Abstract
The objective of this work was to determine the shifts in the PCR-DGGE profiles of bacterial communities associated with the rhizosphere soil of ginseng at varying age levels. Differences in the dominance of intense DNA bands in the DGGE profile was observed over the age of the plants indicating the fluctuation in the microbial community structure. The bacterial orders of actinomycetales of Actinobacteria and Spingomonadales and Rhizobiales of α-Proteobacteria were predominant in the ginseng soil.
Keywords: Korean ginseng soil, Bacterial community, DGGE
Introduction
Denaturing gradient gel electrophoresis (DGGE) is perhaps the most commonly used among the culture-independent fingerprinting techniques. It is usually employed to assess the structure of microbial communities in environment samples and to determine the community dynamics in response to environment and other variations [1]. Ginseng (Panax ginseng C.A. Meyer) has been regarded as one of the important remedies in oriental medicine for more than thousand years [2]. The objective of this work was to determine the shifts on the PCR-DGGE profiles of bacterial communities associated with the rhizosphere soil of ginseng at varying age levels.
The rhizosphere soil samples were collected from I to IV year old ginseng plants at Okcheon, Korea. The soil samples were taken from five healthy plants and pooled into one for each year and kept at 4°C prior to use. Since, 4 year old ginseng plants are economically important, we analyzed the endophytic diversity from 1 to 4 year old plants. They were designated as S I to S IV according to the age of the plants. Total community DNA from the rhizosphere soil of ginseng plants of varied age level was extracted with the Fast DNA Spin Kit (Q-BIO Gene, USA) according to the manufacturer’s instructions. The V-3 region (E. coli position 341–534) of bacterial 16S rDNA was amplified by PCR using primers GC341F/518R [3]. The expected size of the PCR product with these primers was 218 bp. The PCR products were separated on 10% (w/v) polyacrylamide gels (acrylamide: bisacrylamide = 37.5:1) with a 35 to 65% linear gradient formamide. Gels were run for 16 h at 60 V in 1× TAE buffer (40 mM Tris, 20 mM acetic acid of denaturant, where 100% was defined as 7 M urea with 40% (v/v), and 50 mM EDTA, pH 8.0) at 60°C. Denaturing gels were run using the DCode Universal Mutation Detection System (Bio-Rad, USA). The gels were then stained for 30 min in Gel-red (100 μl/l) in 1× TAE buffer and visualized by UV illumination. Gel images were acquired with the ChemDoc (Bio-Rad, USA) gel documentation system. Individual bands that are unique in each sample were selected for sequencing analysis, excised and incubated overnight in distilled water. PCR amplification was performed using the primers (MG3F and 518R) described above but with a non-GC clamp, and the PCR products were purified and sequenced. Nucleotide sequences were compared with sequences in GenBank using BLAST search.
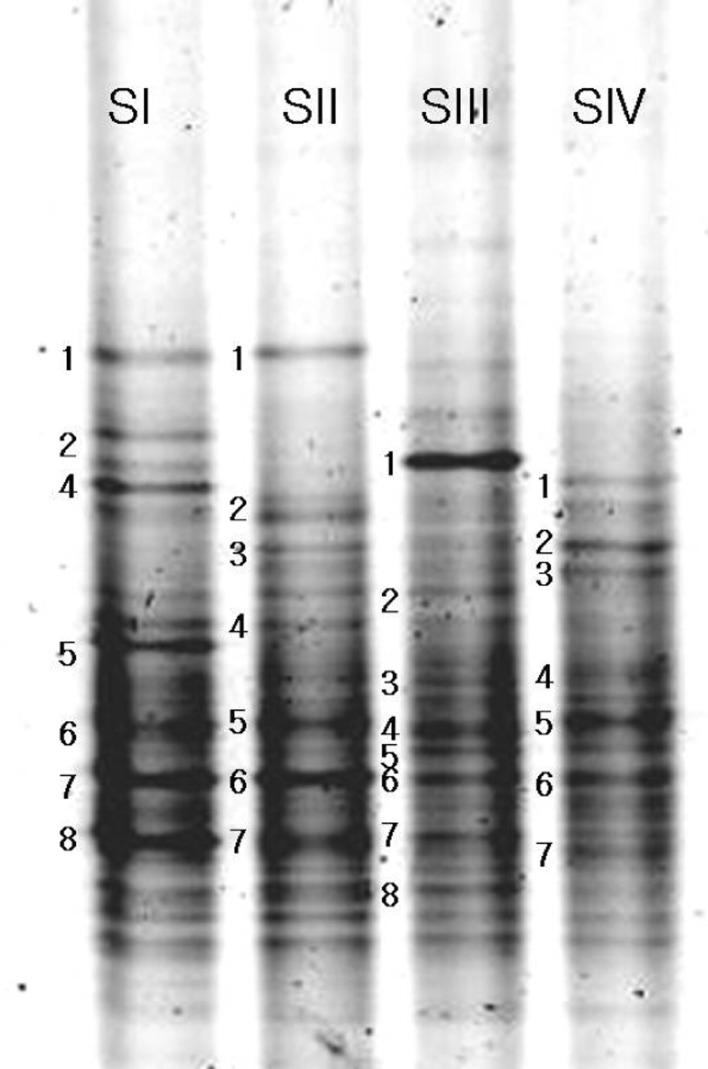
Differences in the dominance of intense DNA bands in the DGGE profile was observed over the age of the plants indicating the fluctuation in the microbial community structure (Fig. 1). Some of the bands were intense in the profile of the rhizosphere soil and others were very faint indicating their population level. It was observed that the rhizosphere soil of one as well as 2 year old plants harboured more of Actinobacteria and α-Proteobacteria. Mixed composition of organisms was noticed in the 3 year old plant rhizosphere sample and Actinobacteria was the predominant class of organisms in the 4 year old ginseng soil. The bacterial community in the rhizosphere is usually dynamic and can be highly affected by the plant developmental stage. Age of the plants is one of the important factors that affect the microbial community associated to the rhizosphere [4]. Rhizosphere bacterial communities of potato cultivars were evaluated through PCR-DGGE profiles and found differences among communities due to age [5]. From this study it is more evident that the age of the plant could largely induce a shift in the bacterial communities in the rhizosphere soil of ginseng.
Fig. 1.
Fingerprinting of the bacterial community from the rhizosphere soil of ginseng by DGGE separation. Lanes S I–S IV: soil collected from 1 to 4 year old plants rhizosphere, respectively
Sequence analysis of the DGGE bands indicated that the dominant bacterial groups in the Korean ginseng soil were Actinobacteria and α-Proteobacteria (Table 1). Kim et al., [6] analyzed the bacterial community in Korean rice field through DGGE and found that members belonging to Actinobacteria division dominated in the field after flooding, followed by the α-Proteobacteria. In the class Actinobacteria, the order Actinomycetales was predominant, whereas, in the class α-Proteobacteria, Spingomonadales and Rhizobiales were dominant and found to be present in all age levels. In addition, the low similarity values suggest that the bacteria may be identified to be new species. Our study showed that ginseng rhizosphere supported more number of Actinomycetales, Spingomonadales and Rhizobiales and better utilization of these organisms in a potential way like antibiotic production, bioremediation and so on requires separate study.
Table 1.
Similarity values of 16S rDNA sequences of bacteria from the rhizosphere soil of ginseng
| Isolate | Accession no. | Closest relative | Phylum | Similarity |
|---|---|---|---|---|
| SI 1 | EF409966 | Bifidobacteriales | Actinobacteria | 86 |
| SI 2 | EF445491 | Uncultured bacterium clone KF-312 | – | 90 |
| SI 4 | EU730917 | Sphingomonadales | α-Proteobacteria | 92 |
| SI 5 | AY963413 | Uncultured bacterium clone AS49 | – | 86 |
| SI 6 | AY661551 | Uncultured alpha Proteobacterium isolate | α-Proteobacteria | 88 |
| SI 7 | AM932264 | Actinomycetales | Actinobacteria | 83 |
| SI 8 | EU834254 | Actinomycetales | Actinobacteria | 94 |
| SII 1 | EU647702 | Pseudomonadales | α-Proteobacteria | 90 |
| SII 2 | EF599674 | Uncultured bacterium clone zzg11 | – | 85 |
| SII 3 | AB027691 | Sphingomonadales | α-Proteobacteria | 92 |
| SII 4 | Y14312 | Rhizobiales | α-Proteobacteria | 83 |
| SII 5 | AY367014 | Rhizobiales | α-Proteobacteria | 86 |
| SII 6 | EF516421 | Uncultured bacterium clone FCPN751 | – | 95 |
| SII 7 | EU834254 | Actinomycetales | Actinobacteria | 94 |
| SIII 1 | CP000724 | Clostridiales | Firmicutes | 88 |
| SIII 2 | D32231 | Rhizobiales | α-Proteobacteria | 87 |
| SIII 3 | AF530968 | Uncultured alpha proteobacterium isolate | α-Proteobacteria | 90 |
| SIII 4 | AF410927 | Sphingomonadales | α-Proteobacteria | 91 |
| SIII 5 | EU730917 | Sphingomonadales | α-Proteobacteria | 96 |
| SIII 6 | EU581867 | Rhizobiales | α-Proteobacteria | 88 |
| SIII 7 | EF601829 | Actinomycetales | Actinobacteria | 93 |
| SIII 8 | DQ521405 | Actinomycetales | Actinobacteria | 90 |
| SIV 1 | AJ971910 | Actinomycetales | Actinobacteria | 91 |
| SIV 2 | AY017137 | Actinomycetales | Actinobacteria | 92 |
| SIV 3 | AM889059 | Actinomycetales | Actinobacteria | 93 |
| SIV 4 | AB362778 | Sphingomonadales | α-Proteobacteria | 83 |
| SIV 5 | DQ114945 | Clostridiales | Firmicutes | 94 |
| SIV 6 | DQ659084 | Actinomycetales | Actinobacteria | 96 |
| SIV 7 | DQ158005 | Actinomycetales | Actinobacteria | 97 |
References
- 1.Ercolini D. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Methods. 2004;56:297–314. doi: 10.1016/j.mimet.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Yu WJ, Lee BJ, Nam SY, Yang DC, Yun YW. Modulating effects of Korean ginseng saponins on ovarian function immature rats. Biol Pharm Bull. 2003;26:2574–2580. doi: 10.1248/bpb.26.1574. [DOI] [PubMed] [Google Scholar]
- 3.Muyzer G, Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths BS, Ritz K, Ebblewhite N, Dobson G. Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem. 1999;31:145–153. doi: 10.1016/S0038-0717(98)00117-5. [DOI] [Google Scholar]
- 5.EPdeB Ferreira, Dusi AN, Xavier GR, Rumjanek NG. Rhizosphere bacterial communities of potato cultivars evaluated through PCR-DGGE profiles. Pesq agropec bras Brasília. 2008;43:605–612. doi: 10.1590/S0100-204X2008000500008. [DOI] [Google Scholar]
- 6.Kim MS, Ahn JH, Jung MK, Yu JH, Joo DH, Kim MC, Shin HC, Kim TS, Ryu TH, Kweon SJ, Kim TS, Kim DH, Ka JO. Molecular and cultivation-based characterization of bacterial community structure in rice field soil. J Microbiol Biotechnol. 2005;15:1087–1093. [Google Scholar]