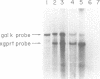
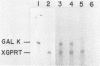
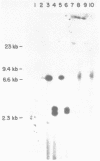
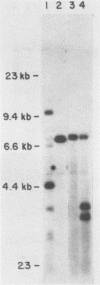
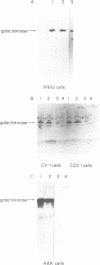
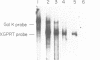
Abstract
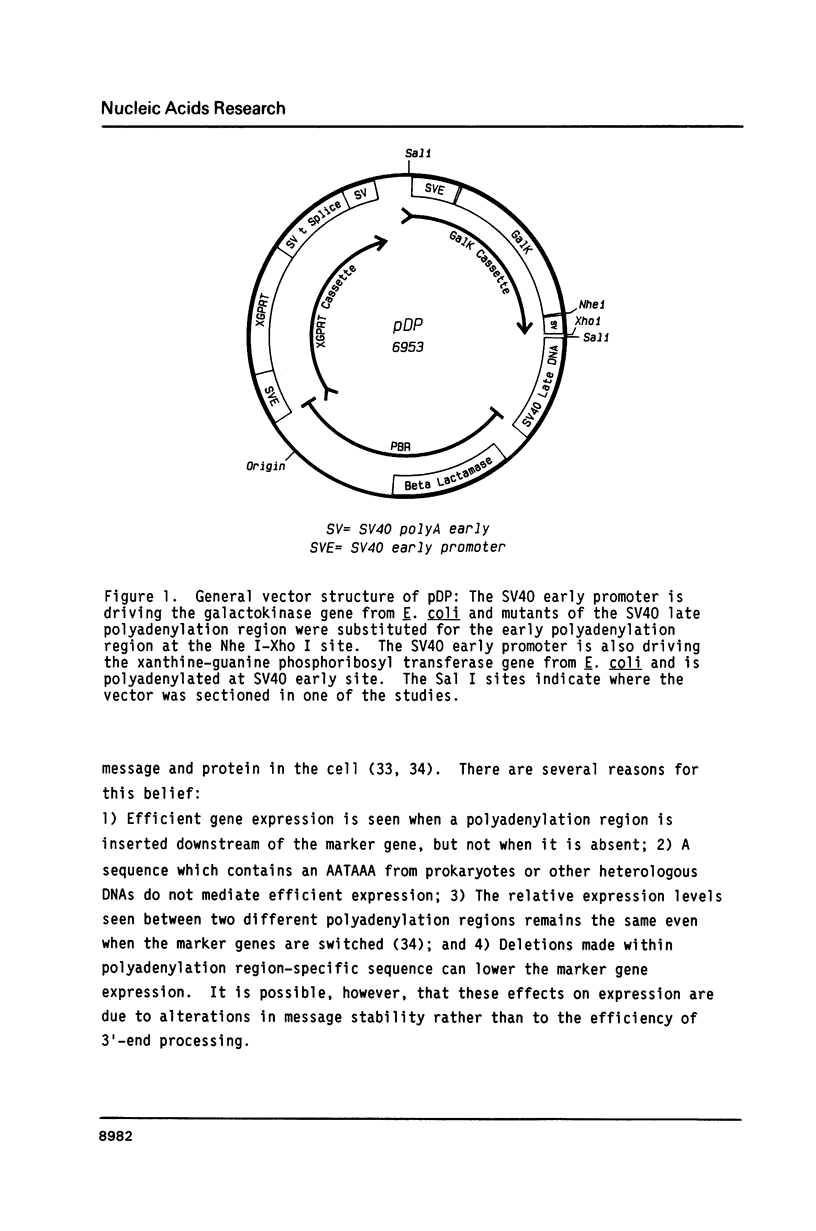
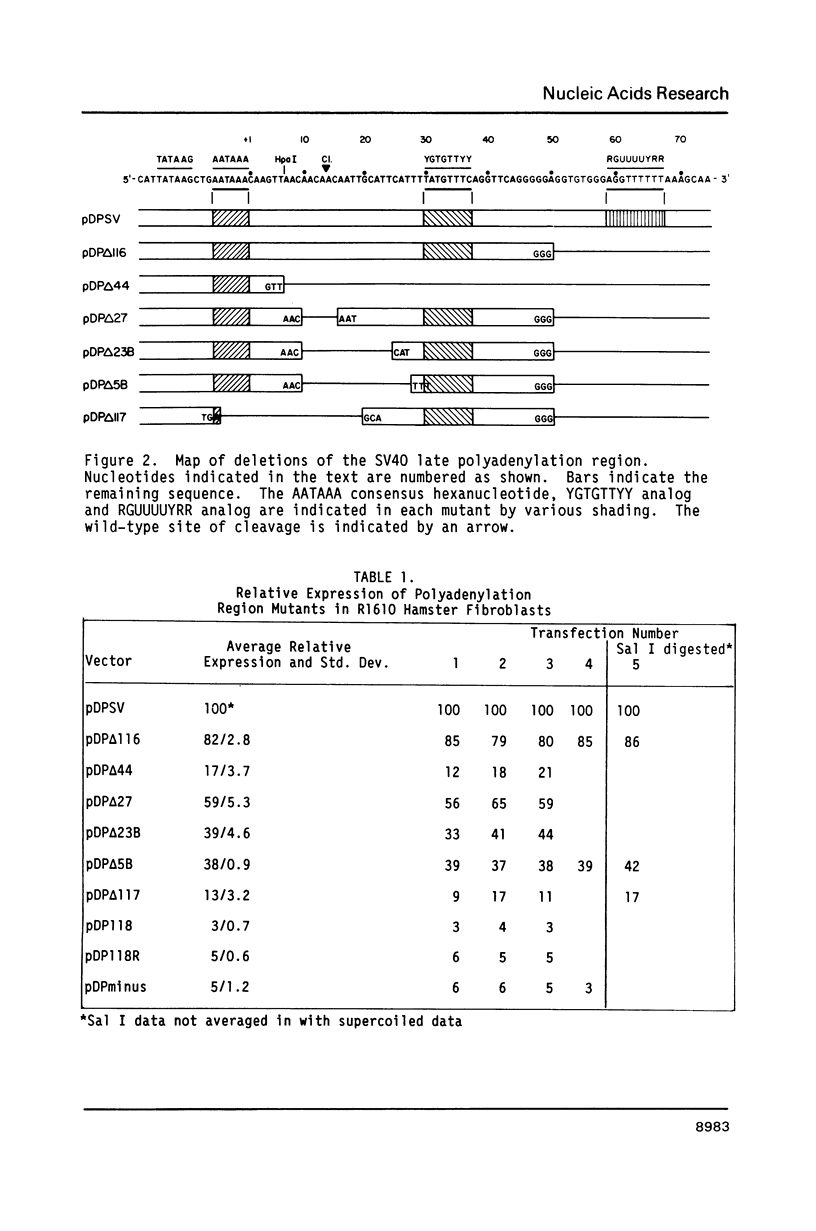
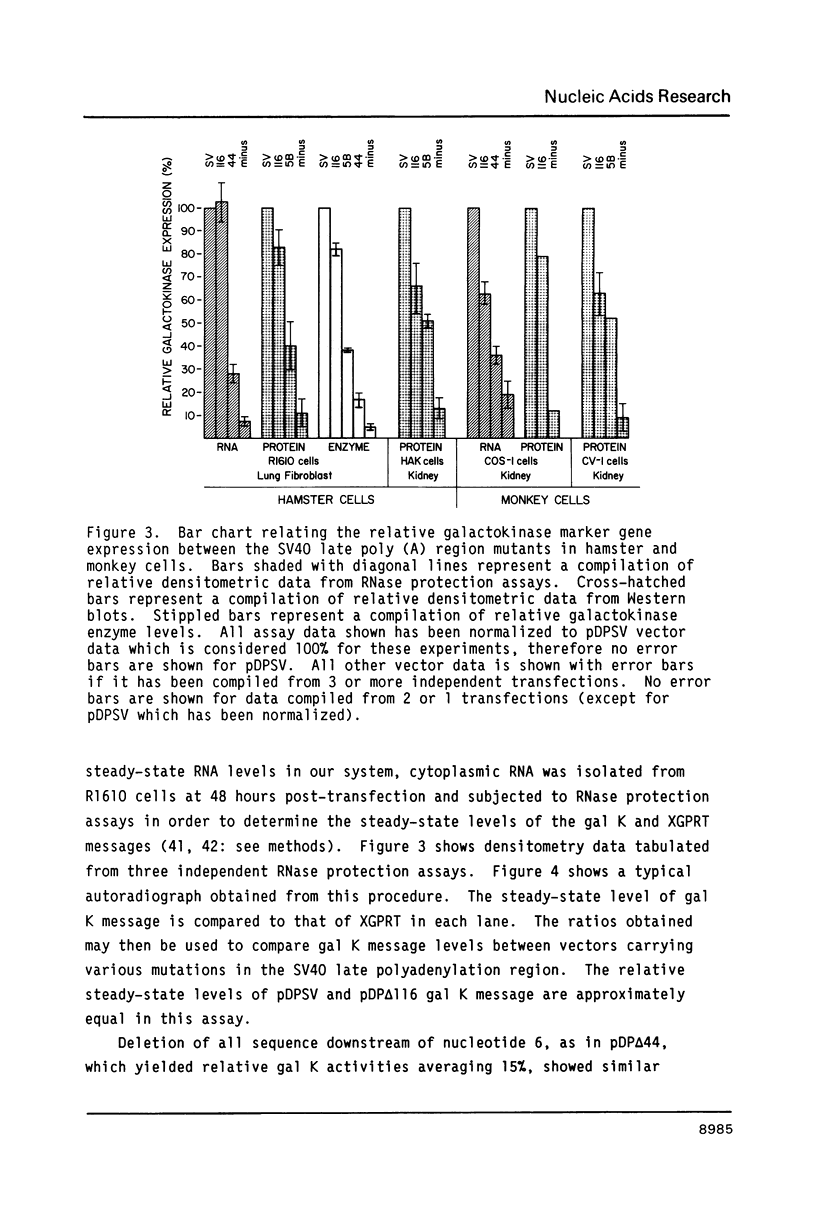
A series of deletions in the SV40 late polyadenylation region was assayed by transient expression in a hamster fibroblast cell line. Because of differences in expression data between our results and the published results of another laboratory using a similar set of deletions introduced into a monkey kidney cell line, we studied our deletions in cells of different tissue-types and species (1). Deletion of the SV40 late polyadenylation region to 49 nucleotides downstream of the hexanucleotide AATAAA showed a small effect on gene expression, while further truncation of the region to 6 nucleotides downstream of the AATAAA showed an 85% drop in marker enzyme activity, protein levels and steady-state message levels. Another deletion in the same region, from base pair 10 to 15 past the AATAAA, which removes the wild-type site of RNA cleavage, showed a 50% drop in marker gene expression. The effects of these mutants on gene expression were similar in all of the cell lines tested and agree with other studies that DNA downstream of the AATAAA plays a role in efficient gene expression.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhat B. M., Wold W. S. ATTAAA as well as downstream sequences are required for RNA 3'-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985 Nov;5(11):3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawerman G. The Role of the poly(A) sequence in mammalian messenger RNA. CRC Crit Rev Biochem. 1981;10(1):1–38. doi: 10.3109/10409238109114634. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Santangelo G. M. Analysis in Cos-1 cells of processing and polyadenylation signals by using derivatives of the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Feb;3(2):267–279. doi: 10.1128/mcb.3.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Stacy T. P. Identification of sequences in the herpes simplex virus thymidine kinase gene required for efficient processing and polyadenylation. Mol Cell Biol. 1985 Aug;5(8):2104–2113. doi: 10.1128/mcb.5.8.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. A sequence downstream of A-A-U-A-A-A is required for formation of simian virus 40 late mRNA 3' termini in frog oocytes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987 May 8;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Goins W. F., Stinski M. F. Expression of a human cytomegalovirus late gene is posttranscriptionally regulated by a 3'-end-processing event occurring exclusively late after infection. Mol Cell Biol. 1986 Dec;6(12):4202–4213. doi: 10.1128/mcb.6.12.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T. L., Hart R. P. Mutations in poly(A) site downstream elements affect in vitro cleavage activity. Mol Cell Biol. 1988 Apr;8(4):1839–1841. doi: 10.1128/mcb.8.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Ali H., Nevins J. R. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol Cell Biol. 1985 Nov;5(11):2975–2983. doi: 10.1128/mcb.5.11.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Nevins J. R. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985 Dec;43(3 Pt 2):677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Johansen H., Schümperli D., Rosenberg M. Affecting gene expression by altering the length and sequence of the 5' leader. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7698–7702. doi: 10.1073/pnas.81.24.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M. M., Beckendorf R. C., Westhafer M. A., Nordstrom J. L. Requirement of A-A-U-A-A-A and adjacent downstream sequences for SV40 early polyadenylation. Nucleic Acids Res. 1986 Jun 25;14(12):4939–4952. doi: 10.1093/nar/14.12.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler M. M., Westhafer M. A., Carson D. D., Nordstrom J. L. Polyadenylation at a cryptic site in the pBR322 portion of pSV2-neo: prevention of its utilization by the SV40 late poly(A) signal. Nucleic Acids Res. 1987 Jan 26;15(2):631–642. doi: 10.1093/nar/15.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. C., Stein J. P., Catterall J. F., Woo S. L., Mace M. L., Means A. R., O'Malley B. W. Molecular structure and flanking nucleotide sequences of the natural chicken ovomucoid gene. Cell. 1979 Nov;18(3):829–842. doi: 10.1016/0092-8674(79)90135-1. [DOI] [PubMed] [Google Scholar]
- Le Moullec J. M., Akusjärvi G., Stålhandske P., Pettersson U., Chambraud B., Gilardi P., Nasri M., Perricaudet M. Polyadenylic acid addition sites in the adenovirus type 2 major late transcription unit. J Virol. 1983 Oct;48(1):127–134. doi: 10.1128/jvi.48.1.127-134.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. J., Elkington J. A., Lloyd M. M., Jones M. B., Williams J. G. Mutations downstream of the polyadenylation site of a Xenopus beta-globin mRNA affect the position but not the efficiency of 3' processing. Cell. 1986 Jul 18;46(2):263–270. doi: 10.1016/0092-8674(86)90743-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McGeady M. L., Wood T. G., Maizel J. V., Vande Woude G. F. Sequences upstream from the mouse c-mos oncogene may function as a transcription termination signal. DNA. 1986 Aug;5(4):289–298. doi: 10.1089/dna.1986.5.289. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds L., O'Malley B. W., Nisbet A. D., Fothergill J. E., Givol D., Fields S., Robertson M., Brownlee G. G. Sequence of chicken ovalbumin mRNA. Nature. 1978 Jun 29;273(5665):723–728. doi: 10.1038/273723a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Perricaudet M., le Moullec J. M., Tiollais P., Pettersson U. Structure of two adenovirus type 12 transforming polypeptides and their evolutionary implications. Nature. 1980 Nov 13;288(5787):174–176. doi: 10.1038/288174a0. [DOI] [PubMed] [Google Scholar]
- Perucho M., Hanahan D., Wigler M. Genetic and physical linkage of exogenous sequences in transformed cells. Cell. 1980 Nov;22(1 Pt 1):309–317. doi: 10.1016/0092-8674(80)90178-6. [DOI] [PubMed] [Google Scholar]
- Peterson J. L., McBride O. W. Cotransfer of linked eukaryotic genes and efficient transfer of hypoxanthine phosphoribosyltransferase by DNA-mediated gene transfer. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1583–1587. doi: 10.1073/pnas.77.3.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr D. S., Rieser L. A., Woychik R. P., Rottman F. M., Rosenberg M., Reff M. E. Differential effects of polyadenylation regions on gene expression in mammalian cells. DNA. 1986 Apr;5(2):115–122. doi: 10.1089/dna.1986.5.115. [DOI] [PubMed] [Google Scholar]
- Pfarr D. S., Sathe G., Reff M. E. A highly modular cloning vector for the analysis of eukaryotic genes and gene regulatory elements. DNA. 1985 Dec;4(6):461–467. doi: 10.1089/dna.1985.4.461. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Robins D. M., Ripley S., Henderson A. S., Axel R. Transforming DNA integrates into the host chromosome. Cell. 1981 Jan;23(1):29–39. doi: 10.1016/0092-8674(81)90267-1. [DOI] [PubMed] [Google Scholar]
- Sadofsky M., Connelly S., Manley J. L., Alwine J. C. Identification of a sequence element on the 3' side of AAUAAA which is necessary for simian virus 40 late mRNA 3'-end processing. Mol Cell Biol. 1985 Oct;5(10):2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Scangos G. A., Huttner K. M., Juricek D. K., Ruddle F. H. Deoxyribonucleic acid-mediated gene transfer in mammalian cells: molecular analysis of unstable transformants and their progression to stability. Mol Cell Biol. 1981 Feb;1(2):111–120. doi: 10.1128/mcb.1.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnik-David H., Moore C. L., Sharp P. A. Electrophoretic separation of polyadenylation-specific complexes. Genes Dev. 1987 Sep;1(7):672–682. doi: 10.1101/gad.1.7.672. [DOI] [PubMed] [Google Scholar]
- Sperry A. O., Berget S. M. In vitro cleavage of the simian virus 40 early polyadenylation site adjacent to a required downstream TG sequence. Mol Cell Biol. 1986 Dec;6(12):4734–4741. doi: 10.1128/mcb.6.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swimmer C., Shenk T. Selection of sequence elements that substitute for the standard AATAAA motif which signals 3' processing and polyadenylation of late simian virus 40 mRNAs. Nucleic Acids Res. 1985 Nov 25;13(22):8053–8063. doi: 10.1093/nar/13.22.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion J. P., Banville D., Noel H. Galactokinase mutants of Chinese hamster somatic cells resistant to 2-deoxygalactose. Genetics. 1976 May;83(1):137–147. doi: 10.1093/genetics/83.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscani A., Soprano D. R., Cosenza S. C., Owen T. A., Soprano K. J. Normalization of multiple RNA samples using an in vitro-synthesized external standard cRNA. Anal Biochem. 1987 Sep;165(2):309–319. doi: 10.1016/0003-2697(87)90274-0. [DOI] [PubMed] [Google Scholar]
- Tushinski R. J., Sussman P. M., Yu L. Y., Bancroft F. C. Pregrowth hormone messenger RNA: glucocorticoid induction and identification in rat pituitary cells. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2357–2361. doi: 10.1073/pnas.74.6.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Stephenson P., Sheets M., Wickens M. The AAUAAA sequence is required both for cleavage and for polyadenylation of simian virus 40 pre-mRNA in vitro. Mol Cell Biol. 1986 Jul;6(7):2317–2323. doi: 10.1128/mcb.6.7.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3'-end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988 Apr 25;263(12):5780–5788. [PubMed] [Google Scholar]
- Zhang F., Denome R. M., Cole C. N. Fine-structure analysis of the processing and polyadenylation region of the herpes simplex virus type 1 thymidine kinase gene by using linker scanning, internal deletion, and insertion mutations. Mol Cell Biol. 1986 Dec;6(12):4611–4623. doi: 10.1128/mcb.6.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]