Abstract
Mastitis is a serious problem in dairy sector and among various aetiological agents, the incidence of staphylococci and streptococci remains high in milking animal. The present study was focused on detection of staphylococci and streptococci in winter season. Milk samples (117) of mastitic animals were tested for presence of staphylococci and streptococci using biochemical and PCR based assays. The testing revealed majority of animals (90.6%) were infected with more than one causative agent. Amongst 117 sample, 109 and 90 comprised of staphylococci and 90 streptococci, respectively. Distribution proportion of S. aureus, S. epidermidis, S. agalactiae, S. uberis and S. dysgalactiae among the mastitic cases was found as 64.9, 7.7, 5.1, 1.7, 48.7, 65.8 and 0.8%, respectively. Streptococci and staphylococci were observed in different combinations and the frequent were S. aureus/S. agalactiae/S. uberis, S. aureus/S. uberis, S. aureus/S. agalactiae and S. agalactiae/S. uberis which were accounted for 23.9, 19.7, 5.9 and 2.6%, respectively. Approximately half of the (52.1%) cases were observed for reoccurrence of mastitis. Reoccurrence of mastitis in winter season among these cases was significantly low as compared to summer (cattle-5 cases; buffaloes-2 cases). In addition, prevalence of S. aureus, S. agalactiae, S. uberis, and S. epidermidis in reoccurring mastitic cases was 73.7, 63.9, 45.9 and 6.6%, respectively. The observations revealed mastitis causing pathogens remains in hidden phase in winter season; however, cannot be neglected. The observation might be helpful in culling or segregation of cows for mastitis reduction programmes.
Keywords: Mastitis, Staphylococci, Streptococci, Winter, Reoccurrence
Introduction
Mastitis is the most common and the quite damaging disease in dairy cattle throughout the world. It is a versatile disease in milch animals and is caused either by pathological, genetical, physiological or environmental factors. In spite of the concerted efforts to control or reduce the incidence of mastitis for decades, still remains a major threat to the dairy industry causing huge economic loss. Worldwide losses due to mastitis has been estimated approximately US $35 billion [1]. The average cost of a clinical mastitis case is US $179 per lactating season [2]. In India 50% of milch animals are affected with mastitis, out of which clinical mastitis accounted for 10%, and this causes an annual economic loss of approximately six thousand crores [3]. A wide range of reasons exist including environmental and management aspects for the failure of treatment and eradication of microbial mastitis from herds; mainly the incomplete perfusion of the antibiotics in bovine neutrophils and associated resistance [4, 5] and hidden capability of microbes in udder abscesses [6]. Complete eradication of mastitic pathogens from herd is very difficult and greater chances for the recurrence of mastitis.
Different types of microbes, namely Staphylococcal sp., Streptococcal sp., Escherichia coli, Klebsiella sp., Enterobacter sp., Corynebacteriumbovis, Corynebacterium pyogenes and Micrococcus sp. are common aetiological agents of mastitis in dairy animals. Amongst various causative agents, streptococci and staphylococci are mainly responsible for mastitis infections in lactating animals. Generally, the frequency of staphylococcal incidences is observed more as compared to streptococcal mastitis [7–10]. Frequently, the most common cause of mastitis is Staphylococcus aureus (S. aureus) followed by Streptococcus dysgalactiae (S. dysgalactiae), Streptococcus uberis (S. uberis), Streptococcus agalactiae (S. agalactiae) and other non-coagulase staphylococci [11]. In tropical conditions like India, staphylococci and streptococci have been also reported the major aetiological agents of mastitis; this disease still remains at peak in summer and rainy season between May and September [10].
Early detection of mastitic infection is quite helpful in disease control, treatment and epidemiological studies. Molecular identification of pathogens is being adopted by clinical sector, as the techniques are quite quick, easy and reproducible. Several PCR based methods have been reported for detection of mastitis pathogens [12, 13]. Among various PCR based assay, multiplex is quiet useful for in vitro detection of more than one pathogen within a single tube reaction. Multiplex PCR has been explored for detection of mastitis pathogens [14, 15]. The information is scanty regarding the detection of occurrence of mastitis pathogens (especially staphylococci and streptococci) in the tropical region like India. Hence, the present study was conducted to determine the prevalence of pathogens (streptococci and staphylococci) in non-reported and reported (in earlier calving season and cured cases) mastitic cattle and buffalo in winter season.
Materials and Methods
Collection of Milk Samples
Milk of 168 animals was tested for mastitic infection and 117 samples were collected from infected cattle and buffaloes from November 2008 to February 2009 for this study. All the animals were screened with California mastitis test (CMT). Prior to collection of sample, teats were cleaned with 70% ethanol and initial few streams of milk were discarded. Then samples were aseptically collected in 15 ml sterile tube. Samples were immediately transferred to ice bucket and transported to laboratory.
Microbiological Analysis of Milk Samples
Milk samples were first allowed to come at room temperature (20–25°C). Then 100 μl aliquot of milk was spread over the blood agar media and incubated at 37°C for 24 h for growth of colonies. These colonies were selected and examined for morphological appearance (colony shape and haemolytic characteristics), gram staining and catalase test. Gram staining and catalase positive cocci were further tested for coagulase and mannitol fermentation as per standard procedure [16] to confirm as staphylococci. Gram-positive cocci and catalase negative test isolates cocci were further biochemically tested with Christie, Atkins and Munch-Petersen reaction (CAMP) and esculin hydrolysis [15] to distinguish the types of streptococci.
DNA Isolation
DNA was extracted as per procedure of Phuektes et al. [15] with modification. In brief, milk samples were enriched in BHI broth by incubating at 37°C for 12 h. Bacterial cells were harvested by centrifugation at 5,000 rpm at 4°C for 15 min. Pellet of cells was re-suspended in 250 μl of 10 mM Tris–HCl (pH 8) and added 25 mg/ml of lysozyme followed by incubation at 37°C for 1 h. Afterwards, 500 μl of lysis buffer (50 mM Tris buffer, 100 mM EDTA, 1% SDS, pH-8), and 10 mg/ml of proteinase-K was added and incubated at 50°C for 2 h. Subsequently the proteinase-K was inactivated in boiling water bath for 10 min. Then, digested cells content was gently mixed with 500 μl of phenol: chloroform (1:1), and centrifuged at 10,000 rpm at 4°C for 15 min. This step was repeated one more time. DNA was precipitated with 50 μl of 5 M NaCl and twice volume of 95% of ethanol. Dried pellet was dissolved in Tris–EDTA buffer (pH-8). DNA quality was checked in 0.8% agarose (containing 0.5 μg/ml ethidium bromide) electrophoresis in 0.5 X Tris borate-EDTA (TBE) buffer.
Multiplex PCR Detection of Pathogens
DNA samples were screened using multiplex PCR programmes. First multiplex PCR was carried out for staphylococci group (S. aureus, S. epidermidis and Staphylococcus sp.) and second multiplex PCR reaction for streptococci group (S. agalactiae, S. dysgalactiae and S. uberis). The oligonucleotide primers used in this study for staphylococcal group and streptococcal group were used as explained by Morot-Bizot et al. [17] and Phuektes et al. [15], respectively. The oligonucleotide sequences and amplification conditions are shown in Table 1. Total volume of 25 μl amplification reaction mixture contained 2 μl dNTPs (200 μm/μl), 2.5 μl of 10X Taq buffer, 1.5 μl 25 mM MgCl2, 0.5 μl of each oligonucleotide primer forward and reverse (20 pm/μl), 0.70 μl of Taq DNA polymerase (3 U/μl), 5 μl DNA (approximately, 25 ng/μl) and rest part was sterile water. Amplification conditions for PCR programmed used for staphylococci were pre-denaturation at 94°C for 3 min, followed by 40 cycles of denaturation at 94°C for 0.5 min, annealing at 55°C for 0.5 min, extension at 72°C for 0.5 min and reaction was completed with final extension at 72°C for 3 min. PCR for streptococci was carried out using the conditions as with pre-denaturation at 95°C for 5 min, followed by 36 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 0.5 min, extension at 72°C for 0.5 min, and then a final extension at 72°C for 7 min. The amplification was carried out in 0.2 ml PCR tube in a thermal cycler. Amplified PCR products were separated by electrophoresis at 90 V for 45 min through 2.0% agarose gel containing 0.5 μg/ml ethidium bromide and visualized under UV light.
Table 1.
List of oligonucleotide primers used in multiplex PCR for staphylococci and streptococci
| Name of microorganism | Primer used | Oligo nucleotide sequence | Amplicon size |
|---|---|---|---|
| Staphylococcus sp. (Genus specific) | SP | F-GGCCGTGTTGAACGTGGTCAAATCA R-TIACCATTTCAGTACCTTCTGGTAA |
370 |
| S. aureus | SA | F-AATCTTTGTCGGTACACGATATTCTTCACG R-CGTAATGAGATTTCAGTAGATAATACAACA |
108 |
| S. epidermidis | SE | F-ATCAAAAAGTTGGCGAACCTTTTCA R-CAAAAGAGCGTGGAGAAAAGTATCA |
124 |
| S. uberis | SU | F-TAAGGAACACGTTGGTTAAG R-TTCCAGTCCTTAGACCTTCT |
330 |
| S. agalactiae | SG | F-AAGGAAACCTGCCATTTG R-TTAACCTAGTTTCTTTAAAACTAGAA |
270 |
| S. dysgalactiae | SD | F-GAACACGTTAGGGTCGTC R-AGTATATCTTAACTAGAAAAACTATTG |
264 |
Data Collection and Analysis
Mastitis history of animals was collected from the treatment record on incidences of infection per lactation or in different lactations, recurrence and seasons. Comparative analysis was carried out on multiplex PCR observations with these parameters and to find out relationship.
Results
In a process of assessment of the mastitic infection, 168 animals comprising of cattle (Sahiwal, 86) and buffaloes (Murrah, 82) were screened with a preliminary CMT test. Milk samples of 117 animals were found mastitic. Amongst the 117 animal, 60 were from Sahiwal cattle and 57 from Murrah buffalo. Microbiological evaluation of samples revealed 85.5% animals were infected with mixed culture of staphylococci, streptococci and other microbes (with single or more types). Gram staining and catalase test of 117 sample revealed 109 were found positive for staphylococci, and 90 for streptococci. Amongst 109 staphylococci coagulase and mannitol tests, 76 isolates were found positive and others remained negative. Streptococci (90) subjected to CAMP and esculin hydrolysis test (shown in parenthesis), revealed that 57 isolates were positive for CAMP positive and esculin negative), 77 were CAMP negative and esculin positive, and one was CAMP and esculin, negative. Based on these biochemical assays, distribution proportion of S. aureus and S. epidermidis was 64.9 and 7.7, whereas S. agalactiae, S. uberis and S. dysgalactiae was observed in 48.7, 65.8 and 0.8%, respectively.
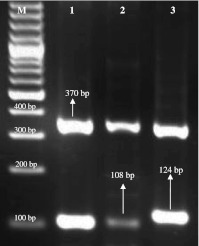
Identification of isolates in samples was confirmed using genus and species specific primers using multiplex PCR. Amplification of DNA samples revealed, 109 positive for Staphylococcal sp. as these samples yielded the amplicon of 370 bp, expected fragment size for staphylococcal genus. Though, eight samples did not show any amplified fragment with the same pair of primer. Among these 109 samples, 76 revealed amplicon of 108 bp (S. aureus species specific-primers; Fig. 1). Of 76 samples, 12 were found solely positive for S. aureus, whereas 64 were positive for S. aureus multiple microorganisms as listed in Table 2. Amplicons of 124 bp (S. epidermidis) showed by nine samples (Fig. 1). Out of 109 staphylococcal strains, 29 could not be identified at the species level, as these DNA samples did not amplified with both the species-specific primes.
Fig. 1.
Multiplex PCR amplicons of different staphylococci. Lane M 100-bp-molecular-size DNA ladder; Lane 1 370 bp (Staphylococcus sp.), Lane 2 108 bp (S. aureus), Lane 3 124 bp (S. epidermidis)
Table 2.
Microorganisms detected by multiplex PCR from mastitic milk
| Causative microorganisms | Observations in Sahiwal (n = 60) | Observations in Murrah (n = 57) | Total observations (n = 117) |
|---|---|---|---|
| S. aureus | 7 | 5 | 12 |
| S. aureus/S. epidermidis | 1 | 1 | 2 |
| S. aureus/S. agalactiae | 7 | – | 7 |
| S. aureus/S. dysgalactiae | 1 | – | 1 |
| S. aureus/S. uberis | 2 | 21 | 23 |
| S. aureus/S. epidermidis/S. uberis | 1 | – | 1 |
| S. aureus/S. epidermidis/S. agalactiae/S. uberis | 2 | – | 2 |
| S. aureus/S. agalactiae/S. uberis | 22 | 6 | 28 |
| S. epidermidis | – | 2 | 2 |
| S. epidermidis/S. agalactiae | 1 | – | 1 |
| S. epidermidis/S. agalactiae/S. uberis | 1 | – | 1 |
| S. agalactiae | – | 1 | 1 |
| S. uberis | 1 | 1 | 2 |
| S. agalactiae/S. uberis | 1 | 2 | 3 |
| Other staphylococcal sp. | 4 | 5 | 9 |
| Other staphylococcal sp./S. agalactiae | 1 | 2 | 3 |
| Other staphylococcal sp./S. uberis | – | 6 | 6 |
| Other staphylococcal sp./S. uberis/S. agalactiae | 6 | 5 | 11 |
| Microbes other than streptococci and staphylococci | – | 2 | 2 |
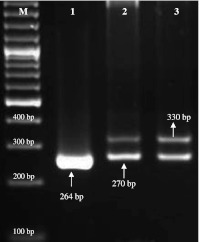
Multiplex PCR (Fig. 2) using species-specific primers of streptococci showed 57 samples positive for S. agalactiae (amplicon size 270 bp) and 77 for S. uberis (amplicon 330 bp). Only, one sample was found infected with S. dysgalactiae. Different combinations of streptococci and staphylococci were observed as depicted in Table 2. The most frequently encountered in samples combinations were S. aureus/S. agalactiae/S. uberis, S. aureus/S. uberis, S. aureus/S. agalactiae and S. agalactiae/S. uberis which were accounted for 23.9, 19.7, 5.9 and 2.6%, respectively.
Fig. 2.
Multiplex PCR amplicons of different streptococci. Lane M 100-bp-molecular-size DNA ladder; Lane 1 264 bp (S. dysgalactiae), Lane 2 270 bp (S. agalactiae), Lane 3 1050 bp (S. uberis)
The treatment record maintained in present herd animal hospital (mastitic cases history) revealed 61 cases (cattle and buffalos) of 117 had got infected earlier in different season (Table 3). Among these mastitic 61 cases reoccurrence proportion of mastitis was more (shown in parenthesis) in cattle (76.6%) as compared to buffaloes (26.3%) through out the year. In addition, reoccurrence of mastitis in winter season of these cases of buffaloes (2cases) and cattle (5cases) was lower compared to summer and rainy seasons (Table 4). The prevalence of S. aureus, S. agalactiae, S. uberis, and S. epidermidis in reoccurring mastitic cases was 73.7, 63.9, 45.9, and 6.6%, respectively. Different combinations of microorganisms in reoccurring mastitic animals are given in Table 3.
Table 3.
Distribution and combinations of microorganisms in animals with reoccurrence of mastitis
| Combination of microorganisms | Observations in Sahiwal (n = 46) | Observations in Murrah (n = 15) | Total observations (n = 61) |
|---|---|---|---|
| S. epidermidis/S. uberis | 1 | – | 1 |
| S. agalactiae | – | 1 | 1 |
| S. agalactiae/S. uberis | 1 | 1 | 2 |
| S. aureus/S. uberis | 2 | 7 | 9 |
| S. aureus/S. epidermidis/S. uberis | 1 | – | 1 |
| S. aureus/S. agalactiae | 7 | – | 7 |
| S. aureus/S. agalactiae/S. uberis | 17 | 1 | 18 |
| S. aureus/S. epidermidis/S. agalactiae/S. uberis | 2 | – | 2 |
| S. aureus | 6 | 2 | 8 |
| S. agalactiae/Other staphylococcal sp. | 1 | – | 1 |
| S. agalactiae/S. uberis/Other staphylococcal sp. | 4 | 2 | 6 |
| Other staphylococcal sp. | 4 | 1 | 5 |
| Type of microorganisms | |||
| S. aureus | 35 | 10 | 45 |
| S. agalactiae | 32 | 5 | 37 |
| S. uberis | 28 | 11 | 39 |
| S. epidermidis | – | – | 4 |
| Other staphylococcal sp. | 9 | 3 | 12 |
Table 4.
Incidences in different seasons in animals with reoccurrence of mastitis
| Season | Observations in Sahiwal (n = 46) | Observations in Murrah (n = 15) | Total observation (n = 61) |
|---|---|---|---|
| Winter | 5 | 2 | 7 |
| Summer | 22 | 5 | 27 |
| Rainy | 7 | 3 | 10 |
| Autumn | 4 | 4 | 8 |
| Winter and summer and autumn | 1 | – | 1 |
| Summer and rainy and autumn | 2 | – | 2 |
| Winter and rainy | 1 | – | 1 |
| Summer and autumn | 3 | – | 3 |
| Summer and rainy | 1 | 1 | 2 |
Discussion
Molecular and biochemical analysis of isolates obtained from various mastitic animals categorically showed staphylococci and streptococci, the main etiological agent of mastitis in this study. In addition, identification of isolates was confirmed by molecular assay as some time false positive and negative biochemical alone could mislead the observations. However, in present study both types of methodology demonstrated the equal results.
Earlier reports supported the results as staphylococci and streptococci have been observed prevalent in mastitic animals compared to other microbial species [7–10, 18, 19]. The relative prevalence rates of various staphylococcal strains isolated in this study showed that S. aureus was the major causative agent in the herd. The major streptococcal strains isolated from this study was S. uberis found to be highest followed by S. agalactiae. Results are in agreement with previous studies [7, 8, 18]. The predominance of microbes varies with mastitic cases, geographical areas, breeds and climatic conditions as reported previously [8–10, 18, 20]. Occurrence of mixed culture among the mastitic animals has been reported previously [21]. In this study, a huge number of samples revealed mixed cultures and various combinations of staphylococci and streptococci were observed (Table 2). Combination of S. aureus and S. agalactiae predominated among the mastitic cases. The proportion of mixed cultures in this study was high when compared to previous findings [10, 21–23].
Data analysis shows high percent of reoccurrence of mastitis in Sahiwal than Murrah buffaloes (Table 3 and Table 4). Increased incidences of isolates showed the elevated mastitis rate among the animals when data was analyzed with previous record of herd. Incidences of mastitis have been studied and reported in literature by several workers. Lower incidence of mastitis in buffaloes has been reported previously [23, 24]. The present study is in agreement as the rate of mastitis incidence was more in cattle cows as compared to buffaloes. Present study showed the increased proportion of infected animals when compared to similar earlier reports [10, 22–24] as results showed the increase in mastitis percent in cattle cows and buffaloes.
Mastitis reoccurrence in different seasons was found higher in Sahiwal cattle than Murrah buffaloes (Table 3). In present study, records showed high incident of mastitis in summer followed by rainy season. However, the PCR results revealed a significant prevalence of staphylococci and streptococci in these cases of winter. It is apparent from the PCR observations that pathogens remain in udder in dormant phase and express the disease under suitable climatic conditions. Significant seasonal variation in the prevalence of mastitis has been reported by different investigators. Bishop et al. [25] found in Holstein–Friesian breed that cases of clinical mastitis were high in summer and slightly elevated in winter as compared to spring and fall. Joshi and Shrestha [24] reported that 75.9% of the cases were during summer and only 5.4% in winter in Murrah buffaloes. Joshi and Gokhale [26] observed subclinical mastitis incidences were more in monsoon season compared to winter. However, Shathele [27] reported that incidence of mastitis was higher in colder months in Holstein–Friesian and showed significant effect of season on incidence of mastitis on new cases. Thus, the results are in concurrences with previous studies.
In conclusion, high occurrence of S. aureus, S. agalactiae and S. uberis was revealed in this study. Biochemical and molecular assays were able to identify these causative agents. The isolates also seemed to have well adaption and hidden capability inside udder during the winter season as a significant number infected animal was observed. These observations could be helpful in implementation of mastitis programmes for control of these pathogens by culling of animal or treatment.
Acknowledgments
This study was supported by Institutional research grant from the National Dairy Research Institute, Karnal, India. Authors are also highly thankful to Rakesh Kumar, Kapil Dev, Anil Kumar and Nanku Singh of Livestock Genome Analysis Laboratory and Milking section of institute for their help in samples collection and support.
References
- 1.Wellenberg GJ, Poel WH, Oirschot JT. Viral infections and bovine mastitis: a review. Vet Microbiol. 2002;88:27–45. doi: 10.1016/S0378-1135(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 2.Bar D, Tauer LW, Bennett G, González RN, Hertl JA, Schukken YH, et al. The cost of generic clinical mastitis in dairy cows as estimated by using dynamic programming. J Dairy Sci. 2008;91:2205–2214. doi: 10.3168/jds.2007-0573. [DOI] [PubMed] [Google Scholar]
- 3.Dua K. Incidence, etiology and estimated loss due to mastitis in India: an update. Ind Dairyman. 2001;53:41–48. [Google Scholar]
- 4.Watts JL, Salmon SA. Activity of selected antimicrobial agents against strains of Staphylococcus aureus isolated from bovine intra-mammary infections that produce β-lactamase. J Dairy Sci. 1997;80:788–791. doi: 10.3168/jds.S0022-0302(97)75999-X. [DOI] [PubMed] [Google Scholar]
- 5.White DG (1999) Use and misuse of antimicrobials in veterinary medicine. Proceeding of 38th Annual Meeting, Arlington (National Mastitis Council, Inc., Madison), p 9
- 6.Bayles KW, Wesson CA, Liou LE, Fox LK, Bohach GA, Trumble WR. Intracellular Staphylococcus aureus escapes the endosome and induces apoptosis in epithelial cells. Infect Immun. 1998;66:336–342. doi: 10.1128/iai.66.1.336-342.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haltia L, Honkanen-Buzalski T, Irina Spiridonova I, Arvi O, Myllys V. A study of bovine mastitis, milking procedures and management practices on 25 Estonian dairy herds. Acta Vet Scand. 2006;48(22):1–6. doi: 10.1186/1751-0147-48-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawari AD, Fawzi A. Prevalence and distribution of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Jordon. Am J Anim Vet Sci. 2008;3:36–39. doi: 10.3844/ajavsp.2008.36.39. [DOI] [Google Scholar]
- 9.Lakew M, Tolosa T, Tigre W. Prevalence and major bacterial causes of bovine mastitis in Asella, South Eastern Ethiopia. Trop Anim Health Prod. 2009;41:1525–1530. doi: 10.1007/s11250-009-9343-6. [DOI] [PubMed] [Google Scholar]
- 10.Patil RL, Kumar SP, Shettar VB, Sudhindra, Honnappagol SS. Epidemiology of subclinical mastitis in buffaloes under field conditions of Bidar, Karnataka state (India) Buffalo Bull. 2005;24:91–97. [Google Scholar]
- 11.Ericsson Unnerstad H, Lindberg A, Persson Waller K, Ekman T, Artursson K, Nilsson-Östa M, et al (2009) Microbial aetiology of acute clinical mastitis and agent-specific risk factors. Vet Microbiol. doi:10.1016/j.vetmic.2008.12.005 [DOI] [PubMed]
- 12.Chotar M, Vidova B, Godany A. Development of specific and rapid detection of bacterial pathogen in dairy products by PCR. Folia Microbiol. 2006;51:639–646. doi: 10.1007/BF02931632. [DOI] [PubMed] [Google Scholar]
- 13.Riffon RE, Sayasith K, Khalil H, Dubreuil P, Drolet M, Lagace′ J. Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J Clin Microbiol. 2001;39:2584–2589. doi: 10.1128/JCM.39.7.2584-2589.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie BE, Oliver SP. Simultaneous detection of mastitis pathogens, Staphylococcus aureus, Streptococcus uberis, and Streptococcus agalactiae by multiplex real-time polymerase chain reaction. J Dairy Sci. 2005;88:3510–3518. doi: 10.3168/jds.S0022-0302(05)73036-8. [DOI] [PubMed] [Google Scholar]
- 15.Phuektes P, Mansell PD, Browning GF. Multiplex polymerase chain reaction assay for simultaneous detection of Staphylococcus aureus and streptococcal causes of bovine mastitis. J Dairy Sci. 2001;84:1140–1148. doi: 10.3168/jds.S0022-0302(01)74574-2. [DOI] [PubMed] [Google Scholar]
- 16.Kloos WE, Schleifer KH (1986) Genus Staphylococcus. In: Sneath PHA, Mair NS, Sharpe ME and Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 2, Williams & Wilkins, Baltimore, pp 1013–1035
- 17.Morot-Bizot SC, Talon R, Leroy S. Development of a multiplex PCR for the identification of Staphylococcus genus and four staphylococcal species isolated from food. J Appl Microbiol. 2004;97:1087–1094. doi: 10.1111/j.1365-2672.2004.02399.x. [DOI] [PubMed] [Google Scholar]
- 18.Getahun K, Kelay B, Bekana M, Lobago F. Bovine mastitis and antibiotic resistance patterns in Selalle smallholder dairy farms, central Ethiopia. Trop Anim Health Prod. 2008;40(4):261–268. doi: 10.1007/s11250-007-9090-5. [DOI] [PubMed] [Google Scholar]
- 19.Kerro DO, Tareke F. Bovine mastitis in selected areas of southern Ethiopia. Trop Anim Health Prod. 2003;35:197–205. doi: 10.1023/A:1023352811751. [DOI] [PubMed] [Google Scholar]
- 20.Sori H, Zerihun A, Abdicho S. Dairy cattle mastitis in and around Sebeta, Ethiopia. Int Appl ResVet Med. 2005;3:332–338. [Google Scholar]
- 21.Sumathi BR, Veeregowda BM, Gomes AR. Prevalence and antibiogram profile of bacterial isolates from clinical bovine mastitis. Vet World. 2008;1:237–238. [Google Scholar]
- 22.Ghose B, Sharda R, Chhabra D, Sharma V. Subclinical bacterial mastitis in cows of Malwa region of Madhya Pradesh. Indian Vet Sci J. 2003;80:499–501. [Google Scholar]
- 23.Shinde SS, Kulkarni GB, Gangane GR, Degloarkar NM (2001) Incidence of mastitis in buffaloes in Parbhani district, Maharashtra. Proc Indian vet Cong, Ludhiana, pp 35–38
- 24.Joshi HD, Shrestha HK (1995) Study on the prevalence of clinical mastitis in cattle and buffaloes under different management systems in the western hills of Nepal. Working paper, Lumle Regi Agric Res Cent 4:95–64
- 25.Bishop JR, Bodine AB, Janzen JJ. Sensitivities to antibiotics and seasonal occurrence of mastitis pathogens. J Dairy Sci. 1980;63:1134–1137. doi: 10.3168/jds.S0022-0302(80)83058-X. [DOI] [PubMed] [Google Scholar]
- 26.Joshi S, Gokhale S. Status of mastitis as an emerging disease in improved an Periurban dairy farms in India. Ann NY Acad Sci. 2006;1081:74–83. doi: 10.1196/annals.1373.007. [DOI] [PubMed] [Google Scholar]
- 27.Shathele MS. Weather effect on bacterial mastitis in dairy cows. Int J dairy sci. 2009;4:57–66. doi: 10.3923/ijds.2009.57.66. [DOI] [Google Scholar]