 1990. pp. 315–322. [Google Scholar]
- 12.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian protein 242 metabolism. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 13.Morton DT, Stroube NH. Antagonistic and stimulatory effect of microorganism upon Sclerotium rolfsii. Phytopathology. 1955;45:419–420. [Google Scholar]
- 14.Reissing JL, Strominger JL, Leloir LF. A modified colorimetric method for estimation of N-acetylamine sugars. J Biol Chem. 1955;217:959–966. [PubMed] [Google Scholar]
- 15.Rifai MA. A revision of the genus Trichoderma. Mycol Pap. 1969;116:1–56. [Google Scholar]
- 16.Bhagat S (2008) Biocontrol potential of Trichoderma spp. from Andaman and Nicobar Islands. PhD thesis, BCKV, West Bengal, India
- 17.Kuhls K, Lieckfeldt E, Samuels GJ, Kovacs W, Meyer W, Petrini O, Gams W, Borner T, Kubicek CP. Molecular evidence that the asexual industrial fungus Trichoderma reesei is a clonal derivate of the ascomycete Hypocrea jecorina. Proc Natl Acad Sci USA. 1996;93:7755–7760. doi: 10.1073/pnas.93.15.7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhls K, Lieckfeldt E, Samuels GJ, Meyer WC, Kubicek P, Borner T. Revision of Trichoderma sec. Longibrachiatum including related teleomorphs based on analysis of ribosomal DNA internal transcribed spacer regions. Mycologia. 1997;89:442–460. doi: 10.2307/3761038. [DOI] [Google Scholar]
- 19.Ospina-Giraldo MD, Royse DJ, Thon MR, Chen X, Romaine CP. Phylogenetic relationships of Trichoderma harzianum causing mushroom green mold in Europe and North America to other species of Trichoderma from world-wide sources. Mycologia. 1998;90:76–81. doi: 10.2307/3761014. [DOI] [Google Scholar]
- 20.Blakeman JP, Fokemma NJ. Potential for biological control of plant disease on the phylloplane. Ann Rev Phytopathol. 1982;20:167–192. doi: 10.1146/annurev.py.20.090182.001123. [DOI] [Google Scholar]
- 21.Bankole SA (1990) Micro flora associated with cowpea phylloplane and their role in disease development and control. MSc thesis, University of Ibadan, Ibadan
- 22.Bhagat S, Pan S. Mass multiplication of Trichoderma harzianum on agricultural byproducts and their evaluation against seedling blight of mungbean and collar rot of groundnut. Ind J Agric Sci. 2007;77:583–588. [Google Scholar]
- 23.Bhagat S, Pan S. Variability in production of extra cellular hydrolytic enzymes by Trichoderma spp. and induction of disease resistance in gram (Ciccer arietinum) J Biol Conserv. 2008;22:57–66. [Google Scholar]
- 24.Carasolio CA, Gutierrez A, Jimenez B, Monkngu M, Herrera Estrella A. Primary structures and expression pattern of the 33 kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum. Proc Natl Acad Sci USA. 1994;91:10903–10907. doi: 10.1073/pnas.91.23.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulhoa CJ, Peberdy JF. Regulation of chitinase synthesis in Trichoderma harzianum. J Gen Microbiol. 1991;137:2163–2169. doi: 10.1099/00221287-137-9-2163. [DOI] [PubMed] [Google Scholar]
- 26.Elad Y, Chet I. Improved selective media for isolation of Trichoderma spp. and Fusarium spp. Phytoparasitica. 1983;11:55–58. doi: 10.1007/BF02980712. [DOI] [Google Scholar]